SUMMARY
Major histocompatibility complex (MHC)-restriction is the cardinal feature of T cell antigen recognition and is thought to be intrinsic to αβ T cell receptor (TCR) structure because of germline-encoded residues which impose MHC specificity. Here, we analyzed TCRs from T cells that had not undergone MHC-specific thymic selection. Instead of recognizing peptide-MHC complexes, the two αβTCRs studied here resembled antibodies in recognizing glycosylation-dependent conformational epitopes on a native self-protein, CD155, and they did so with high affinity independently of MHC molecules. Ligand recognition was via the αβTCR combining site and involved the identical germline-encoded residues that have been thought to uniquely impose MHC specificity, demonstrating that these residues do not only promote MHC binding. Thus, this study demonstrates that, without MHC-specific thymic selection, αβTCRs can resemble antibodies in recognizing conformational epitopes on MHC-independent ligands.
INTRODUCTION
The adaptive immune system is composed of T and B lymphocytes bearing antigen receptors generated by gene recombination to recognize a huge diversity of different antigens. Although generated by the same recombination machinery, antigen receptors on T and B cells recognize fundamentally different kinds of antigenic ligands. Antigen receptors on B cells recognize conformational epitopes on native proteins, whereas antigen receptors on mature αβT cells (αβTCRs) only recognize linear peptides of antigenic proteins bound to products of the major histocompatibility complex (MHC) (Davis and Bjorkman, 1988). The unique recognition characteristic of mature T cells is referred to as `MHC-restriction' because they are restricted to only recognizing peptides of antigenic proteins bound to MHC glycoproteins as antigenic peptide-MHC (pMHC) complexes. MHC-restriction focuses T cell recognition on cell bound MHC molecules that display peptides derived from proteins either synthesized within the cell or pinocytosed from extracellular fluids. MHC-restricted antigen recognition is the cardinal feature of αβTCR recognition and is central to αβT cell function, but its basis is not known.
One perspective proposes that MHC-restriction is germline-encoded and intrinsic to αβTCR structure (Feng et al., 2007; Huseby et al., 2005; Merkenschlager et al., 1997; Zerrahn et al., 1997). The germline concept is supported by structural analyses of αβTCRs which reveal that αβTCR binding to pMHC complexes not only involves amino acid residues encoded in the highly variable complementary determining region (CDR) 3 that directly contact antigenic peptides in the MHC groove, but also involves evolutionarily conserved amino acid residues encoded in the invariant CDR2 region that directly contact MHC α-helices (Garcia et al., 2009; Marrack et al., 2008; Rudolph et al., 2006). Based on these structural analyses, it has been proposed that germline encoded amino acid residues in the invariant CDR2 region specifically promote MHC binding and account for the preferential binding of αβTCRs to pMHC complexes (Garcia et al., 2009; Marrack et al., 2008). Notably, the germline basis of MHC restriction is not contradicted by reports of rare αβTCRs cloned from conventional T cell populations that bind ligands independently of MHC molecules (Barnd et al., 1989; Hanada et al., 2011; Rao et al., 1984; Siliciano et al., 1985) because their MHC-independent ligand is bound with such low apparent affinity that it is likely not to be their TCR's primary recognition specificity (Garcia et al., 2009).
An alternative to the germline concept is that MHC restriction is imposed by thymic selection (Collins and Riddle, 2008; Van Laethem et al., 2007). The thymic selection concept proposes that αβTCRs specific for MHC-independent ligands exist and are expressed on preselection thymocytes, but fail thymic selection and so are excluded from the mature αβT cell repertoire (Van Laethem et al., 2007). A key presumption of this perspective is that thymic selection distinguishes MHC-specific from MHC-independent αβTCRs, but a potential mechanism for distinguishing MHC-specific from MHC-independent ligand engagements was only recently proposed (Van Laethem et al., 2007). Extending observations in mature T cells (Haughn et al., 1992) to preselection thymocytes, we proposed that Lck, the kinase necessary for most TCR signaling, is sequestered away from αβTCRs on preselection thymocytes by CD4 and CD8 coreceptor proteins which bind to MHC molecules, with the result that immature thymocytes can only be signaled to undergo selection by αβTCRs that access Lck by co-engaging pMHC complexes together with CD4 or CD8 coreceptors (Van Laethem et al., 2007). However, if preselection thymocytes were deficient in both CD4 and CD8 coreceptor proteins, Lck would be available to all αβTCRs which would signal thymic selection upon engagement of any intrathymic ligand. Thus CD4 and CD8 coreceptor proteins impose MHC specificity on thymic selection and impose MHC restriction on the mature αβTCR repertoire. In this perspective, every αβTCR that has been analyzed to date possesses structural features that promote binding to MHC molecules because each αβTCR had been pre-screened for MHC-specificity in the thymus (Marrack et al., 2008; Rudolph et al., 2006).
We now characterize αβTCRs obtained from mature T cells that had not undergone MHC-specific selection in the thymus. We analyzed two autoreactive αβTCRs cloned from T cells in the lymphoid periphery of mice deficient in both MHC and coreceptor proteins (so-called `Quad-deficient' mice) (Van Laethem et al., 2007). Remarkably, instead of pMHC complexes, these two αβTCRs recognized conformational epitopes on the self-protein CD155 and did so with an apparent affinity that was substantially higher than the affinity with which conventional αβTCRs bind to cognate pMHC complexes. Binding to their MHC-independent ligand depended on specific CDR3α sequences in the TCR combining site and also depended on the identical CDR2β amino acid residues (Y46, Y48, E54) that have been proposed to confer germline-encoded MHC specificity (Scott-Browne et al., 2009). Thus, our study demonstrates that, without MHC-specific thymic selection, αβTCRs on post-thymic T cells can resemble antibodies in recognizing conformational epitopes on MHC-independent ligands.
RESULTS
Specificity of αβTCRs that did not undergo MHC-specific thymic selection
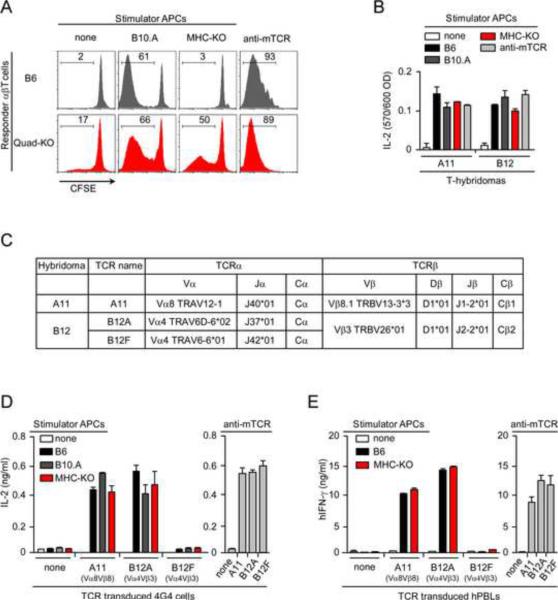
To obtain αβT cells that had not been screened in the thymus for MHC-specificity, we utilized mice deficient in both MHC and coreceptor proteins (B2m−/−H-2-Ab1−/−Cd4−/−Cd8a−/− mice) that we refer to as Quad-deficient mice. Quad-deficient mice contain peripheral αβT cells whose numbers are further increased by the anti-apoptic Bcl-2 transgene (Bcl-2Tg) that prevents strongly signaled Quad-deficient thymocytes from undergoing clonal deletion (Van Laethem et al., 2007). Because MHC complexes are not expressed in Quad-deficient.Bcl2 mice, their T cells express αβTCR that have not been screened for peptide-MHC specificity and so might display unique recognition specificities (Figure S1). Indeed, Quad-deficient.Bcl2 αβT cells recognized and responded against cell surface ligands on MHC-deficient stimulator cells, whereas conventional MHC-selected αβT cells from B6 or B10.A mice did not (Figure 1A).
Figure 1. αβTCR from Quad-deficient mice recognize ligands independently of MHC molecules.
(A) Reactivity of αβT cells from B6 and Quad-deficient mice. Proliferation of CFSE-labeled LN αβT cells (1×105) was measured after 4d culture with 2×105 stimulator cells (T depleted, LPS-stimulated, and irradiated splenocytes). Stimulation with soluble anti-TCRβ (1 μg/ml) in the presence of FcR+ cells from MHC-deficient mice served as a positive control. Data are representative of six independent experiments.
(B) Specificity of A11 and B12 T-hybridomas generated from Quad-deficient.Bcl2 αβTcells. T-hybridomas (1×105) were cultured with stimulators (2×105) or platebound anti-TCRβ (5 μg/ml) for 16hr and assayed for IL-2 production by proliferation of the IL-2 dependent cell line CTLL. Each data point represents the mean (±SEM) of triplicate cultures. Data are representative of five independent experiments.
(C) Characterization of αβTCR from A11 and B12 T-hybridomas. The A11 T-hybridoma expressed a TCR-Vα8 chain and a TCR-Vβ8.1 chain and we refer to this TCR pair as `A11'. The B12 T-hybridoma contained a TCR-Vβ3 chain and two TCR-Vα4 chains that were from different subfamilies (TRAV6D-6*02 and TRAV6-6*01) and contained different Jα regions (J37*01 and J42*01). The `B12A' TCR consists of the TCR-Vβ3 chain paired to the TRAV6D-6*02 J37*01 chain, and the `B12F' TCR consists of the TCR-Vβ3 chain paired to the TRAV6-6*01 J42*01 chain.
(D) Reactivity of untransduced or TCR-transduced 4G4 cells (4×104) against stimulators (2×105) or platebound anti-TCRβ (5 μg/ml) after 16hr stimulation as determined by ELISA quantification of IL-2 secretion. Data are representative of five independent experiments.
(E) Reactivity of hPBL transduced with murine TCR (2×105) against stimulators (5×105) or platebound anti-TCRβ (murine, 5 μg/ml) as determined by ELISA quantification of hIFN-γ secretion. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two independent experiments.
To study the ligand specificities of MHC-unselected αβTCRs, we generated multiple T-hybridoma cell lines from Quad-deficient.Bcl2 T cells which reacted against MHC-deficient stimulators. We selected two T-hybridomas, A11 and B12, to be studied in detail (Figure 1B) and sequenced their TCR chains (Figure 1C and Figure S2). A11 T-hybridoma cells expressed one TCRα and one TCRβ chain to form a TCR that was Vα8Vβ8.1 and was designated `A11'. B12 T-hybridoma cells expressed two TCRα chains that were both Vα4 and expressed one TCRβ chain that was Vβ3 to form two different Vα4Vβ3 TCRs referred to as B12A and B12F (Figure 1B and Figure S2). The TCRα chains were from different Vα4 subfamilies (TRAV6D-6*02 and TRAV6-6*01) and differed mainly in their Jα regions (J37*01 versus J42*01). The Vα4Vβ3 TCR containing the TRAV6D-6*02 J37*01 TCRα chain was designated `B12A', and the Vα4Vβ3 TCR that uses the TRAV6-6*01 J42*01 TCRα chain was designated `B12F'.
To determine the ligand specificities of A11, B12A, and B12F TCRs, we retrovirally transduced each TCR into the TCR-negative 4G4 T cell line which is also coreceptor negative (Hong et al., 1992). Untransduced 4G4 cells lacked TCRs and so could not be stimulated by anti-TCR (Figure 1D). However, 4G4 cells transduced with A11, B12A, and B12F TCRs did respond to anti-TCR stimulation, indicating that they expressed functional αβTCRs (Figure 1D). Importantly, 4G4 cells expressing A11 and B12A TCRs also responded against MHC-deficient and MHC-sufficient stimulators, whereas 4G4 cells expressing B12F TCRs did not (Figure 1D). Thus, A11 and B12A TCRs recognized MHC-independent ligands expressed on MHC-deficient stimulator cells but B12F TCR displayed no observable ligand specificity. We then retrovirally transduced A11, B12A, and B12F TCRs into human peripheral blood lymphocytes (hPBLs) to assess ligand recognition without contribution from species-specific adhesion molecules (Figure1E). The murine TCRs were functional in hPBL as they signaled human interferon-γ (hIFN-γ) secretion in response to anti-murine TCR. More importantly, A11 and B12A TCRs on hPBL conferred reactivity to MHC-deficient murine stimulators (Figure 1E), demonstrating that these TCRs recognized murine ligands with sufficiently high affinity to signal responses without contribution from species-specific adhesion molecules.
CD155 is a self-protein recognized by A11 and B12A TCR independently of MHC
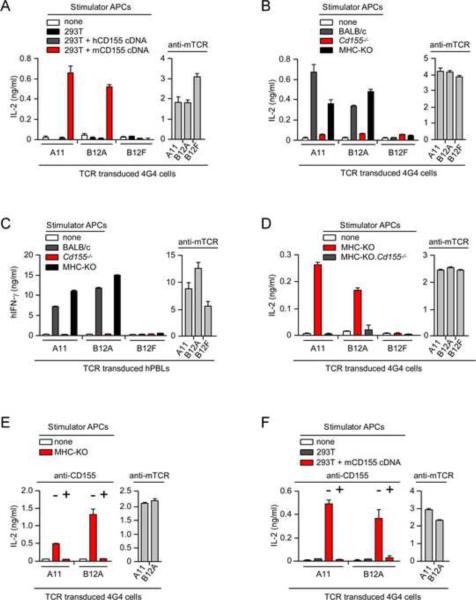
To identify the stimulatory ligand(s) recognized by A11 and B12A TCRs, we performed limiting dilution cDNA expression cloning. We generated a cDNA library from the murine L1210 cell line which potently stimulated both A11 and B12A TCRs (Figure S3), and introduced the L1210 cDNA library into the human 293T cell line which was non-stimulatory for both TCRs (Figure S3). We ultimately identified three cDNA clones each of which encoded stimulatory ligand(s) for both A11 and B12A TCRs. To our surprise, all three stimulatory cDNA clones encoded the same protein, namely murine CD155 (mCD155) which is the murine analog of the human poliovirus receptor and is expressed in varying amounts on hematopoietic and non-hematopoietic cells (Chadeneau et al., 1996a; Chadeneau et al., 1996b; Maier et al., 2007).
To confirm that mCD155 was indeed the ligand recognized by both A11 and B12A TCRs, we transfected bona fide mCD155 cDNA into 293T cells and used the transfected cells to stimulate TCR transduced 4G4 cells. 4G4 transduced cells expressing A11 and B12A TCR responded against 293T cells transfected with mCD155 cDNA but did not respond against 293T cells transfected with human CD155 (hCD155) cDNA, documenting that mCD155 was the stimulatory ligand for both A11 and B12A TCRs (Figure 2A). In addition, A11 and B12A TCRs which signaled 4G4 and hPBL to respond against wildtype murine stimulator cells failed to signal responses against CD155-deficient stimulator cells from Cd155−/− (CD155-deficient) mice (Maier et al., 2007), confirming that both TCRs recognized and reacted against mCD155 proteins on murine stimulator cells (Figure 2B,C).
Figure 2. Murine CD155 is the ligand recognized by A11 and B12A TCRs.
(A) IL-2 production by TCR-transduced 4G4 cells (4×104) after 16hr stimulation with 293Tcells transfected with cDNA encoding either hCD155 or mCD155. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of five experiments.
(B) TCR-transduced 4G4 cells (4×104) do not respond against CD155-deficient stimulators (5×105). Data are representative of three experiments.
(C) TCR-transduced hPBL (2×105) do not respond against CD155-deficient stimulators. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two experiments.
(D) TCR-transduced 4G4 cells (4×104) do not respond against CD155-deficient stimulators (5×105). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
(E) Anti-mCD155 (10 μg/ml) blocks stimulation of TCR-transduced 4G4 cells (4×104) by MHC-deficient (MHC-KO) stimulators (5×105). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
(F) Anti-mCD155 (10 μg/ml) blocks stimulation of TCR-transduced 4G4 cells (4×104) by 293T cells transfected with cDNA encoding mCD155. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
Before concluding that mCD155 was the MHC-independent ligand recognized by A11 and B12A TCRs, we documented that mCD155 was the ligand specifically recognized by A11 and B12A TCRs on MHC-deficient stimulator cells. Indeed, A11 and B12A TCRs responded against MHC-deficient stimulator cells but did not respond against MHC-deficient stimulator cells that were additionally CD155-deficient, documenting that mCD155 was the stimulatory ligand on MHC-deficient cells (Figure 2D).
TCR recognition of native mCD155 proteins in the absence of antigen processing
Ligand recognition by conventional αβTCRs requires antigen processing, which involves degradation of antigenic proteins into peptide fragments that are subsequently loaded onto MHC molecules and expressed on the cell surface. As a consequence, ligand recognition by conventional αβTCRs cannot be blocked by antibodies directed against native antigenic proteins (Schwartz et al., 1976). However, because antigen recognition by A11 and B12A TCRs is MHC-independent, we considered that their ligand recognition might be susceptible to blockade by antibodies directed against the native antigenic protein. In fact we observed that A11 and B12A responses were blocked by anti-mCD155 (Figure 2E and Figure 2F).
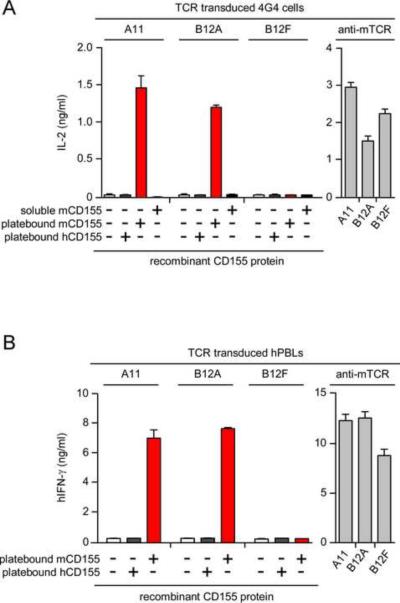
Blockade of CD155-specific responses by anti-mCD155 suggested that ligand recognition by A11 and B12A TCRs did not require antigen processing. Consequently, we wondered if these TCRs could respond to recombinant mCD155 proteins that had not undergone antigen processing and in the absence of antigen presenting cells (APC). We observed that TCR-transduced 4G4 and hPBL expressing A11 and B12A TCRs both responded to platebound recombinant mCD155 protein (Figure 3A and Figure 3B). These responses were TCR-specific because transduced cells expressing A11 and B12A TCR responded but those expressing B12F TCR did not; and these responses were antigen-specific because they were specific for immobilized mCD155 but not immobilized hCD155 (Figure 3A and Figure 3B).
Figure 3. A11 and B12A TCR directly recognize recombinant mCD155 protein immobilized on plastic.
(A) Responses of TCR-transduced 4G4 cells (4×104) to immobilized (10 μg/ml) or soluble (10 μg/ml) CD155 protein. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of four experiments.
(B) Responses of TCR-transduced hPBL to immobilized CD155 protein (10 μg/ml). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two independent experiments.
We conclude that, unlike conventional αβTCRs, A11 and B12A TCRs recognize native mCD155 proteins and do so without antigen processing.
Frequency of T-hybridomas with CD155 specificity
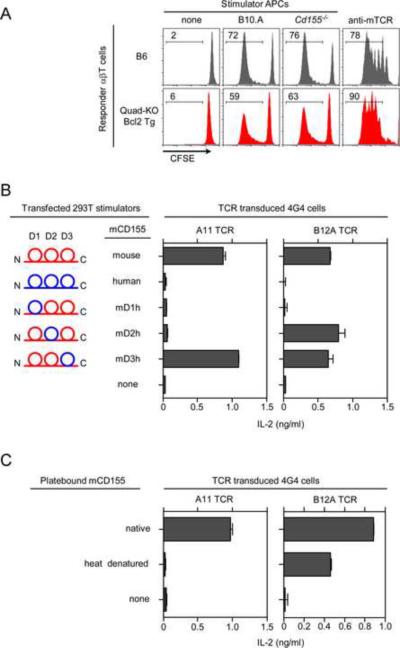
Although A11 and B12 T-hybridomas were generated from the same Quad-deficient.Bcl2 T cell population, we were surprised that their αβTCRs recognized the same protein ligand, i.e. mCD155, so we decided to assess whether CD155 specificity was frequent among newly generated Quad-deficient T-hybridomas. We stimulated Quad-deficient.Bcl2 primary T cells with immobilized anti-TCR+anti-CD28 and fused them with BW5147 thymoma cells to generate a large panel of Quad-deficient T hybridomas. Remarkably, we found that ~40% of newly generated TCR+ Quad-deficient T-hybridomas displayed CD155-specificity which might either reflect preferential survival of T-hybridomas bearing CD155-specific TCRs or might infer that CD155 was a predominant ligand for MHC-independent Quad-deficient T cells. Contrary to this latter possibility, polyclonal Quad-deficient primary T cells proliferated as vigorously against CD155-deficient as against CD155-sufficient stimulators (Figures 4A and S4). Consequently, we think that Quad-deficient T-hybridomas display a markedly skewed sample of the MHC-independent ligand specificities present among Quad-deficient primary T cells.
Figure 4. Ligand specificity of MHC-independent αβTCRs.
(A) αβT cells. Proliferative responses were measured by CFSE dye dilution of primary αβT cells from Quad-deficient.Bcl2 (Quad-KO.Bcl2) mice against various stimulators. Data are representative of two experiments.
(B) CD155 epitopes recognized by MHC-independent TCRs. Schematic representation of chimeric mouse-human CD155 molecules are displayed (left). IL-2 response of TCR-transduced 4G4 cells (4×104) stimulated with 293T cells transfected with cDNA encoding various CD155 chimeric constructs (right). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two experiments.
(C) IL-2 response of TCR transduced 4G4 cells (1×105) stimulated with either native or heat denatured recombinant mCD155 protein. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three independent experiments.
Recognition of different conformational epitopes on mCD155
To determine if A11 and B12A TCRs which were both CD155-specific actually recognized different antigenic epitopes on CD155, we stimulated A11 and B12A TCRs with 293T cells that had been transfected with cDNA encoding murine-human CD155 chimeric proteins in which each of the three mCD155 domains (D1, D2, and D3) was replaced by its hCD155 counterpart (Figure 4B). We found that stimulation of B12A TCR required the murine D1 domain as they did not respond against mCD155 chimeric proteins containing the human D1 domain (Figure 4B), and that stimulation of A11 TCR required both murine D1 and D2 domains as they did not respond against mCD155 chimeric proteins containing either human D1 or D2 domains (Figure 4B). Thus A11 and B12A TCRs recognize different mCD155 epitopes, with B12A recognizing an epitope confined to the D1 domain of mCD155 and A11 recognizing a complex epitope composed of both D1 and D2 domains.
Because conventional αβTCRs do not recognize conformational epitopes on native antigenic proteins, denaturation of the antigenic protein does not affect their recognition by conventional αβTCRs. In marked contrast, we found that heat denaturation of recombinant mCD155 proteins abolished A11's recognition of its complex D1–D2 epitope and reduced B12A's recognition of its D1 epitope (Figure 4C). These results demonstrate that A11 and, to a lesser extent, B12A recognize conformational epitopes on mCD155 proteins.
TCR recognition of CD155 depends on glycosylation
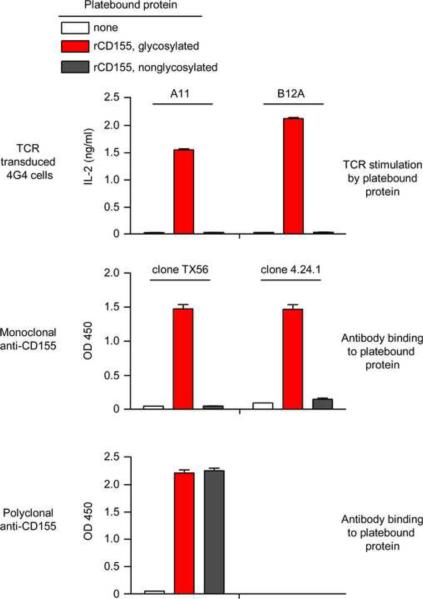
Because conventional αβTCRs recognize linear peptides of antigenic proteins presented by MHC, glycosylation of the antigenic protein from which the peptides are derived does not usually enhance T cell immunogenicity. In contrast A11 and B12A TCRs recognized conformational antigenic epitopes, and conformational epitopes are often affected by glycosylation. Consequently, we compared antibody recognition and TCR responses to platebound recombinant CD155 proteins that were either glycosylated or non-glycosylated (Fig. 5). Polyclonal CD155-specific antibodies did not distinguish glycosylated and non-glycosylated CD155 proteins as they recognized both (Fig. 5 bottom), but two different CD155-specific monoclonal antibodies detected only glycosylated CD155 and failed to detect non-glycosylated CD155 (Fig. 5 middle), indicating that the conformational epitopes recognized by the mAbs were dependent on CD155 glycosylation. Interestingly, CD155-specific responses by A11 and B12A TCRs resembled the two mAbs in discriminating between glycosylated and nonglycosylated CD155 proteins. Indeed, A11 and B12A TCRs both responded to glycosylated CD155 but failed to respond to non-glycosylated CD155 proteins (Fig. 5 top). Thus, antigen recognition by A11 and B12A TCRs resembled monoclonal antibodies in being dependent on glycosylation and differed fundamentally from that by conventionally MHC-selected αβTCRs.
Figure 5. TCR recognition of CD155 depends on glycosylation.
IL-2 response of TCR transduced 4G4 cells (1×105) stimulated with either glycosylated or nonglycosylated recombinant CD155 (10 μg/ml) in medium (top panel), in the presence of 10 μg/ml monoclonal anti-CD155 (clones TX56 and 4.24.1) (middel panel), or in the presence of 10 μg/ml polyclonal anti-CD155 (bottom panel). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two experiments.
Role of CDR3α TCR residues in ligand recognition
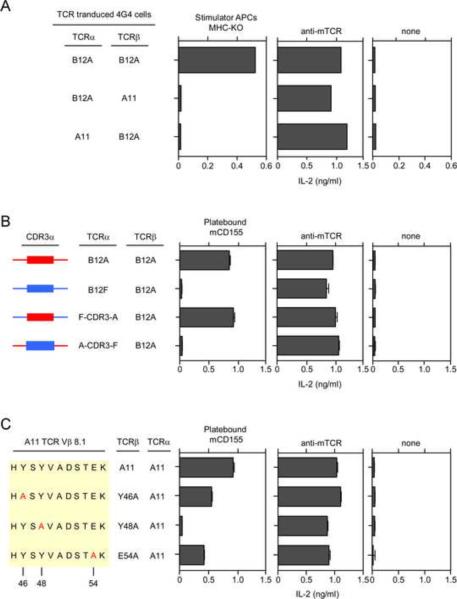
To better understand the basis for αβTCR recognition of mCD155, we considered the possibility that A11 and B12A TCRs might bind to mCD155 outside of their antigen combining sites, in a manner resembling superantigen binding to TCRβ chains (Li et al., 1999). Consequently, we performed mix-and-match experiments in which we transduced 4G4 cells with mismatched α and β TCR chains from A11 and B12A. Mismatched αβTCR chains were functionally expressed on the 4G4 cell surface as they signaled interleukin-2 (IL-2) secretion in response to anti-TCR stimulation (Figure 6A). However, mismatched αβTCR chains failed to respond to MHC-deficient stimulators, indicating that correctly matched αβTCR pairs were required for mCD155 recognition.
Figure 6. Ligand Recognition by A11 and B12A TCR.
(A) Ligand recognition by A11 and B12A TCRs requires specific αβ pairing. IL-2 responses of 4G4 cells (4×104) transduced with the indicated TCR chains. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of two experiments.
(B) Importance of CDR3α-encoded TCR sequences for CD155 recognition. CDR3α sequences were exchanged between B12A and B12F to generate chimeric F-CDR3-A and A-CDR3-F TCRα chains. F-CDR3-A designates B12F TCRα chains with B12A CDR3α sequences, and A-CDR3-F designates B12A TCRα chains with B12F CDR3α sequences. The indicated TCRα chains were transduced together with B12 TCRβ chains into 4G4 cells, and the TCR-transduced 4G4 cells (1×105) were stimulated with mCD155 (10 μg/ml). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
(C) Role of CDR2β-encoded Y46, Y48, E54 TCR residues in mCD155 recognition. Y46, Y48, E54 amino acid residues in the TCR-Vβ8.1 chain of the A11 TCR were each mutated to alanine, were transduced into 4G4 cells along with the A11 TCRα chain, and stimulated with mCD155 protein (10 μg/ml). Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
Next, we examined if the CDR3 TCR region, a major component of the TCR combining site, was important for mCD155 recognition. To pursue this possibility, we made use of the fact that B12A and B12F TCRs differed dramatically in their recognition of mCD155 but were structurally similar to one another, with the major difference being Jα-encoded CDR3α sequences (Figure 1B and Figure S2). Consequently, we swapped CDR3α sequences (identified in Figure S2) between B12A and B12F TCRα chains and assessed responses to immobilized recombinant mCD155 (Figure 6B). Impressively, replacing CDR3α sequences in B12F with those from B12A (designated F-CDR3-A) resulted in acquisition of mCD155 responsiveness (Figure 6B), and replacing CDR3α sequences in B12A with those from B12F (designated ACDR3-F) resulted in loss of mCD155 responsiveness (Figure 6B). Because both gain and loss of function were observed in these CDR3α swaps, we conclude that MHC-independent recognition of mCD155 by B12A TCR critically depends on CDR3α TCR sequences and so is mediated by the TCR combining site.
Role of germline-encoded CDR2β residues in ligand recognition
Structural analyses of conventional αβTCRs with peptide-MHC complexes have identified germline-encoded CDR2 TCR residues that contact MHC and promote MHC-specific thymic selection (Scott-Browne et al., 2009). It has been argued that such germline-encoded CDR2 residues provide a simple and compelling demonstration that MHC specificity is intrinsic to the germline-encoded structure of αβTCRs. However, the presumption underlying this argument is that germline-encoded CDR2 residues only promote TCR binding to peptide-MHC complexes, a presumption that could not previously be tested. We could now directly test this presumption because A11 TCRβ chains contain the canonical Vβ8.1 CDR2β residues (Y46, Y48, E54) that have been identified as MHC contact points (Marrack et al., 2008; Scott-Browne et al., 2009). To determine if these CDR2β residues affected MHC-independent TCR recognition of mCD155, we mutated to alanine each of the three canonical residues (Y46A, Y48A, E54A) and retrovirally transduced mutant A11 TCRβ and wildtype TCRα chains into 4G4 cells (Figure 6C). All three mutant A11 TCRs were functionally expressed in 4G4 cells as they responded to anti-TCR stimulation, but they differed in their recognition of recombinant mCD155 protein (Figure 6C). Strikingly, mutation of Y48 in the TCRβ chain completely abrogated A11 responsiveness to mCD155 protein, and TCRβ mutations of Y46 and E54 reduced A11 responsiveness (Figure 6C). These findings unexpectedly reveal that germline-encoded CDR2β residues are critically important for MHC-independent ligand recognition of CD155 conformational epitopes even though the structure of CD155 is distinct from that of MHC (He et al., 2000; Zhang et al., 2008).
A11 and B12A TCRs are not alloreactive and have no apparent peptide-MHC specificity
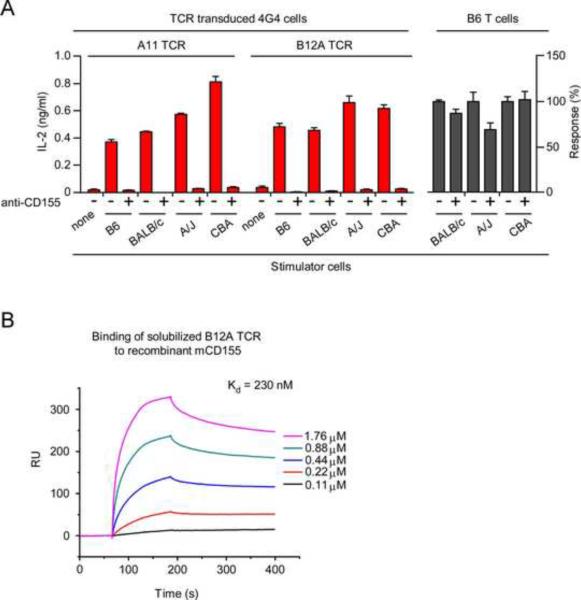
We then wondered if either A11 or B12A TCR could recognize syngeneic or allogeneic peptide-MHC complexes in addition to mCD155. To assess this possibility, 4G4 cells expressing A11 and B12A TCR were stimulated by cells expressing a variety of MHC and background genes, and responses were assessed in the presence and absence of blocking mCD155 mAb (Figure 7A). Notably, anti-CD155 did not interfere with the responses of conventional MHC-selected T cells from B6 mice against allogeneic BALB/c (H-2d), A/J (H-2a), or CBA (H-2k) stimulator cells (Figure 7A). However, anti-CD155 abrogated responses by A11 and B12A TCRs to the entire panel of stimulator cells, regardless of their MHC and background genes (Figure 7A). Thus, A11 and B12A TCR did not respond to any of the peptide-MHC complexes that either syngeneic or allogeneic stimulator cells expressed (Figure 7A).
Figure 7. A11 and B12A TCR specificities.
(A) A11 and B12A TCRs do not display alloreactivity against allogeneic pMHC complexes. Anti-mCD155 (10 μg/ml) blocked responses of TCR-transduced 4G4 cells (4×104) to a panel of allogeneic stimulator cells (5×105), but did not block alloreactive responses of primary B6 T cells. IL-2 responses of primary B6 T cells (2×105) in the presence of anti-mCD155 were expressed relative to those in the absence of anti-mCD155 which were set equal to 100%. Actual amounts of IL-2 secreted in response to each stimulator strain were BALB/c=292 pg/ml; A/J=277 pg/ml; CBA= 1234 pg/ml. Each data point represents the mean (±SEM) of triplicate culture. Data are representative of three experiments.
(B) Binding of soluble B12A TCR to recombinant mCD155 protein. Surface Plasmon Resonance (SPR) measurements of binding between solubilized B12A αβTCR heterodimers and immobilized recombinant mCD155. The analytes consisted of serial dilutions of soluble αβTCR heterodimers ranging from 0.11–1.76 μM. The dissociation constants were obtained by kinetic curve fitting using BIAevaluation. Data are representative of two experiments.
High affinity binding of soluble B12A TCR to recombinant mCD155
Finally, we generated soluble B12A α and β extracellular domains that refolded together to form soluble B12A αβ heterodimers whose binding to recombinant mCD155 protein could be determined by surface plasmon resonance (Figure 7B). In an entirely cell-free assay containing only B12A TCR and recombinant mCD155 protein, we observed that soluble B12A TCR did directly bind to recombinant mCD155 proteins and did so with the surprisingly high affinity of ~200 nanomolar (Figure 7B). The observed binding affinity is substantially higher than the 10–200 micromolar affinities with which conventional αβTCRs bind to peptide-MHC ligands (Margulies, 1997; Matsui et al., 1991). Indeed, the high binding affinity of B12A TCR to mCD155 was consistent with our observations that B12A TCR on intact cells (4G4 and hPBL) signaled mCD155-specfic responses without contributions from CD4 or CD8 coreceptor proteins or from species-specific adhesion molecules. These results indicate that mCD155 is the primary ligand for B12A αβTCR and perhaps for A11 αβTCR as well, although we have not yet successfully obtained soluble A11 αβ heterodimers.
DISCUSSION
The present study reveals that MHC-restriction is extrinsically imposed on the developing αβTCR repertoire. We have shown that T cells unscreened for MHC specificity in the thymus can express αβTCRs that recognize conformational antigenic epitopes independently of MHC molecules. The two αβTCRs analyzed in detail here (A11 and B12A) recognized different epitopes on CD155, a self-protein whose structure does not resemble that of MHC molecules (He et al., 2000; Rudolph et al., 2006; Takai et al., 2003; Zhang et al., 1992). Importantly, TCR recognition of CD155 was dependent on glycosylation and involved CDR3-encoded amino acids in the TCR antigen combining site, demonstrating that αβTCR recognized CD155 as a `conventional' antigen and not as a superantigen. In addition, MHC-independent TCR recognition of CD155 involved CDR2-encoded amino acids which were the identical CDR2-encoded residues required for recognition of pMHC complexes by conventional αβTCRs (Scott-Browne et al., 2009). We conclude that MHC specificity is not intrinsic to the germline-encoded structure of all αβTCRs and that, in the absence of MHC-specific thymic selection, ligand recognition by αβTCR can resemble that of antibodies.
The present study was based on the concept that thymic selection was constrained by CD4 and CD8 coreceptors to be specific for MHC molecules, so that in Quad-deficient mice thymic selection would be MHC-independent and result in mature T cells expressing αβTCRs with unknown ligand recognition specificities. A11 and B12A TCRs were obtained from T-hybridoma cells made from peripheral αβT cells generated in Quad-deficient.Bcl2 mice. The particular αβT cell from which the B12 T-hybridoma was constructed contained one TCRβ chain but two different, yet closely related, TCR-Vα4 chains: one TCR-Vα4 chain which formed the B12F TCR and was likely a passenger TCR chain during thymic selection, and one TCR-Vα4 chain which formed the B12A TCR and likely promoted thymic selection. The Vα4Vβ3 B12F TCR which had no discernible ligand recognition specificity provided an excellent negative TCR control for our present studies because it was nearly identical to the Vα4Vβ3 B12A TCR and primarily differed in Jα-encoded CDR3α sequences. Unlike the pMHC ligands recognized by conventional αβTCRs, A11 and B12A TCRs recognized glycosylation-dependent conformational epitopes on native CD155 proteins which neither required antigen processing nor required antigen presenting cells. As a result, antibodies specific for conformational epitopes on mCD155 blocked ligand recognition by A11 and B12A TCRs, whereas ligand recognition by conventional αβTCRs was only blocked by antibodies directed against MHC molecules. Moreover, the ability of platebound recombinant mCD155 to trigger A11 and B12A TCRs to signal both murine 4G4 cells and human PBL in the absence of any antigen presenting cells indicated that A11 and B12A bound mCD155 with sufficient affinity to trigger TCR signal transduction without contribution from intercellular adhesion molecules. Moreover, we were able to generate solubilized B12A αβTCR heterodimers and to determine that they bound recombinant mCD155 protein with ~ 200 nanomolar affinity which was 10–100 times higher than the micromolar affinities with which conventional αβTCRs bind antigenic pMHC complexes (Manning and Kranz, 1999; Margulies, 1997).
A few αβTCRs capable of MHC-independent ligand binding have been previously reported but, in contrast to the TCRs in the present study, all such αβTCRs were obtained from T cells that had undergone MHC-specific thymic selection and displayed low apparent affinity for their MHC-independent ligand (Barnd et al., 1989; Hanada et al., 2011; Rao et al., 1984). As the most recent example, an αβTCR obtained from T cells in human PBL bound a self-ligand independently of MHC, but it did so with low apparent affinity as it was unable to bind its MHC-independent ligand without additional CD2-CD58 adhesive interactions (Hanada et al., 2011). It has been argued that such low affinity binding to MHC-independent ligands can be a property of MHC-specific αβTCRs because they would only bind MHC-independent ligands when their preferred pMHC ligands were unavailable (Garcia et al., 2009). However, this argument is directly contradicted by the αβTCRs in the present study which bind mCD155 with high affinity. Moreover, A11 and B12A TCRs recognized their MHC-independent ligand even when confronted by a vast array of potential pMHC complexes on a panel of syngeneic and allogeneic stimulator cells, indicating that these MHC-independent αβTCRs were not alloreactive and that CD155 was likely their primary ligand. In fact the absence of A11 and B12A cross-reactivity contrasts with the extensive cross-reactivity displayed by conventional αβTCRs, suggesting that extensive cross-reactivity derives from MHC-specific thymic selection.
Analysis of CD155 recognition by MHC-independent TCRs revealed that it was mediated by the TCR antigen combining site and not by another site on the TCR. Experiments involving reciprocal exchanges of CDR3α sequences between B12A and B12F TCRs documented that B12A CDR3α sequences induced CD155 recognition by B12F TCR, and that B12F CDR3α sequences abrogated CD155 recognition by B12A TCR. The fact that MHC-independent recognition of CD155 is mediated by the TCR antigen combining site proves that CD155 is specifically recognized as an antigen, albeit an MHC-independent antigen, but is not recognized non-specifically as either a superantigen or an adhesion molecule.
Further analysis of CD155 recognition by A11 TCR revealed that it involved the same evolutionarily conserved CDR2β-encoded amino acid residues (Y46, Y48, E54) that, in conventional Vβ8 TCRs, contact MHC and are thought to confer MHC-specificity (Scott-Browne et al., 2009; Yin et al., 2011). Indeed, these CDR2β-encoded residues were required for thymic selection of at least one MHC-restricted transgenic TCR (Scott-Browne et al., 2009). However, the proposal that these germline-encoded CDR2β amino acid residues imposed MHC specificity was only tested on MHC-selected TCRs. The present study now documents that the identical CDR2β residues which promote TCR recognition of pMHC ligands (Garcia et al., 2009; Scott-Browne et al., 2009; Yin et al., 2011) also promote TCR recognition of the MHC-independent ligand CD155. In other words, this study indicates that germline-encoded CDR2β residues promote ligand recognition by αβTCRs regardless of whether the ligand is MHC-dependent or MHC-independent. Consequently, we think that these germline-encoded MHC contact residues neither impose MHC specificity nor prevent αβTCR from binding MHC-independent ligands.
Nevertheless, we do think that evolutionary pressures have created a germline bias toward MHC recognition, but the bias is not strong enough to have eliminated MHC-independent αβTCRs. Indeed, the current study supports a potential mechanism by which evolutionary pressures create such a bias. Specifically, we think that the evolutionary pressure for germline bias toward MHC recognition is exerted during thymic selection because Lck in immature thymocytes is sequestered away from the TCR by CD4 and CD8 coreceptor proteins (Van Laethem et al., 2007). It is our perspective that the dual specificity of CD4 and CD8 coreceptors for extracellular MHC and intracellular Lck provides the molecular basis for thymic selection of MHC-specific T cells and provides the evolutionary pressure for a germline bias toward recognition of MHC molecules (Van Laethem et al., 2007).
Murine CD155 is the murine analog of the human polio virus receptor and is a member of the nectin-like family of adhesion molecules. CD155 is bound by a number of counter-receptors (Miyoshi and Takai, 2007) which could theoretically either promote or impair TCR binding to CD155. Indeed, it is conceivable that CD155 counter-receptors might be able to contribute to CD155-specific αβTCRs in a manner analogous to CD4 and CD8 coreceptors which contribute to the pMHC specificity of conventional αβTCRs. Notably, B12A TCR recognition of CD155 was not dependent on CD155 counter-receptors because, in a cell-free assay devoid of all other proteins, soluble B12A TCR directly bound to recombinant CD155 protein with high affinity. In addition, when expressed on human PBL bearing human counter-receptors that bind strongly to human but not murine CD155, A11 and B12A TCRs nevertheless signaled hPBL to respond to platebound CD155 proteins.
It was surprising that two different αβTCRs recognized the same self-protein, albeit different epitopes on that protein, raising the possibility that CD155 might be a unique ligand for MHC-independent TCRs, perhaps analogous to MHC for conventional αβTCRs. While intriguing, such a possibility is contradicted by the fact that polyclonal αβT cells from Quad-deficient mice reacted against CD155-deficient stimulators, indicating that polyclonal αβT cells recognized additional MHC-independent ligands other than CD155. Instead, we think that the apparent over-representation of MHC-independent αβTCRs from Quad-deficient mice with specificity for CD155 may have resulted from fusing Quad-deficient αβT cells with BW5147 cells to construct T-hybridoma cell lines. The fusion of αβT cells with BW5147 extinguishes expression of a number of genes, such as Cd8 (Lee et al., 1994), and Cd155 may also be one of those genes because CD155 expression was markedly reduced on T-hybridoma cells. Because its expression is reduced on T-hybridomas, T-hybridomas with αβTCR specific for CD155 would expand because they would not be constitutively signaled; whereas T-hybridoma cells with αβTCR specific for MHC-independent ligands that T-hybridoma cells express would be continuously stimulated and signaled to commit fratricide. As a result, MHC-independent αβTCRs derived from T-hybridoma cells would give the appearance of being markedly skewed toward CD155 specificity. Experiments to generate MHC-independent αβTCRs from Quad-deficient mice with other ligand specificities are in progress.
In conclusion, this study demonstrates that, in the absence of MHC-specific thymic selection, T cells can resemble B cells in their expression of antigen-specific receptors that recognize glycosylation-dependent conformational epitopes on unprocessed protein antigens independently of MHC molecules. This study contradicts the perspective that MHC-specificity is intrinsic to the structure of all αβTCRs, and suggests the existence of a repertoire of αβTCRs with specificity for MHC-independent ligands that has never been examined. Thus, this study indicates that αβTCRs need to be screened for MHC-specificity in the thymus in order to insure the generation of an MHC-restricted peripheral αβTCR repertoire.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6 (B6), BALB/c and B10.A mice were obtained from NCI Frederick (Frederick, MD). MHC-deficient (B2m−/−H-2-Ab1−/−), Quad-deficient (B2m−/−H-2-Ab1−/−Cd4−/−Cd8a−/−)(Van Laethem et al., 2007), and Quad-deficient.Bcl2 mice containing the hBcl2 transgene (Linette et al., 1996) were bred in our own animal colony. Mice deficient in CD155 (Cd155−/−) were generated as described (Maier et al., 2007). Animal care was in accordance with National Institutes of Health (NIH) guidelines.
Generation of T Cell Hybridomas
αβ lymph node T (LNT) cells from Quad-deficient.Bcl-2 mice were stimulated with platebound anti-TCRβ (10 μg/ml) for 48 hr, fused to TCR null BW5147 cells and subcloned at <1 cell/well (Van Laethem et al., 2007). T-hybridomas reactive against MHC-deficient stimulator cells were selected for further study. IL-2 secretion was measured by the IL-2-dependent CTLL-2 cell line using the Alamar Blue colorimetric assay (Ahmed et al., 1994). The TCR-deficient 4G4 cell line (Hong et al., 1992) was kindly provided by Alexander Chervonsky (University of Chicago).
Reagents
MAbs with the following specificities were used: murine TCRβ (clone H57-957; Becton Dickinson, San Diego, CA); murine CD155 (clones TX56 and 4.42.1; Biolegend, San Diego, CA). Glycosylated recombinant human and mouse CD155 were purchased from Sino Biologicals (Beijing, China). Non-glycosylated recombinant CD155 protein was made in bacteria. Polyclonal rabbit anti-CD155 was purchased from Sino Biologicals. LEAF purified antibody (4.24.1) against mouse CD155 was obtained from Biolegend. Murine IL-2 was measured either using the CTLL-2 bioassay or by enzyme-linked immunosorbent assays (ELISA) (R&D systems, Minneapolis, MN). Human IFN-γ was measured by ELISA (R&D systems). Mouse CD155 cDNA cloned into pCMV6Entry vector was obtained from OriGene (Rockville, MD).
αβTCR constructs and retroviral transductions
Full-length TCR cDNAs for TCRα and TCRβ were cloned using PCR primers specific to sequences 5' of the start and 3' of the stop of rearranged TCR chains. TCR expression constructs were cloned in murine stem cell virus (MSCV)-based retroviral plasmids and transfected into PlatE cells to produce retrovirus containing supernatants. TCRβ was cloned into pMX-IRES-GFP (provided by Remy Bosselut, NCI) and TCRα was cloned into MSV-IRES-tNGFR (provided by Warren Pear, University of Pennsylvania). TCRs were expressed by retroviral transduction in TCR-deficient 4G4 cells using 2μg/ml of polybrene. For stimulation assays, 4×104 retrovirally transduced 4G4 cells were incubated with stimulators for 16hr at 37°C, 5% CO2.
TCR transduction to human PBL was done as described (Frankel et al., 2010). Briefly, retrovirus was prepared by co-transfecting the 293gp retrovirus producer cell line with retrovirus vectors that encode TCR genes and the RD-114 envelope plasmid. hPBLs were stimulated with anti-CD3 (OKT3) at 50 ng/ml in AIM-V with human IL-2 (300 IU/ml) for 2 days prior to retroviral transduction. Retroviral supernatant was spin-loaded onto RetroNectin (Takara Bio, Otsu, Japan) coated plates (10 μg/ml in PBS, overnight) by centrifugation at 2000× g for 2 hours at 32°C. hPBLs were then loaded onto plates, centrifuged at 1000× g for 10 min, and incubated overnight at 37°C, 5% CO2, and retrovirally transduced a second time using the second set of plates loaded with retrovirus. During stimulation assays, 2×105 retrovirally transduced hPBL were incubated with stimulators at 37°C, 5% CO2.
Construction and screening of the cDNA library
Total RNA was obtained from the murine L1210 cell line using RNeasy Maxi (Qiagen Inc., Valencia, CA) and was purified using FastTrack MAG Maxi mRNA isolation kit (Invitrogen, Carlsbad, CA) to obtain poly (A)+ RNA. cDNA was synthesized with the SuperScript system (Invitrogen) and was cloned into SPORT6 vector using SalI and NotI restriction sites. ElectroMAX DH10B competent cells (Invitrogen) were transformed by electroporation and after titration, E.coli (~150 clones/well) were inoculated overnight into 96-well format culture blocks (10 blocks). Plasmids were purified using a Qiaprep 96 Turbo miniprep kit (Qiagen) and were transfected to HEK-293T cells using Lipofectamine 2000 (Invitrogen) in 96-well flat bottom plates and left overnight.
Transduced 4G4 cells were co-cultured with cDNA transfected 293T cells for 24 hours, after which mIL-2 amounts in the supernatants were determined. Positive clones were selected for secondary and tertiary screenings. Sub-pool libraries (~20 clones/well, 48 wells) and clone libraries (1 clone/well, 96 wells) were prepared and screened. Positive clones were sequenced to identify the specificity of the transfected cDNA.
Stimulation with platebound ligands
Flat bottom 96-well plates were coated overnight with intact or heat denatured recombinant hCD155 or mCD155 in 50 μl of PBS. Transduced 4G4 cells (4×104 cells) or hPBLs (2×105 cells) were added overnight after which supernatants were assessed for IL-2 or hIFN-γ.
Generation of soluble αβTCR heterodimers
DNA encoding extracellular portions of TCR α and β chains were cloned into pET30a vectors (GenScript, Piscataway, NJ) (Dai et al., 2008; Tynan et al., 2007). Receptors were expressed as inclusion bodies in BL21 (DE3) cells. Functional and soluble TCR heterodimers were produced by a rapid dilution refolding procedure as previously described (Clements et al., 2002).
Surface Plasmon Resonance
Surface Plasmon Resonance (SPR) measurements were performed using a BIAcore 3000 instrument and analyzed with BIAeveluation 4.1 software (GE Healthcare, Piscataway, NJ). To measure the affinity of the TCRs, CD155 was immobilized on caboxylated dextran CM5 chips (GE Healthcare) to 200–500 response units (RU) using a primary amine-coupling and 5 μl/min flow rate in 10mM sodium acetate (pH 5.5). The analytes consisted of serial dilutions of soluble TCR heterodimers from 0.31 to 2.5 μM in a buffer containing 10mM Hepes (pH7.4) and 0.15M NaCl. The dissociation constants were obtained by kinetic curve fitting using BIAevaluation (GE Healthcare).
Epitope Mapping
Chimeric CD155 molecules between human and mouse were constructed. The chimeric constructs were synthesized (GenScript) and cloned into pIRES2-ZsGreen1 expression vector. Transduced 4G4 cells (4×104 cells) were co-cultured with 293T cells transfected with chimeric cDNA for 24 hours, after which supernatant IL-2 was measured.
Exchange of CDR3α-encoded TCR sequences
B12A TCRα containing B12F TCRα CDR3 region (A-CDR3-F) and B12F TCRα containing B12A TCRα CDR3 region (F-CDR3-A) were constructed. The CDR3α TCR mixand-match constructs were synthesized (GenScript) and cloned into MSV-IRES-tNGFR. B12 TCRβ was cloned into pMX-IRES-GFP. TCRs were expressed by retroviral transduction in TCR-deficient 4G4 cells. For stimulation assays, 1×105 transduced 4G4 cells were stimulated overnight with platebound mCD155 (5 μg/ml) or anti-TCRβ (5 μg/ml) in flat bottom 96-well plate for 16hr at 37°C, 5% CO2.
TCR Mutagenesis
Constructs containing A11 TCRVβ with Y46A, Y48A, or E54A mutations in CDR2 region were synthesized (GenScript) and cloned into pMX-IRES-GFP. A11 TCRα was cloned into MSV-IRES-tNGFR. TCRs were expressed by retroviral transduction in TCR-deficient 4G4 cells.
Supplementary Material
HIGHLIGHTS
Without MHC-specific thymic selection, TCRs can recognize non-MHC ligands
Two of these TCRs recognize conformational epitopes on CD155 with high affinity
CD155 recognition is mediated by CDR3 residues in the TCR combining site
CD155 recognition requires the same CDR2 residues that conventional TCR use for MHC
ACKNOWLEDGEMENTS
We thank R. Bosselut, W. Pear, and A. Chervonsky for providing reagents; and N. Taylor, J. Di Santo, M. Kimura, T. McCaughtry, X. Tai and D. Margulies for critically reading the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmed SA, Gogal RM, Jr., Walsh JE. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170:211–224. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Barnd DL, Lan MS, Metzgar RS, Finn OJ. Specific, major histocompatibility complex-unrestricted recognition of tumor-associated mucins by human cytotoxic T cells. Proc Natl Acad Sci U S A. 1989;86:7159–7163. doi: 10.1073/pnas.86.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadeneau C, LeCabellec M, LeMoullac B, Meflah K, Denis MG. Over-expression of a novel member of the immunoglobulin superfamily in Min mouse intestinal adenomas. Int J Cancer. 1996a;68:817–821. doi: 10.1002/(SICI)1097-0215(19961211)68:6<817::AID-IJC21>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Chadeneau C, LeMoullac B, LeCabellec M, Mattei M, Meflah K, Denis MG. Isolation and chromosomal location of mE4, a novel murine gene of the immunoglobulin superfamily. Mamm Genome. 1996b;7:636–637. doi: 10.1007/s003359900194. [DOI] [PubMed] [Google Scholar]
- Clements CS, Kjer-Nielsen L, MacDonald WA, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr D Biol Crystallogr. 2002;58:2131–2134. doi: 10.1107/s0907444902015482. [DOI] [PubMed] [Google Scholar]
- Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- Frankel TL, Burns WR, Peng PD, Yu Z, Chinnasamy D, Wargo JA, Zheng Z, Restifo NP, Rosenberg SA, Morgan RA. Both CD4 and CD8 T cells mediate equally effective in vivo tumor treatment when engineered with a highly avid TCR targeting tyrosinase. J Immunol. 2010;184:5988–5998. doi: 10.4049/jimmunol.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Wang QJ, Inozume T, Yang JC. Molecular identification of an MHC-independent ligand recognized by a human {alpha}/{beta} T-cell receptor. Blood. 2011;117:4816–4825. doi: 10.1182/blood-2010-11-317743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn L, Gratton S, Caron L, Sekaly RP, Veillette A, Julius M. Association of tyrosine kinase p56lck with CD4 inhibits the induction of growth through the alpha beta T-cell receptor. Nature. 1992;358:328–331. doi: 10.1038/358328a0. [DOI] [PubMed] [Google Scholar]
- He Y, Bowman VD, Mueller S, Bator CM, Bella J, Peng X, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci U S A. 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SC, Chelouche A, Lin RH, Shaywitz D, Braunstein NS, Glimcher L, Janeway CA., Jr An MHC interaction site maps to the amino-terminal half of the T cell receptor alpha chain variable domain. Cell. 1992;69:999–1009. doi: 10.1016/0092-8674(92)90618-m. [DOI] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Lee WH, Banan M, Harriss JV, Hwang I, Woodward E, Youn HJ, Gottlieb PD. Cis-acting DNA elements and cell type-specific nuclear proteins which may play a role in regulation of mouse CD8 alpha (Lyt-2) gene transcription. Int Immunol. 1994;6:1307–1321. doi: 10.1093/intimm/6.9.1307. [DOI] [PubMed] [Google Scholar]
- Li H, Llera A, Malchiodi EL, Mariuzza RA. The structural basis of T cell activation by superantigens. Annu Rev Immunol. 1999;17:435–466. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- Linette GP, Li Y, Roth K, Korsmeyer SJ. Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc Natl Acad Sci U S A. 1996;93:9545–9552. doi: 10.1073/pnas.93.18.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MK, Seth S, Czeloth N, Qiu Q, Ravens I, Kremmer E, Ebel M, Muller W, Pabst O, Forster R, et al. The adhesion receptor CD155 determines the magnitude of humoral immune responses against orally ingested antigens. Eur J Immunol. 2007;37:2214–2225. doi: 10.1002/eji.200737072. [DOI] [PubMed] [Google Scholar]
- Manning TC, Kranz DM. Binding energetics of T-cell receptors: correlation with immunological consequences. Immunol Today. 1999;20:417–422. doi: 10.1016/s0167-5699(99)01508-x. [DOI] [PubMed] [Google Scholar]
- Margulies DH. Interactions of TCRs with MHC-peptide complexes: a quantitative basis for mechanistic models. Curr Opin Immunol. 1997;9:390–395. doi: 10.1016/s0952-7915(97)80086-6. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Boniface JJ, Reay PA, Schild H, Fazekas de St Groth B, Davis MM. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science. 1991;254:1788–1791. doi: 10.1126/science.1763329. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Graf D, Lovatt M, Bommhardt U, Zamoyska R, Fisher AG. How many thymocytes audition for selection? J Exp Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi J, Takai Y. Nectin and nectin-like molecules: biology and pathology. Am J Nephrol. 2007;27:590–604. doi: 10.1159/000108103. [DOI] [PubMed] [Google Scholar]
- Rao A, Ko WW, Faas SJ, Cantor H. Binding of antigen in the absence of histocompatibility proteins by arsonate-reactive T-cell clones. Cell. 1984;36:879–888. doi: 10.1016/0092-8674(84)90037-0. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Schwartz RH, David CS, Sachs DH, Paul WE. T lymphocyte-enriched murine peritoneal exudate cells. III. Inhibition of antigen-induced T lymphocyte Proliferation with anti-Ia antisera. J Immunol. 1976;117:531–540. [PubMed] [Google Scholar]
- Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, Keegan AD, Dintzis RZ, Dintzis HM, Shin HS. The interaction of nominal antigen with T cell antigen receptors. I. Specific binding of multivalent nominal antigen to cytolytic T cell clones. J Immunol. 1985;135:906–914. [PubMed] [Google Scholar]
- Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655–667. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, Adoro S, Adams A, Sharrow SO, Feigenbaum L, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Yin L, Huseby E, Scott-Browne J, Rubtsova K, Pinilla C, Crawford F, Marrack P, Dai S, Kappler JW. A Single T Cell Receptor Bound to Major Histocompatibility omplex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers. Immunity. 2011;35:23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci U S A. 2008;105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Young AC, Imarai M, Nathenson SG, Sacchettini JC. Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc Natl Acad Sci U S A. 1992;89:8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.