SUMMARY
The repression of transcription, through the concerted actions of tissue specific DNA binding proteins, Polycomb repressor complexes, and DNA methylation, is essential for maintaining stem cell pluripotency and for cell fate specification in development. In this report, we show that recruitment of the co-repressor protein Grg4 to a Pax DNA binding site displaces the adaptor protein PTIP and a histone H3K4me complex. Grg4 recruits the arginine methyltransferase PRMT5 to chromatin resulting in symmetric H4R3 dimethylation. PRMT5 is essential for recruiting Polycomb proteins, in a Pax2/Grg4 dependent manner, which results in H3K27 methylation. These data define the early epigenetic events in response to Pax/Grg mediated gene repression and demonstrate that a single DNA binding protein can recruit either an activator or a repressor complex depending on the availability of Grg4. These data suggest a model for understanding the initiation of Groucho/Grg/TLE mediated gene silencing.
INTRODUCTION
Transcriptional repression or gene silencing is essential for maintaining embryonic stem cell pluripotency and for the differentiation of stem cells to more specialized fates, such that inappropriate genes are not expressed (Hirabayashi and Gotoh, 2010; Lunyak and Rosenfeld, 2008). At the level of chromatin, gene silencing is achieved and maintained through the concerted efforts of the Polycomb Group of histone modification complexes, the DNA CpG methylation machinery, and tissue specific DNA binding and corepressor proteins. The Polycomb proteins KMT1A/1B (SUV39H1/2) and KMT6 (EZH2) are responsible for methylating histone H3 at lysine 9 (H3K9me) and 27 (H3K27me) respectively to promote gene silencing and chromatin compaction (Ringrose and Paro, 2007; Schwartz and Pirrotta, 2008). In contrast, the KMT2s (MLLs) and the KMT3s (Set2) proteins of the Trithorax Family methylate Histone H3 at lysine 4 (H3K4me) or Lysine 36 (H3K36me) to maintain gene expression and prevent silencing (Schuettengruber et al., 2007; Shilatifard, 2008). Critical developmental control genes in embryonic stem (ES) cells have neither high levels of H3K4me3 nor H3K27me3 on promoters, rather they exhibit bivalent domains with both types of epigenetic marks (Bernstein et al., 2006; O'Neill et al., 2006). As ES cells differentiate along specific lineages, these bivalent chromatin domains are resolved into either active or silent domains, as reflected by high levels of H3K4me3 or H3K27me3 respectively. Yet, how the tissue and locus specificity of these changing epigenetic imprints are controlled during differentiation remain poorly understood.
The Polycomb and Trithorax complexes must recognize the right genes at specific developmental stages by responding to or interacting with DNA binding proteins and associated co-factors that determine cell lineages. One of the first corepressor proteins identified in metazoans was the Drosophila Groucho protein, whose mammalian homologues are called Groucho related proteins (Grg) in the mouse or Transducin-Like Enhancer of split (Tle) in humans (Cinnamon and Paroush, 2008; Jennings and Ish-Horowicz, 2008). The Grg/TLE proteins have nuclear localization signals and multiple WD40 repeat domains but no intrinsic DNA binding activity. Rather, Grg proteins complex with specific DNA binding proteins (Jennings et al., 2006) and promote gene repression, at least in part, by inhibiting the basal transcription machinery and recruiting histone deacetylases (Chen and Courey, 2000). Recently, Grg3 was shown to bind nucleosomal arrays to promote condensation into higher order chromatin that was resistant to transcription (Sekiya and Zaret, 2007). Binding of Grg3 to chromatin required the DNA binding protein FoxA1, although Grg3 binding and nucleosome condensation extended beyond the FoxA1 binding site. These data suggest that Grg recruitment by a DNA binding protein is a key step in specifying a region of chromatin for silencing.
The Grg proteins interact with bHLH proteins to specify neuronal cell types and embryonic segmentation in the fly (Heitzler et al., 1996; Paroush et al., 1994; Schrons et al., 1992). In vertebrates, Grg proteins interact with the Nkx family of homeodomain proteins to specify motor neurons along the dorsal-ventral axis of the spinal cord (Muhr et al., 2001). Grg4 also specifies the neuronal cell types in the laminae of the optic tectum (Sugiyama et al., 2000; Sugiyama and Nakamura, 2003) and coordinates the midbrain-hindbrain boundary, which requires the Pax2/5/8 proteins (Nakamura and Sugiyama, 2004; Ye et al., 2001). Grg4 binds directly to Pax2/5/8 through the conserved octapeptide domain (Eberhard et al., 2000). In developing B-cells, Grg4 interacts with Pax5 and PU.1 to repress subsets of B-cell specific genes (Linderson et al., 2004). In the spinal cord and the developing kidney (Cai et al., 2003), Grg4 expression overlaps with Pax2, which specifies the renal epithelial cells from the intermediate mesoderm (Bouchard et al., 2002; Torres et al., 1995) and subsets of neurons in the CNS (Burrill et al., 1997; Cheng et al., 2004; Torres et al., 1996). In cell culture, Grg4 suppresses activation by Pax2 and blocks phosphorylation of the Pax2 activation domain (Cai et al., 2003). Thus, Pax proteins can function as repressors in specific contexts. Despite the evidence linking Grg/Tle proteins to developmental transcription factors, how Grg/Tle proteins impact nucleosome modifications and determine chromatin structure remains obscure.
In this report, we defined the effects of Grg4 on Pax2 mediated histone modifications and gene expression. Pax2/5/8 interact with the adaptor protein PTIP, which is part of the KMT2C/D (Mll3/4) histone H3 lysine 4 (H3K4) methylation complex (Daniel et al., 2010; Patel et al., 2007). Grg4 is able to displace PTIP and inhibit H3K4 methylation, thereby completely suppressing Pax mediated gene activation. Grg4 is part of a complex that contains the arginine methyltransferase PRMT5, the protein phosphatase PPM1B, and the adaptor protein WDR77. Expression of Grg4 recruits these proteins to a Pax2 DNA binding site resulting in symmetric histone H4 arginine 3 (H4R3) dimethylation and further recruitment of the Polycomb proteins KMT6 (Ezh2) and Suz12. The data provide a molecular, epigenetic basis for Grg/Tle mediated repression at sites of Pax protein binding that may drive chromatin condensation and gene silencing.
RESULTS
Recruitment of Grg4 to chromatin displaces PTIP
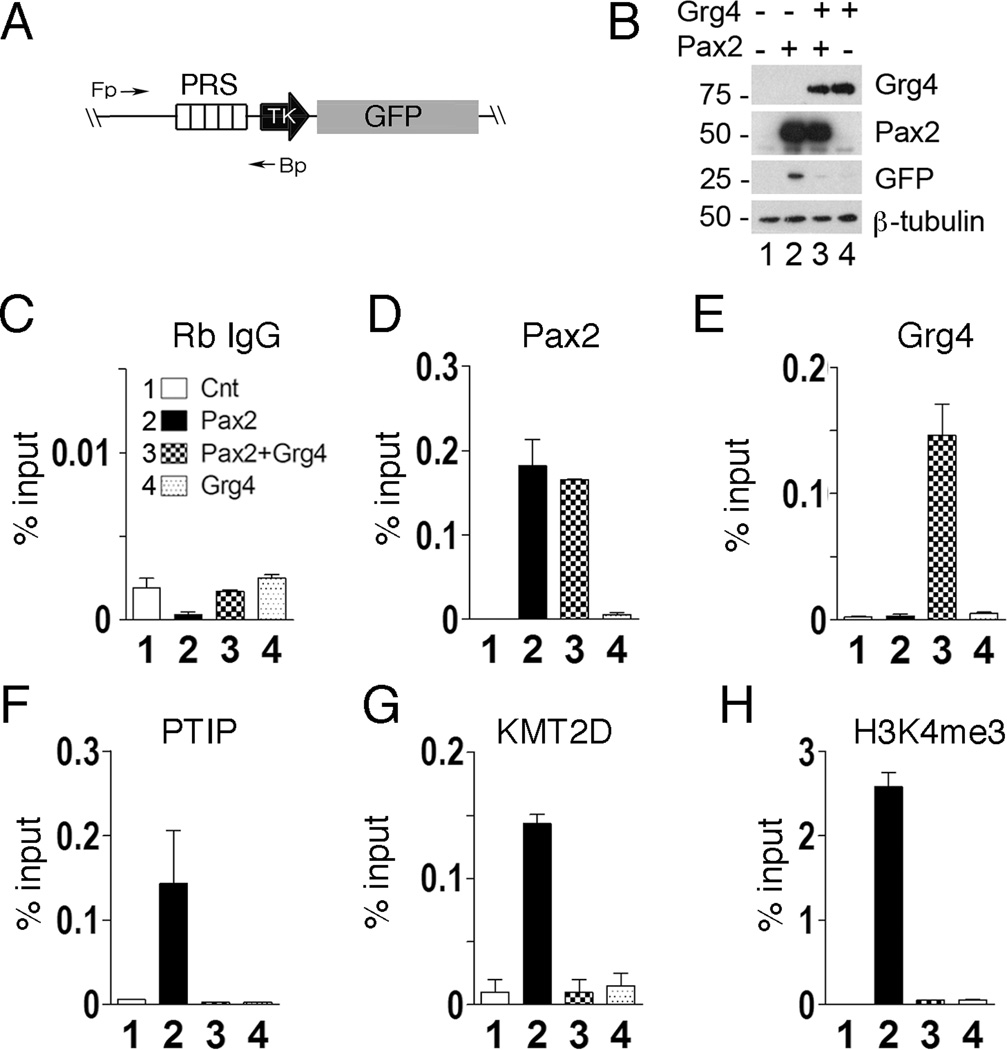
To understand how Grg proteins repress transcription, we first used an integrated, chromatin based reporter gene to characterize histone methylation patterns in response to the Pax2 or Pax5 DNA binding proteins (Patel et al., 2007; Schwab et al., 2011). HEK293 cells carrying a Pax Response Sequence (PRS-GFP) reporter gene were transfected with Pax2 and Grg4 (Fig. 1). Transient expression of Pax2 increased GFP expression and recruited PTIP and a KMT2D (Mll4) methyltransferase complex that resulted in high levels of H3K4me3 at the PRS. However, co-expression of Grg4 with Pax2 inhibited expression of GFP (Fig. 1B). Chromatin immunoprecipitation (ChIP) experiments show that Pax2 binding was unaffected by Grg4 (Fig. 1D) and that Grg4 was only bound to the PRS in the presence of Pax2 protein (Fig. 1E). Grg4 expression was able to displace PTIP (Fig. 1F) and KMT2D (Fig. 1G) from the PRS and suppress the increase in H3K4me3 observed with Pax2 alone (Fig. 1H). Other components of the KMT2D complex, such as Rbbp5, were also displaced and the levels of H3K4me1 and H3K4me2 were reduced in the presence of Grg4 (data not shown).
Figure 1. Grg4 displaces PTIP and the KMT2D complex to inhibit Pax2 dependent reporter gene expression.
A) A schematic of the Pax responsive reporter gene integrated into HEK293 cells: PRS, Pax Response Sequence; TK, minimal HSV-TK promoter; GFP, green fluorescent protein coding. B) HEK293 cells containing the integrated PRS-GFP reporter were transfected with control vectors (lane1), Pax2 alone (lane 2), Pax2 and Grg4 (lane 3) or Grg4 alone (Lane 4). Whole cell lysates were blotted for the proteins indicated. C–H) ChIP assays from HEK293 cells transfected as in B. The antibodies used for each ChIP are indicated above the graph. Primer pairs flank the PRS. Note that Pax2 binds to PRS independent of Grg4 (D) and Grg4 localizes to PRS in a Pax2 dependent manner (E). PTIP (F) and KMT2D (G) bind to PRS in a Pax2 dependent manner, but are displaced upon Grg4 co-expression. Pax2/Grg4 expression suppresses H3K4me3 at the Pax2 binding element (H). Averages from triplicate are shown with error bars representing 1 standard error of the mean (SEM).
Purification of a Grg4 Complex
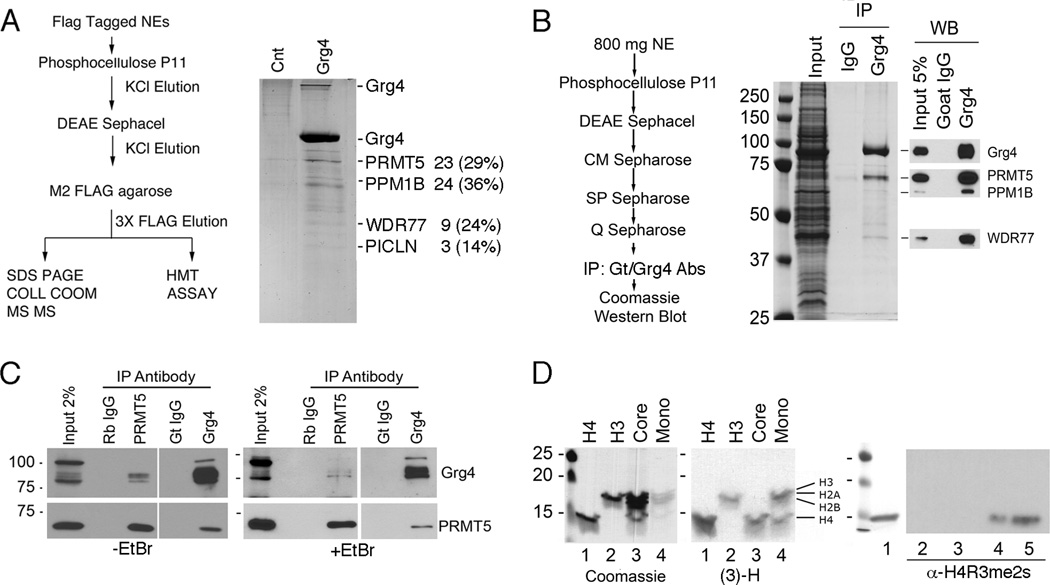
How Grg4 suppressed Pax2 dependent gene activation, displaced the PTIP/KMT2D complex, and prevented H3K4me3 was studied further by purification of Grg4 and associated proteins (Fig. 2). Flag tagged Grg4 was expressed in HEK293 cells and purified by conventional chromatography and M2-flag immunoaffinity (Fig. 2A). Mass spectrometry identified proteins associated with Grg4, including the arginine methyltransferase PRMT5, a protein phosphatase PPM1B, and WDR77 and PICLN proteins. Because PRMT5 may interact with M2-flag agarose directly (Nishioka and Reinberg, 2003), we repeated the purification without the use of M2-flag beads. Nuclear lysates from Grg4 transfected HEK293 cells were fractionated over multiple columns followed by immunoprecipitation (IP) with goat anti-Grg4 antibodies (Fig. 2B). As with the M2-flag purification, endogenous PRMT5 co-purified with Grg4. The interaction with PRMT5 was also confirmed by co-IPs from unfractionated nuclear extracts using either Grg4 or PRMT5 specific antibodies (Fig. 2C). To rule out the possibility that DNA fragments might link the two proteins indirectly, IPs were repeated with EtBr. In all cases, Grg4 and PRMT5 co-precipitated.
Figure 2. Grg4 Co-purifies with an Arginine Methyltransferase.
A) The purification of 3X-flag-Grg4 is outlined and the associated proteins stained with colloidal Coomassie blue. The proteins identified by MS/MS are indicated, with the number of peptides and the percent coverage labeled on the right. The 60 kD protein in the Grg4 lane was a Keratin contaminant. B) An alternate purification of Grg4 is outlined and the associated proteins stained with colloidal comassie (left). Proteins were confirmed by Western blots (right). C) Rabbit anti-PRMT5 or Goat anti-Grg4 were used to IP proteins from nuclear extracts directly in the absence or presence of EtBr and western blotted as indicated. Controls are Rabbit IgG for PRMT5 and Goat IgG for Grg4. D) Histone methyltransferase assay using final 3X FLAG elutions from Grg4 transfected cells, [3]H-SAM, and Histone H4 (lane 1), Histone H3 (lane 2), core histones (lane 3), or mononucleosomes (lane 4) as substrates. Coomassie stained gels are shown (left panel) with the corresponding Fluorograms (middle panel). Note that PRMT5 methylates both Histone H3 and Histone H4 when mononucleosomes are used as substrate. The right panel shows a western blot for H4R3me2s using histone H4 as a substrate (all lanes) and unlabeled SAM: lane 1, Coomassie stained Histone H4; lane 2, no lysate; lane 3, no SAM; lane 4, lysate plus SAM; lane 5, acid extracted histones. Methylation reactions were run on 15% SDS/PAGE gels.
Since PRMT5 is an arginine methyltransferase, we tested the ability of the Grg4 complex to methylate purified histones and core nucleosomes (Fig. 2D). The Grg4 complex methylated histone H4 and histone H3, though less efficiently. When using the four core histones, only H4 was methylated, though a mixture of mononuclesomes showed methylation of H3 and H4 (Fig. 2D). Only the symmetric dimethylation (me2s) of H4R3 was observed (Fig. 2D), as antibodies against the asymmetric H4R3me2 did not react (data not shown). Thus, the PRMT5 in the Grg4 complex is able to methylate H4R3me2s.
Grg4 recruits PRMT5 and PRC2 proteins to chromatin
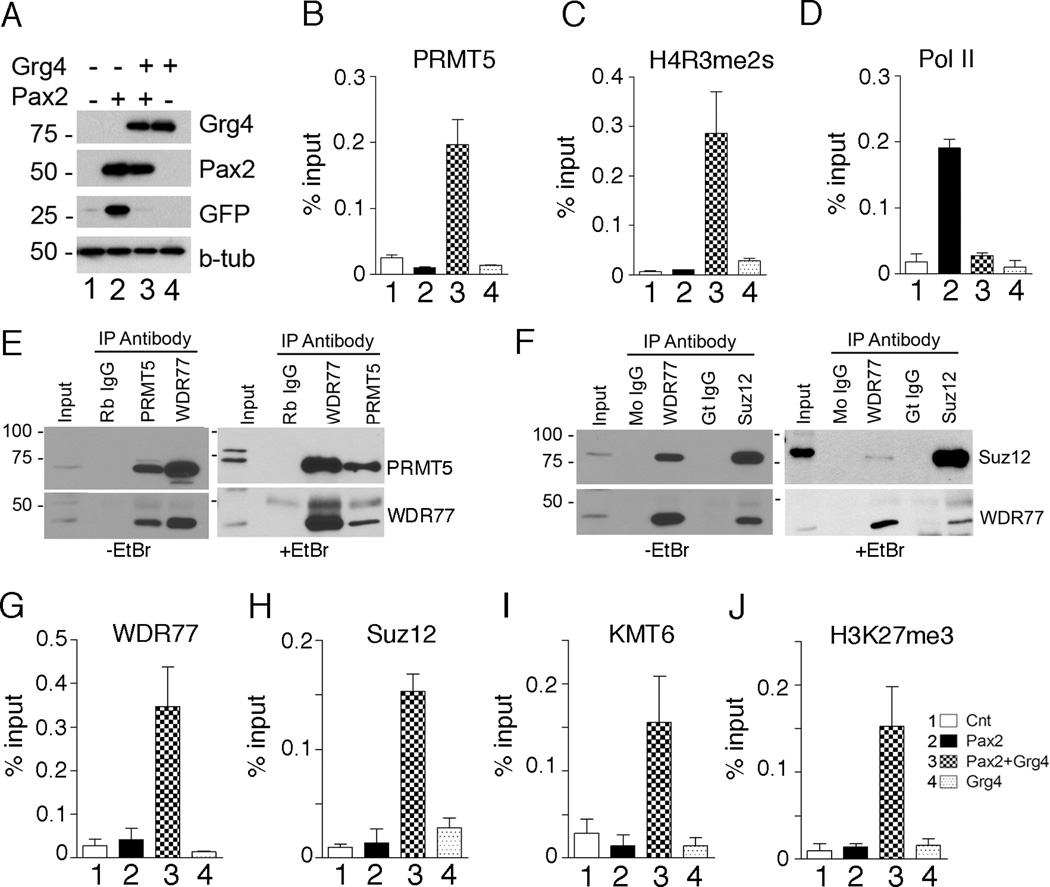
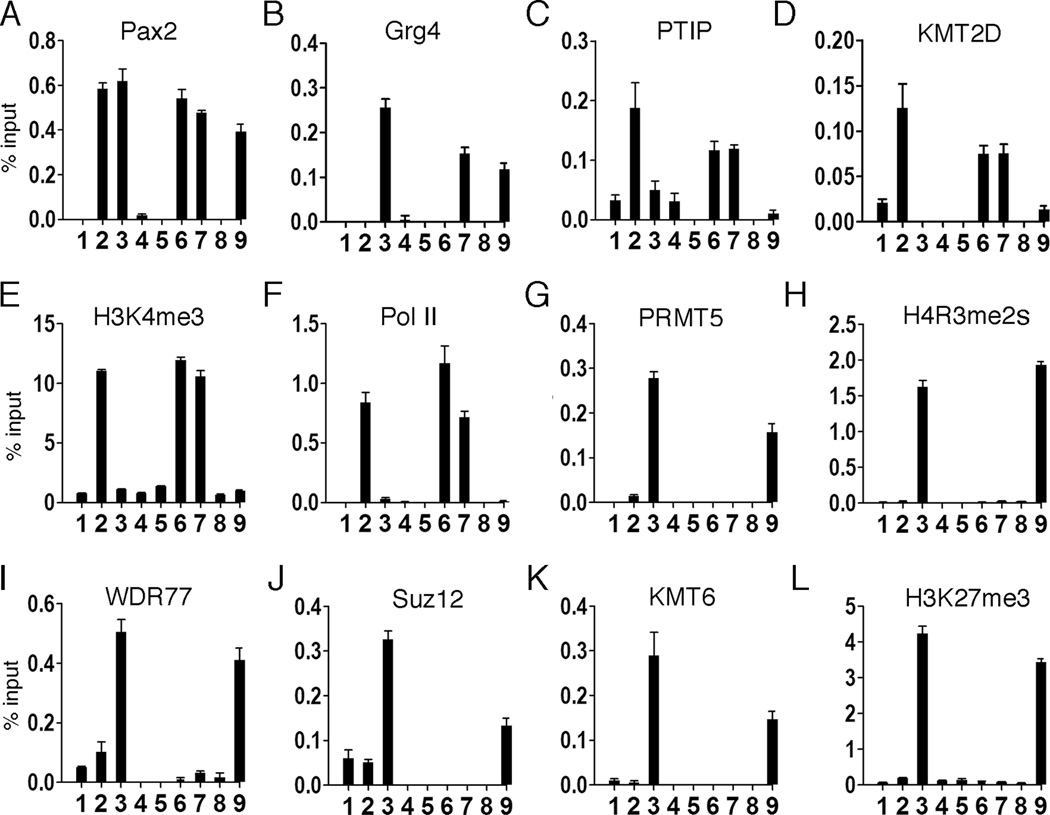
Given the Grg4/PRMT5 interaction, we next used ChIP to determined that Grg4 was able to recruit PRMT5 to the PRS (Fig. 3B), which increased H4R3me2s (Fig. 3C) and inhibited RNA Polymerase II at the promoter region (Fig. 3D). The presence of WDR77 in the Grg4 complex suggested that additional epigenetic silencing marks may be Grg4 and PRMT5 dependent. Co-IPs of endogenous proteins confirmed that PRMT5 can complex with WDR77 (Fig. 3E). Furthermore, WDR77 interacts with the Suz12 protein (Fig. 3F), a component of the Polycomb repressor complex 2 (PRC2) (Furuno et al., 2006). These interactions were confirmed by ChIP assays at the PRS, as WDR77, Suz12, and the H3K27 methylase KMT6 (Ezh2) were all recruited to chromatin in a Grg4/Pax2 dependent manner (Fig. 3 G–J). Thus, the co-expression of Grg4 and Pax2 resulted in increased H3K27me3, a hallmark of PRC2 repression. Neither Pax2 nor Grg4 alone were able to recruit any PRC2 proteins. These data demonstrate multiple effects on patterns of histone methylation are mediated by PRMT5 and PRC2 in a Pax2/Grg4 dependent manner.
Figure 3. Grg4 recruits PRMT5 and PRC2 proteins to a Pax2 binding element.
A) HEK 293 cells were transfected as outlined above the panel and cell lysates were Western blotted for the proteins indicated. B–D) ChIP assays at the PRS from HEK293 cells after transfection with: control vector (lane1), Pax2 alone (lane 2), Pax2 and Grg4 (lane 3) or Grg4 alone Lane 4. The antibodies used for ChIP are indicated above the graph. Note PRMT5 is recruited to the PRS element only when Pax2 and Grg4 are co-expressed (B) and PRMT5 localization at PRS is associated with symmetric dimethylation of H4R3 (C). RNA Polymerase II does not localize to the PRS element when Pax2 and Grg4 are coexpressed (D). E) Rabbit anti-PRMT5 or Rabbit anti-WDR 77 were used to IP proteins from nuclear extracts directly, in the absence or presence of EtBr. Coprecipitated proteins were detected by western blotting as indicated. F) Mouse anti-WDR77 or Goat anti-Suz12 were used to IP proteins from nuclear extracts directly, with or without EtBr. Coprecipitated proteins were detected by western blotting as indicated. G–J) ChIP assays at the PRS from HEK293 cells after transfection with: control vector (lane1), Pax2 alone (lane 2), Pax2 and Grg4 (lane 3) or Grg4 alone (lane 4). WDR77 is recruited to the PRS element only when Pax2 and Grg4 are co-expressed (G). Similarly, Suz12 (H) and KMT6 (I) localize to PRS in a Grg4 dependent manner. Recruitment of KMT6 to PRS results in H3K27 trimethylation in cells transfected with both Pax2 and Grg4 (J). All error bars are 1 SEM.
PRMT5 is required for Pax2/Grg4 mediated silencing
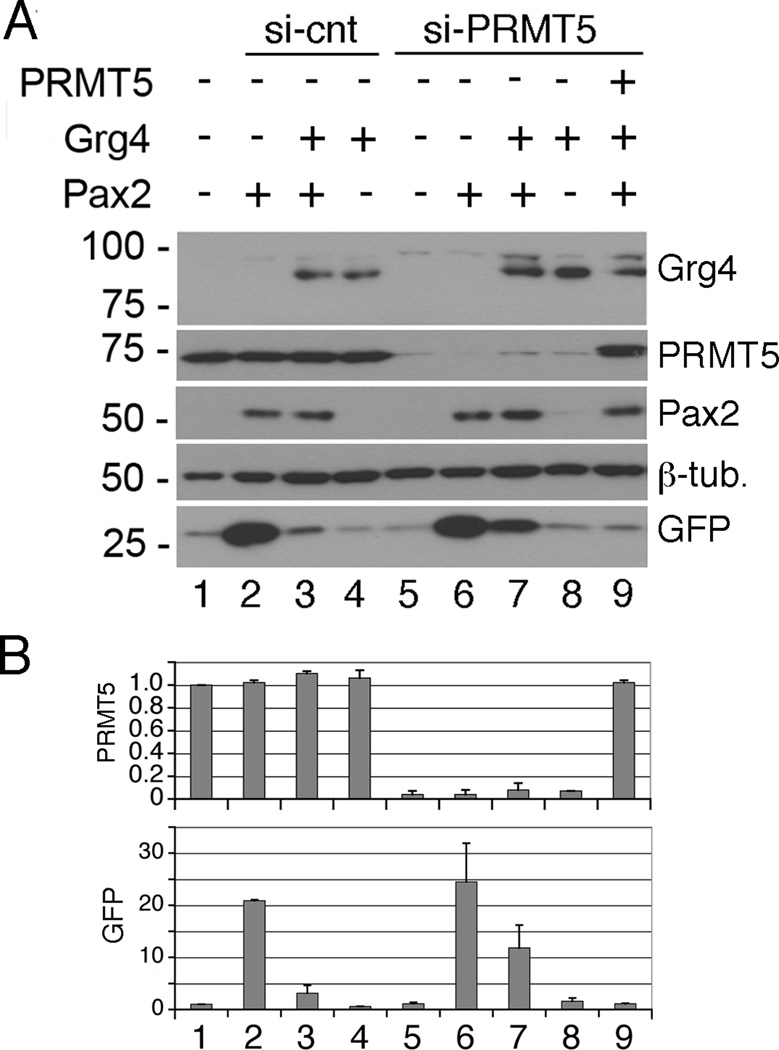
We tested whether PRMT5 is needed for Pax2/Grg4 mediated repression by siRNA knockdown of human PRMT5 in HEK293 cells. Cells were transfected with Pax2, Grg4, or Pax2 and Grg4, in the presence or absence of endogenous PRMT5 (Fig. 4). After PRMT5 knockdown, Pax2 still activated the GFP reporter but Grg4 mediate repression was attenuated (Fig. 4, lane 7). GFP activation by Pax/Grg4 was still 12.5 fold in the absence of PRMT5, though reduced from the 25 fold seen with Pax2 alone (Fig. 4B). This may reflect some residual level of PRMT5, as the knockdown was approximately 90%. With an siRNA resistant PRMT5 vector, the Grg4 mediated repression could be rescued (Fig. 4, lane 9). Thus, PRMT5 was necessary for mediating at least part of the Grg4 repressive activity.
Figure 4. Transcriptional repression by Grg4 is mediated by PRMT5.
A) Cells were transfected with control si oligos (lanes 1–4) or si-PRMT5 specific oligos (lanes 5–9). Subsequently, cells were transfected with Grg4, Pax2, or si resistant PRMT5 as indicated. Whole cell lysates were Western blotted for the indicated proteins. Note that GFP expression was increased upon Pax2 transfection (lane 2) and this activation inhibited by Grg4 (lane 3). In PRMT5 knockdowns, Grg4 mediated inhibition of GFP was less (lane 7). The Grg4 mediated inhibition was rescued with an si resistant PRMT5 vector (lane 9). B) Quantitation of PRMT5 and GFP protein levels by scanning densitometry. Lanes are as in A and error bars represent 1 SEM.
To characterize the role of PRMT5 at the level of chromatin, antibodies were used for ChIPs under the same conditions as in Fig. 4. Pax2 binding to the PRS was unaffected by Grg4 or PRMT5 levels (Fig. 5A). Grg4 associated with chromatin only in the presence of Pax2 (Fig. 5B). Strikingly, PTIP was displaced by Grg4 only in the presence of PRMT5 (Fig. 5C) and this correlated with reduced levels of gene expression and less KMT2D, H3K4me3, and Pol II localization at the promoter (Fig. 5D–F). PRMT5 localization was dependent on Pax2 and Grg4 and rescued by siRNA resistant PRMT5 in the knockdowns (Fig. 5G). The presence of PRMT5 correlated exclusively with H4K3me2s at the PRS (Fig. 5H). Most significantly, PRMT5 was necessary for the Grg4/Pax2 dependent localization of WDR77, Suz12, and KMT6 (Fig. 5I–K) to the PRS and for high levels of H3K27me3 (Fig. 5L). These data show an essential role for PRMT5 in establishing the epigenetic modifications mediated by Grg4 at a Pax DNA binding site and suggest that H4R3me2s is a critical component of gene silencing that may recruit PRC2 for additional histone modifications.
Figure 5. Pax2/Grg4 mediated recruitment of PRC2 proteins is PRMT5 dependent.
Cells were transfected with control si oligos (lanes 1–4) or si-PRMT5 specific oligos (lanes 5–9) as in Fig. 4 followed by transfection with: control vector, lanes 1 & 5; Pax2, lanes 2 & 6; Pax2 and Grg4, lanes 3 & 7; Grg4 only, lanes 4 & 8; Pax2, Grg4, and mouse PRMT5, lane 9. After 48h, ChIP assays were performed with the indicated antibodies. Note Pax2 binds to the PRS element in all cells transfected with Pax2 (A, lanes 2,3,6,7,9). Grg4 localizes to PRS only when co-transfected with Pax2 (B, lanes 3,7,9). PTIP localizes to the PRS when Pax2 is expressed (C, lanes 2,6) and when PRMT5 expression is attenuated in the presence of Pax2 and Grg4 (C, lane 7). Similarly. KMT2D localizes to PRS when PTIP is present (D, lanes 2,6,7), H3K4 is methylated accordingly (E), and RNA Pol II is recruited to this site (F). PRMT5 knockdown (G, lane 6) is rescued by a mouse cDNA that is resistant to the PRMT5 siRNAs (G, lane 9). H4R3me2s is only detected when PRMT5 is localized to PRS (H). Similarly, WDR77 (I), Suz12 (J) and KMT6 (K) are only localized to the PRS element when PRMT5 is expressed (lanes 3,9). H3K27 methylation increases only when KMT6 is localized to the promoter in a Pax2/Grg4/PRMT5 dependent manner (L). All bars are averages with error bars being 1 SEM.
Pax2/Grg4 interactions at the Rap1A locus in HEK293 cells
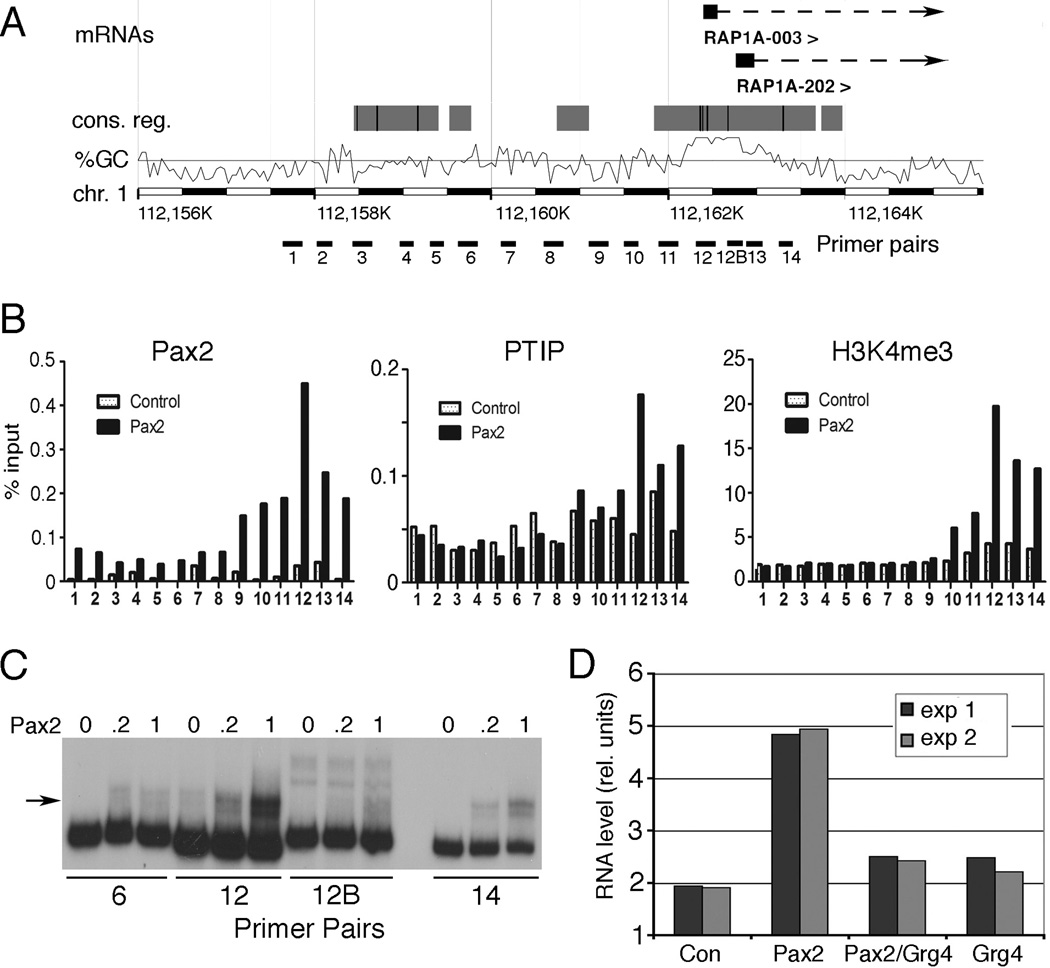
While the PRS-GFP reporter was useful in defining the patterns of histone modification upon Pax2 or Pax2/Grg4 binding, it was not clear whether such modifications can occur at endogenous loci. The HEK293 cells are immortalized embryonic kidney fibroblasts and presumably have a stable pattern of epigenetic marks. These cells do not express Pax2, which is specific for the epithelial lineage in the developing kidney. Transfection of Pax2 did not grossly alter the transcription profile of HEK293 cells, when compared by Affymetrix microarray analyses. Only 9 probe sets showed statistically significant differences in RNA levels, with 5 increasing and 4 decreasing. Of the genes up-regulated by Pax2, two probe sets corresponded to Rap1A, which encodes a GTPase that functions in cell adhesion. Thus, we examined whether Pax2 and Grg4 could establish activating and repressive marks at the Rap1A locus, similar to the PRS-GFP reporter. First, the Rap1A promoter was examined for Pax2 binding sites by ChIP and electrophoretic mobility shift assays using 14 primer pairs spanning 5 kb of the promoter region (Fig. 6). Maximum Pax2 binding by ChIP occurred at primer pair 12, close to the transcription start site (Fig. 6A, B). This correlated with PTIP localization and H3K4me3 (Fig. 6B). In vitro, Pax2 bound best to primer pair 12, consistent with the ChIP data (Fig 6C). While Pax2 transfection increased Rap1A mRNA levels approximately 2.5 fold, expression of Grg4 and Pax2 inhibited this level of activation (Fig. 6D). However, Grg4 alone had no affect on basal levels of Rap1A RNA.
Figure 6. Pax2 recruits PTIP to the Rap1A locus.
A) A schematic of the 5' end of the human Rap1A gene showing position of mRNAs, conserved potential regulatory regions, and GC percentage, according to the ENSEMBL genome browser. PCR primer pairs used for ChIP experiments are indicated below. B) Control or Pax2 transfected HEK293 cells were used for ChIP experiments with the indicated antibodies and primer pairs spanning the Rap1A transcription start sites. Note a peak of Pax2 binding at primer pair 12 corresponds with PTIP and H3K4me3 peaks. C) Electrophoretic mobility shift experiments using DNAs corresponding to the indicated primer pairs and increasing amounts of recombinant Pax2 paired domain. The arrow indicates the Pax2/DNA complex; note strong binding to primer pair 12. D) RNA levels as determined by Affymetrix microarrays from HEK293 cells transfected with Pax2, Pax2/Grg4, or Grg4 alone. Note that Pax2 alone increases Rap1A mRNA levels approximately 2.5 fold, whereas Grg4 inhibits Pax2 activation.
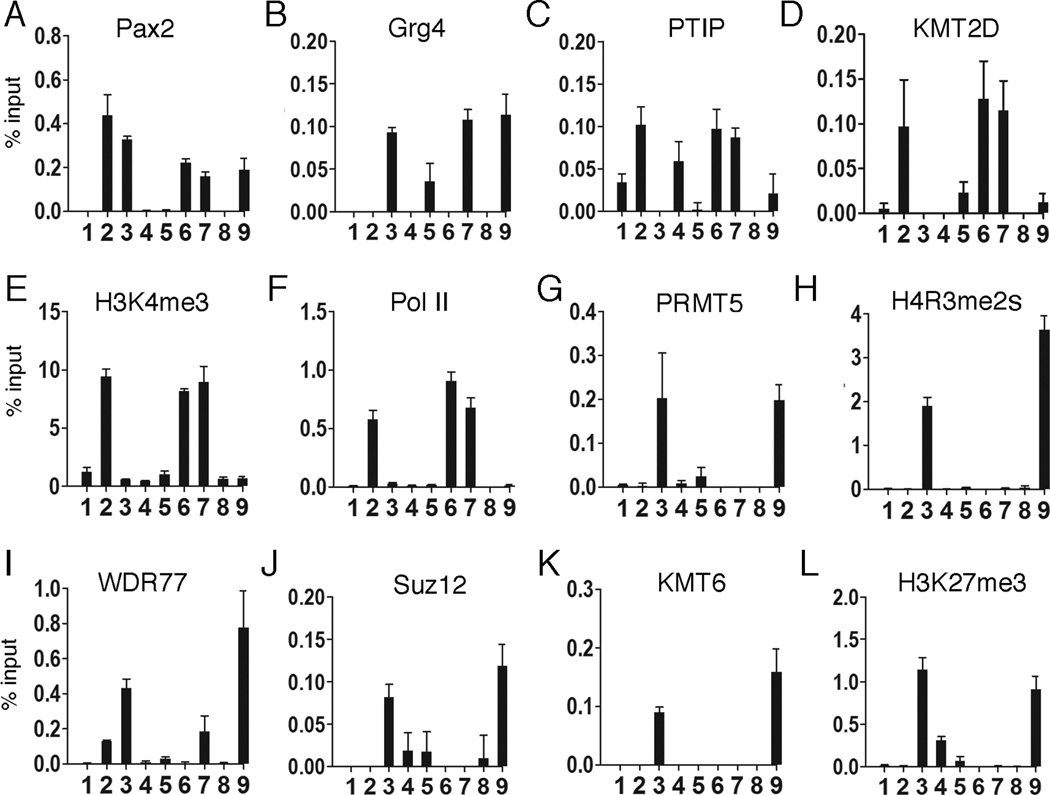
Once we had identified a Pax2 binding site, we characterized the effects of Pax2 and Grg4 at the Rap1A promoter region using primer pair 12 for all ChIP experiments, with and without PRMT5 (Fig. 7). The overall patterns of histone methylation and protein recruitment were similar to that observed with the integrated PRS-GFP reporter, with subtle differences. Pax2 expression increased PTIP and H3K4me3 at the promoter, although base levels of PTIP were slightly higher compared to the GFP reporter likely due to higher basal levels of Rap1A expression. Pax2/Grg4 displaced PTIP and recruited PRMT5, WDR77, KMT6 and Suz12 to the Rap1A promoter, resulting in high levels of H4R3me2s and H3K27me3. In the absence of PRMT5, Pax2/Grg4 could not recruit any of the PRC2 proteins examined and this defect could be fully rescued by the mouse PRMT5 cDNA. These data further underscore the significance of PRMT5 in mediating Grg4 dependent repression at an endogenous gene.
Figure 7. Pax2/Grg4 mediated chromatin remodeling at the Rap1A locus is PRMT5 dependent.
As in Fig. 4 & 5, cells were subject to control si oligos (lanes 1–4) or si-PRMT5 specific oligos (lanes 5–9). Cells were then transfected with the following vectors: controls, lanes 1 & 5; Pax2, lanes 2 & 6; Pax2 and Grg4, lanes 3 & 7; Grg4 only, lanes 4 & 8; Pax2, Grg4, and mouse PRMT5, lane 9. After 48h, ChIPs were performed with the antibodies against the indicated proteins and primer pair 12 of the Rap1A promoter region. Pax2 binding to Rap1A is minimally affected (A, lanes 2,3,6,7,9). Grg4 localizes to Rap1A when co-transfected with Pax2 (B, lanes 3,7,9). PTIP localizes to the Rap1A promoter only when Pax2 is expressed alone (C, lanes 2,6) or when PRMT5 is knocked-down in the presence of Pax2 and Grg4 (C, lane 7). Similarly, KMT2D localizes to Rap1A when PTIP is present (D, lanes 2,6,7), H3K4 is trimethylated accordingly (E), and RNA Pol II is recruited (F). PRMT5 knockdown (G, lane 7) is rescued by the mouse cDNA (G, lane 9) and H4R3me2s is restored (H, lane 9). As in Fig. 5, WDR77 (I), Suz12 (J) and KMT6 (K) are only localized to the Rap1A promoter when PRMT5 is recruited (lanes 3,9). H3K27 methylation increases only when KMT6 is localized to the promoter in a Pax2/Grg4/PRMT5 dependent manner (L). Error bars are 1 SEM.
DISCUSSION
Genetic screens in flies and mutational analyses in mice identified DNA binding proteins essential for embryonic development and cell lineage specification. Among these, the vertebrate Pax genes encode proteins required for development of the eye, central nervous system, kidney, vertebral column, thyroid, and the B cell lineage (Blake et al., 2008; Robson et al., 2006). The Pax2/5/8 subfamily share a DNA binding domain, an octapeptide sequence (YSINGILG) that binds to Grg4 (Eberhard et al., 2000), and a carboxy-terminal activation domain that is serine-threonine phosphorylated (Cai et al., 2002). Pax proteins can activate or repress genes during cell lineage determination, but how they achieve their biological outputs is not well understood. We have identified the critical steps and associated proteins that mediate Groucho dependent gene silencing at a Pax DNA binding site. Furthermore, we identify components of the Grg4 silencing complex, of which PRMT5 is critical for recruiting PRC2.
Grg4 is able to switch Pax2 from an activator to a repressor and to initiate Polycomb mediated gene silencing. In the absence of Grg4, Pax2 recruits the adaptor protein PTIP and associated KMT2C/D complexes to chromatin, which increases H3K4me3. PTIP is required for maintaining high levels of H3K4me3 in ES cells (Kim et al., 2009) and in differentiated tissues (Lefevre et al., 2010; Patel et al., 2007; Stein et al., 2011). PTIP mediated H3K4me3 is observed at the germ line transcript promoters of the heavy chain Ig locus upon activation of B cells (Daniel et al., 2010) where it promotes Pax5 dependent changes in transcription and chromatin looping (Schwab et al., 2011). Thus, the ability of Grg4 to recruit PRMT5 and displace PTIP at the Pax2 binding site is likely to be a critical step towards gene silencing. Since PTIP is widely expressed in development, our data suggests that Pax proteins activate gene expression but can repress in response to increased Grg4 levels. Unlike PTIP, Grg4 levels are regulated in the developing kidney and neural tube where they are low in undifferentiated cells and high in terminally differentiated neurons and glomerular podocytes (Cai et al., 2003).
The arginine methyltransferase PRMT5 associates with Grg4 in a Pax2 independent manner but recruitment to DNA was dependent on Pax2 and Grg4. Upon PRMT5 knockdown, we did not observe H4R3me2s at the PRS, as might be expected. However, we also did not see PTIP displacement even though Grg4 was recruited. This suggests that Grg4 is not simply competing with PTIP for Pax2 binding. The displacement of PTIP either required PRMT5 directly or was a result of the changes in the local histone methylation patterns.
The PRMT5 protein has been linked to transcriptional repression at multiple loci and in response to a variety of inputs (Fabbrizio et al., 2002; Majumder et al., 2010; Pal et al., 2004; Wang et al., 2008). PRMT5 can be recruited to chromatin by other DNA binding proteins, such as Znf224 (Cesaro et al., 2009) and BLIMP1 (Ancelin et al., 2006), and by chromatin binding proteins such as COPR5 (Lacroix et al., 2008) and BRD7 (Tae et al., 2011). Whether PRMT5 was required for Polycomb mediated repression had not been studied in detail. Our data supports a model in which Pax2/Grg4 recruits PRMT5 and promote H4R3me2s, which subsequently leads to PRC2 recruitment and H3K27me3. Without PRMT5, we do not see WDR77, Suz12, or KMT6 (Ezh2) recruited to the Pax2 DNA binding site, suggesting that PRMT5 and/or H4R3me2s is a rate-limiting step for repression. Recent studies linked PRMT5 to DNA methylation, as H4R3me2s marks were able to recruit the de novo methyltransferase DNMT3A (Zhao et al., 2009), although structural analyses does not support direct binding of DNM3TA to a H4R3me2s peptide (Otani et al., 2009). Our data are consistent with the hypothesis that PRMT5 is essential for initiating epigenetic silencing. Indeed, recent bioinformatics analyses suggest that H4R3me2s is one of the most common marks found in silent chromatin (Xu et al., 2011).
Pax2 and Grg4 are co-expressed in the developing kidney and the nervous system, and are implicated in cell fate determination (Cai et al., 2003; Kim et al., 2009; Koop et al., 1996; Sugiyama and Nakamura, 2003; Ye et al., 2001). The effects of Pax2 on chromatin have been difficult to study in vivo because of the small number of renal epithelial progenitor cells, making biochemical and ChIP analysis difficult. We have developed a cell culture assays to analyze the effects of Pax and Grg proteins on chromatin in a controlled system. While transfection of Pax2 into HEK293 cells had dramatic affects on the GFP reporter, the impact on endogenous gene expression was minimal. This suggests that HEK293 cells are not competent to respond to Pax2, perhaps because their chromatin landscape is already fixed and most potential Pax2 target genes are either already on or are silenced and inaccessible. This underscores the utility of the PRS-GFP reporter gene model cell system because it enabled selection for an integrated element that was responsive and accessible. The Rap1A locus was one of few responsive genes in HEK293 cells and showed identical changes in PTIP displacement, PRC2 recruitment, and histone modification upon Pax2 and Pax2/Grg4 expression. These results define a model for the epigenetic basis of Groucho/Grg/TLE mediated gene silencing.
EXPERIMENTAL PROCEDURES
Plasmids
CMV-Pax2HA and FLAG-Grg4 plasmids have been described (Cai et al., 2003). Myc-PRMT5 was made by inserting a PCR amplified full length cDNA of murine PRMT5 (Open Biosystems) into the BamHI-EcoRI site of pCMV-Tag3B (Agilent Technologies). All expression plasmids were sequenced for verification.
Antibodies
Rabbit antibodies to Pax2 and PTIP are described (Patel et al., 2007). Mouse IgG (015-000-003; IP), Rabbit IgG (011-000-003; IP and ChIP) and goat IgG (005-000-003) were from Jackson Immunoresearch. Anti-FLAG (F-3165), anti-M2 FLAG agarose (A-2220), and anti-β-tubulin (T-4026) were from Sigma. Anti-PRMT5 (A300-849A), anti-RBBP5 (A300-109A), anti-WDR77 (A301-562A; A301-561A) were from Bethyl Labs. Goat anti-human KMT2D (ab1596), anti-H3K4me1/2/3 (ab8895, ab7766, and ab8580), anti-H4R3me2s (ab5823), anti-Pol II CTD (ab12073) and anti-Suz12 (ab12073) were purchased from Abcam. Rabbit anti-PRMT5 (07-405), and anti-H3K27me2/3 (07-452, 07-449) were from Millipore. Mouse anti-GFP (sc9996), anti KMT6/Ezh2 (sc17270), anti-Grg4/TLE4 (sc13377), Suz12 (sc46264) and anti-WDR77 (sc100899) were from Santa Cruz.
Cell Culture, Transfection and Cell Lysate Preparation
HEK293 cells were cultured in DMEM (450 mg/dl glucose) supplemented with 10% fetal bovine serum and 100 U/ml penicillin and 100 mg/ml streptomycin in 5%CO2/95% air at 37°C. 10 cm plates were transfected with 1–2 µg of DNA and 12 µl of Fugene 6 as described (Roche Molecular Biochemicals). 40 h after transfection, cells were harvested, and nuclear extracts or whole cell lysates prepared as described (Patel and Dressler, 2004).
HEK293 cells with an integrated PRS-EGFP were transfected with 1 µg CMV-PAX2, 1µg CMV-Pax2 and 2 µg FLAG-Grg4, or FLAG-Grg4 (2 µg) as described (Patel et al., 2007).
Protein Purification and Mass Spectrometry
Nuclear extracts were prepared from 200 plates transfected with pFLAG-Grg4 or controls and dialyzed overnight into Buffer A (20 mM Tris-HCl pH 7.9, 0.2 mM EDTA, 10 mM bME, 10% glycerol, 0.2 mM PMSF, 0.1 M KCl). 40 mg of nuclear extract was loaded on a 10 ml column of phosphocellulose (P11, Whatman) and fractionated stepwise in buffer A with increasing concentrations of KCl (0.1 M wash, 0.3 M, 0.5 M, 0.7 M, and 0.9 M elutions) (Bochar et al., 2000). The P11 0.3 and 0.5 M KCl Grg4 fractions were loaded on a 6 ml DEAE-Sephacel column (Pharmacia) and eluted stepwise with increasing concentrations of KCl in buffer A. The peak Grg4 fractions were pooled and dialyzed against buffer BC (20 mM HEPES pH 7.9, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol, and 0.2% NP-40) with 100 mM KCl (Zhang et al., 2002) and incubated overnight with anti-M2 FLAG agarose at 4°C. The beads were washed twice in BC with 300 mM KCl. The beads were washed four times in BC with 100 mM KCl. Proteins were eluted at 4°C with 150 µg/ml 3X-FLAG peptide (SIGMA). The elutions were concentrated on Microcon columns (Millipore) and resolved on a 12% SDS-PAGE gel. Gels were stained with colloidal Coomassie (Invitrogen) and excised bands sequenced by the Harvard Microchemistry Facility.
For purification with anti-Grg4 antibodies, 800 mg of nuclear extract from flag-Grg4 transfected HEK 293 cells was loaded onto an 80 ml P11 column and fractionated as above. Peak fractions were loaded onto a 40 ml DEAE-sephacel column as above. Grg4 fractions were dialyzed into buffer BC with 0.1M KCl (BC.1), loaded onto a 15 ml CM-sepharose column (Pharmacia) and eluted with increasing concentrations of KCl in BC buffer. Peak Grg4 fractions were dialyzed into BC.1 and fractionated with increasing concentrations of KCL in BC buffer on a 5 ml SP-sephaorse (Pharmacia) column. The final purification was performed on a 5 ml Q-sepharose column with increasing concentrations of KCl in BC buffer. After dialysis into BC.1, protein complexes were IPed overnight with either 20 µg of goat IgG or 20 µg of goat anti-Grg4 that had been coupled to protein G beads. The beads were washed in BC.1 four times for 10 minutes, followed by four washes with BC .3M KCl, followed by four washes with BC.1. Proteins bound to beads were eluted in 0.9 ml 0.1M glycine pH 3.0, neutralized with 0.1 ml 1 M Tris pH 8.0, and concentrated with Strataclean beads (Agilent Technologies, Santa Clara, Ca). 15% of the sample was separated by SDS-PAGE on a 10% gel with 5% input and immunoblotted. The remainder was resolved on a 10% SDS-PAGE Criterion gel (BioRad), stained with colloidal Coomassie, and excised bands sequenced by MS/MS.
Coimmunoprecipitation and Immunoblotting
For IPs, 1 mg of HEK293 nuclear extract was cleared with protein-A agarose or protein-G sepharose (Amersham Biosciences). 2–4 µg of antibody was added and protein complexes were immobilized on agarose beads. After washing with IP buffer, the beads were boiled in 2X Lamelli sample buffer and separated by SDS-PAGE with 2% input. Immunoblotting was performed as previously described (Patel and Dressler, 2004). All the co-IPs were also done with 200 µg/ml ethidium bromide (Lai and Herr, 1992).
Histone Methyltransferase Assay
3H-HMT assay was performed as described (Pal et al., 2004), with 1 µg recombinant histone H3 or H4 (Upstate), 4 µg core histones (Upstate), or 4 µg mononucleosomes (gift from D. Bochar) as substrates and FLAG purified extracts in a 25-ul reaction mixture containing 15 mM HEPES pH 7.9, 100 mM KCl, 5 mM MgCl2, 20% glycerol, 1 mM EDTA, 0.25 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 2.75 uCi of S-(3H) adenosylmethionine (SAM) (Amersham Pharmacia Biotech., Inc.). After 90 min incubations at 30°C, the reaction was boiled in 2X SDS sample buffer, separated on 15% SDS-PAGE gels, and visualized as described (Patel et al., 2007). For unlabeled SAM reactions, gels were transferred to PVDF membrane and immunobloted with antibodies specific for H4R3me2s.
RNA Interference and Rescue of PRMT5 in HEK293 Cells
Three hPRMT5 siRNA sequences were purchased from Dharmacon: GAGGAUUGCAGUGGCUCUUdTdT (n600–618 of human PRMT5 cDNA), GGCCAUCUAUAAAUGUCUGdTdT (n1017–1035), and CCGCUAUUGCACCUUGGAAdTdT (n1599–1617). Cells were transfected with pooled PRMT5 siRNAs at a concentration of 80 nM per siRNA (240 nM total siRNA) using lipofectamine 2000 (Invitrogen). Control used the Dharmacon universal control siRNA at a final concentration of 240 nM. After 96 hr, cells were split 1:4 and retransfected with siRNA. Maximal depletion of PRMT5 was 80–120 hours after the first transfection. 24 h after the second siRNA transfection, cells were transfected with 1 µg mouse CMV-PAX2, 1µg CMV-Pax2 and 2 µg FLAG-Grg4, or FLAG-Grg4 (2µg) as described (Patel et al., 2007). PRMT5 expression was rescued in the knockdown cells with 2 µg of Myc-PRMT5.
ChIP and Real-Time PCR
GFP reporter cells were harvested 48 hr after transfection. ChIP was performed exactly as described previously (Patel et al., 2007). The precipitated DNA was reconstituted in sterile water and real-time PCR quantitation of precipitated genomic DNA relative to inputs was performed in triplicate using IQ SYBR GREEN with ROX mastermix (Bio-rad) in an iCycler (Bio-rad). The sequences of the PRS primer pair are 5’ GCTACCGGACTCAGA TCTCG-3’ and 5’-TGCGAAGTGGACCTCGGACC-3’. The sequences for Rap1A primer set 12 are 5’-CTTTAAGCGGACTCCGGAAC-3’ and 5’-CTCCTCCTCCTCCCTCCTCT-3’. The sequences for the other Rap1A primer sets are available upon request. ChIP data are presented as percent input after subtraction of non-specific binding to either rabbit IgG or Goat IgG. In all experiments, non-specific binding was less than 0.02% of input.
RNA Microarray Analyses
Total RNA was isolated from HEK293 cells 48 h after transfection using TRIZOL (Invitrogen, Carlsbad, CA). Microarray analyses were done by the University of Michigan Comprehensive Cancer Center (UMCCC) Affymetrix and Microarray Core Facility. The FL-Ovation™ cDNA Biotin Module V2 kit (NuGEN Technologies, San Carlos, CA) was used to produce biotin-labeled cRNA, which was then fragmented and hybridized to Human U133 Plus 2.0 gene chips (Affymetrix, Santa Clara, CA). Array hybridization, washes, staining, and scanning procedures were carried out according to standard Affymetrix protocols. Expression data were normalized by the robust multiarray average method and fitted to weighted linear models in R, using the affy and limma packages of Bioconductor, respectively (Irizarry et al., 2006; Smyth, 2004). Only probe sets with a variance over all samples superior to 0.1, a p-value inferior or equal to 0.05 after adjustment for multiplicity using the false discovery rate and a minimum 2-fold difference in expression were deemed significant.
Electrophoretic Mobility Shift Assay
The paired domain from Pax2, amino acids 1–170 was expressed in E. coli and purified as previously described (Brophy et al., 2001). DNAs were end-labeled with γ-(32)P-ATP and polynucleotide kinase. Binding reactions were performed in Z buffer (25 mM Hepes pH 7.8, 20% glycerol, 12.5 mM MgCl2, 0.1 M KCl, 1 mM DTT). Free DNA and DNA-protein complexes were resolved at room temperature on 6% neutral polyacrylamide gels in 0.5 X TBE at 150 V.
Highlights.
Expression of Grg4 displaces PTIP and Mll3/4 on chromatin at a Pax2 binding site.
Grg4 recruits the PRMT5 arginine methyltransferase to symmetrically methylate H4R3.
PRMT5 is necessary for PRC2 recruitment and H3K27 methylation.
Grg4 switches the epigenetic marks at a Pax2 target from activating to silencing.
ACKNOWLEDGMENTS
We thank Dr. William Lane of the Harvard Microsequencing facility for the Mass Spectrometry and Dr. Dan Bochar for mononucleosomes. This work was supported by NIH grant DK073722 (G.R.D.) and DK082409 (S.R.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nature cell biology. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blake JA, Thomas M, Thompson JA, White R, Ziman M. Perplexing Pax: from puzzle to paradigm. Dev Dyn. 2008;237:2791–2803. doi: 10.1002/dvdy.21711. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes & development. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development (Cambridge, England) 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- Burrill JD, Moran L, Goulding MD, Saueressig H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development (Cambridge, England) 1997;124:4493–4503. doi: 10.1242/dev.124.22.4493. [DOI] [PubMed] [Google Scholar]
- Cai Y, Brophy PD, Levitan I, Stifani S, Dressler GR. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. The EMBO journal. 2003;22:5522–5529. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Lechner MS, Nihalani D, Prindle MJ, Holzman LB, Dressler GR. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. The Journal of biological chemistry. 2002;277:1217–1222. doi: 10.1074/jbc.M109663200. [DOI] [PubMed] [Google Scholar]
- Cesaro E, De Cegli R, Medugno L, Florio F, Grosso M, Lupo A, Izzo P, Costanzo P. The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene. The Journal of biological chemistry. 2009;284:32321–32330. doi: 10.1074/jbc.M109.043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nature neuroscience. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Current opinion in genetics & development. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, et al. PTIP Promotes Chromatin Changes Critical for Immunoglobulin Class Switch Recombination. Science (New York, NY. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. The EMBO journal. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO reports. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno K, Masatsugu T, Sonoda M, Sasazuki T, Yamamoto K. Association of Polycomb group SUZ12 with WD-repeat protein MEP50 that binds to histone H2A selectively in vitro. Biochemical and biophysical research communications. 2006;345:1051–1058. doi: 10.1016/j.bbrc.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development (Cambridge, England) 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22:789–794. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome biology. 2008;9:205. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Molecular cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Kim D, Patel SR, Xiao H, Dressler GR. The role of PTIP in maintaining embryonic stem cell pluripotency. Stem Cells. 2009;27:1516–1523. doi: 10.1002/stem.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop KE, MacDonald LM, Lobe CG. Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mechanisms of development. 1996;59:73–87. doi: 10.1016/0925-4773(96)00582-5. [DOI] [PubMed] [Google Scholar]
- Lacroix M, El Messaoudi S, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO reports. 2008;9:452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS genetics. 2010;6:e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderson Y, Eberhard D, Malin S, Johansson A, Busslinger M, Pettersson S. Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell-specific genes. EMBO reports. 2004;5:291–296. doi: 10.1038/sj.embor.7400089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Human molecular genetics. 2008;17:R28–R36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- Majumder S, Alinari L, Roy S, Miller T, Datta J, Sif S, Baiocchi R, Jacob ST. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. Journal of cellular biochemistry. 2010;109:553–563. doi: 10.1002/jcb.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Sugiyama S. Polarity and laminar formation of the optic tectum in relation to retinal projection. Journal of neurobiology. 2004;59:48–56. doi: 10.1002/neu.10339. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nature genetics. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO reports. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Molecular and cellular biology. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Patel SR, Dressler GR. Expression of Pax2 in the intermediate mesoderm is regulated by YY1. Developmental biology. 2004;267:505–516. doi: 10.1016/j.ydbio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Developmental cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development (Cambridge, England) 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Schrons H, Knust E, Campos-Ortega JA. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Molecular and cellular biology. 2011;31:1503–1511. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Current opinion in cell biology. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Molecular cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Current opinion in cell biology. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. The Journal of clinical investigation. 2011;121:2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S, Funahashi J, Nakamura H. Antagonizing activity of chick Grg4 against tectum-organizing activity. Developmental biology. 2000;221:168–180. doi: 10.1006/dbio.2000.9643. [DOI] [PubMed] [Google Scholar]
- Sugiyama S, Nakamura H. The role of Grg4 in tectal laminar formation. Development (Cambridge, England) 2003;130:451–462. doi: 10.1242/dev.00232. [DOI] [PubMed] [Google Scholar]
- Tae S, Karkhanis V, Velasco K, Yaneva M, Erdjument-Bromage H, Tempst P, Sif S. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic acids research. 2011;39:5424–5438. doi: 10.1093/nar/gkr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development (Cambridge, England) 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development (Cambridge, England) 1996;122:3381–3391. doi: 10.1242/dev.122.11.3381. [DOI] [PubMed] [Google Scholar]
- Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Molecular and cellular biology. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC bioinformatics. 2011;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Bouchard M, Stone D, Liu X, Vella F, Lee J, Nakamura H, Ang SL, Busslinger M, Rosenthal A. Distinct regulators control the expression of the mid-hindbrain organizer signal FGF8. Nature neuroscience. 2001;4:1175–1181. doi: 10.1038/nn761. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Molecular cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature structural & molecular biology. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]