Abstract
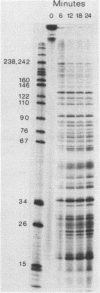
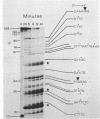
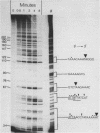
The substrate specificity of micrococcal nuclease (EC 3.1.4.7.) has been studied. The enzyme recognises features of nucleotide composition, nucleotide sequence and tertiary structure of DNA. Kinetic analysis indicates that the rate of cleavage is 30 times greater at the 5' side of A or T than at G or C. Digestion of end-labelled linear DNA molecules of known sequence revealed that only a limited number of sites are cut, generating a highly specific pattern of fragments. The frequency of cleavage at each site has been determined and it may reflect the poor base overlap in the 5' T-A 3' stack as well as the length of contiguous A and T residues. The same sequence preferences are found when DNA is assembled into nucleosomes. Deoxyribonuclease 1 (EC 3.1.4.5.) recognises many of the same sequence features. Micrococcal nuclease also mimics nuclease S1 selectively cleaving an inverted repeat in supercoiled pBR322. The value of micrococcal nuclease as a "non-specific" enzymatic probe for studying nucleosome phasing is questioned.
Full text
PDF





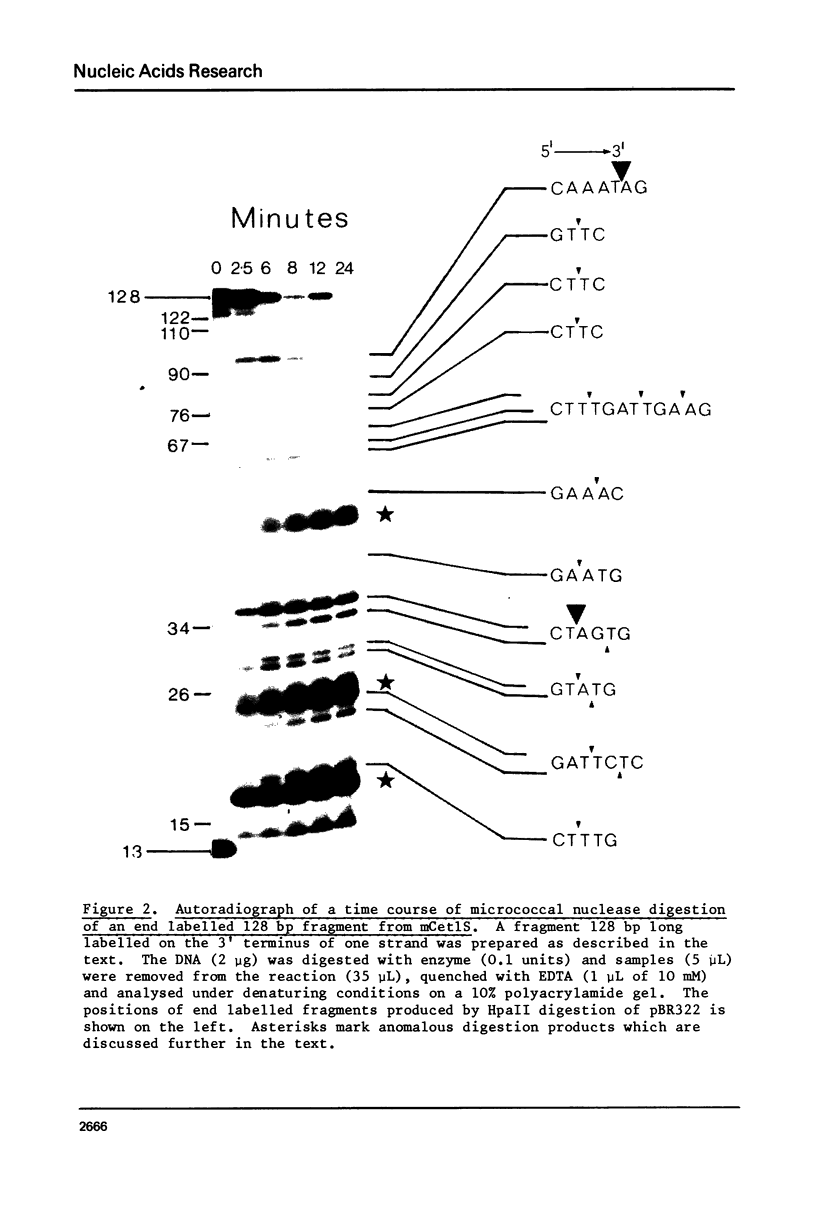
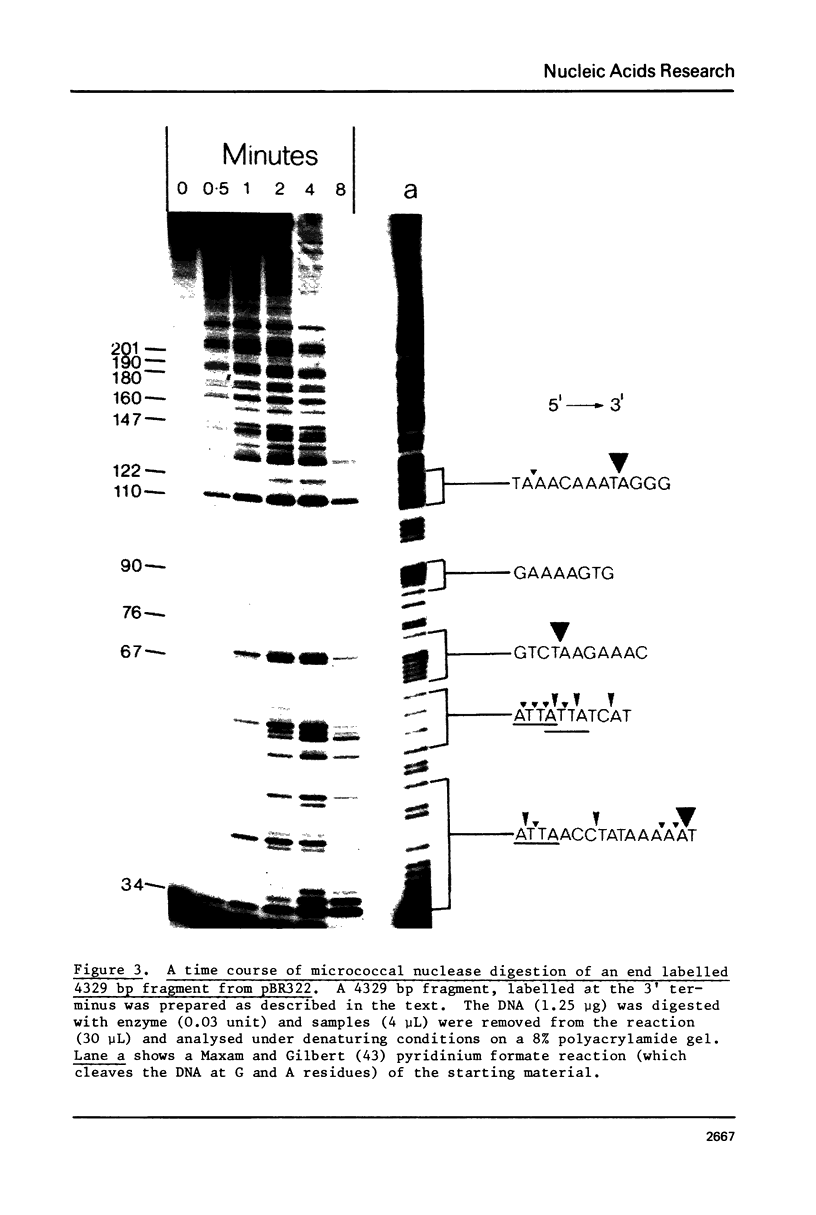






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOLLUM F. J. DEGRADATION OF THE HOMOPOLYMER COMPLEXES POLYDEOXYADENYLATE-POLYDEOXYTHYMIDYLATE, POLYDEOXYINOSINATE-POLYDEOXYCYTIDYLATE, AND POLYDEOXYGUANYLATE-POLYDEOXYCYTIDYLATE BY DEOXYRIBONUCLEASE I. J Biol Chem. 1965 Jun;240:2599–2601. [PubMed] [Google Scholar]
- Cortese R., Harland R., Melton D. Transcription of tRNA genes in vivo: single-stranded compared to double-stranded templates. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4147–4151. doi: 10.1073/pnas.77.7.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEKKER C. A. Nucleic acids. Selected topics related to their enzymology and chemistry. Annu Rev Biochem. 1960;29:453–474. doi: 10.1146/annurev.bi.29.070160.002321. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Honda B. M., Laskey R. A., Thomas J. O. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell. 1980 Sep;21(2):373–383. doi: 10.1016/0092-8674(80)90474-2. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bloomer L. S. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980 Oct;21(3):751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Omori A., Zachau H. G. Non-random arrangement of nucleosomes in satellite I containing chromatin of rat liver. Nucleic Acids Res. 1980 Nov 25;8(22):5377–5390. doi: 10.1093/nar/8.22.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacian D. L., Spiegelman S. Use of micrococcal nuclease to monitor hybridization reactions with DNA. Anal Biochem. 1974 Apr;58(2):534–540. doi: 10.1016/0003-2697(74)90221-8. [DOI] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Multiple phases of nucleosomes in the hsp 70 genes of Drosophila melanogaster. Nucleic Acids Res. 1980 Dec 20;8(24):6059–6068. doi: 10.1093/nar/8.24.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis C., Schedl P., Samal B., Worcel A. Chromatin structure of the 5S RNA genes of D. melanogaster. Cell. 1980 Nov;22(2 Pt 2):387–392. doi: 10.1016/0092-8674(80)90349-9. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Heterogeneity in deoxyribonucleic acids. I. Dependence on composition of the configurational stability of deoxyribonucleic acids. Nature. 1959 May 23;183(4673):1427–1429. doi: 10.1038/1831427a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mikulski A. J., Sulkowski E., Stasiuk L., Laskowski M., Sr Susceptibility of dinucleotides bearing either 3'- or 5'-monophosphate to micrococcal nuclease. J Biol Chem. 1969 Dec 25;244(24):6559–6565. [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- ROBERTS W. K., DEKKER C. A., RUSHIZKY G. W., KNIGHT C. A. Studies on the mechanism of action of micrococcal nuclease. 1. Degradation of thymus deoxyribonucleic acid. Biochim Biophys Acta. 1962 May 14;55:664–673. doi: 10.1016/0006-3002(62)90844-2. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6(5):1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULKOWSKI E., LASKOWSKI M., Sr Mechanism of action of micrococcal nuclease on deoxyribonucleic acid. J Biol Chem. 1962 Aug;237:2620–2625. [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]
- Wingert L., Von Hippel P. H. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim Biophys Acta. 1968 Mar 18;157(1):114–126. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- Wittig B., Wittig S. A phase relationship associates tRNA structural gene sequences with nucleosome cores. Cell. 1979 Dec;18(4):1173–1183. doi: 10.1016/0092-8674(79)90230-7. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]