Abstract
A hallmark of the bacterial twin-arginine translocation (Tat) pathway is its ability to export folded proteins. Here, we discovered that overexpressed Tat substrate proteins form two distinct, long-lived translocation intermediates that are readily detected by immunolabeling methods. Formation of the early translocation intermediate, Ti-1, which exposes the N- and C-termini to the cytoplasm, did not require an intact Tat translocase, a functional Tat signal peptide, or a correctly folded substrate. In contrast, formation of the later translocation intermediate, Ti-2, which exhibits a bitopic topology with the N-terminus in the cytoplasm and C-terminus in the periplasm, was much more particular, requiring an intact translocase, a functional signal peptide, and a correctly folded substrate protein. The ability to directly detect Ti-2 intermediates was subsequently exploited for a new protein engineering technology called MAD-TRAP (membrane-anchored display for Tat-based recognition of associating proteins). Using just two rounds of mutagenesis and screening with MAD-TRAP, the intracellular folding and antigen-binding activity of a human single-chain antibody fragment were simultaneously improved. This approach has several advantages for library screening, including the unique involvement of the Tat folding quality control mechanism that ensures only native-like proteins are displayed, thus eliminating poorly folded sequences from the screening process.
Keywords: antibody engineering, bacterial surface display, directed evolution, protein expression and folding, twin-arginine translocation protein export pathway
Introduction
The bacterial twin-arginine translocation (Tat) system is unique in its ability to export folded proteins or protein domains across the tightly sealed cytoplasmic membrane. This remarkable feat is accomplished by a translocase composed of the TatABC integral membrane proteins that function independently of soluble factors or nucleoside triphosphates 1; 2; 3; 4. The Tat system appears to accommodate at least two broad classes of proteins: globular proteins that fold too rapidly to be handled by the well characterized Sec export pathway and proteins that assemble cofactors or protein subunits in the cytoplasm and necessarily must be exported in a folded form 5; 6; 7. The ability of the Tat pathway to accept these folded substrates has significant implications for the export mechanism and raises key questions about the structure/function of the translocase and whether substrates need to be correctly folded prior to export.
It is now firmly established that the vast majority of Tat substrates are only competent for export if they fold properly in the cytoplasm 8; 9; 10; 11; 12; 13; 14; 15 with rare exceptions 16; 17. On the basis of these observations, it has been speculated that an inbuilt feature of the Tat system is a quality control mechanism that discriminates between folded and unfolded proteins, allowing the export of only the former 8. More recent findings support a model in which the Tat translocase is at the center of an integrated quality control system that involves “sensing” the degree of folding of its protein substrates prior to export 13 and also initiating degradation of those substrates that are rejected due to incomplete folding or assembly 10. Such substrate quality control appears to involve productive interactions between the substrate and the TatBC components 13; 14, suggesting a direct role for the translocase in discriminating between correctly folded and misfolded substrate proteins. Moreover, these findings imply that membrane targeting, quality control, and translocation of Tat substrates are distinct steps that can be analyzed separately from each other.
Therefore, one objective of this work was to dissect the Tat transport process into several discrete steps that are characterized by distinct translocation intermediates. Previous work on the plant thylakoidal Tat system identified two Tat translocation intermediates 18; 19. The first was an early translocation intermediate called Ti-1 that was observed to insert into the membrane in a loop-like conformation with both the N- and C-termini exposed to the chloroplast stroma (the cytoplasm equivalent of chloroplasts). In later stages of the transport process, the C-terminal domain of the substrate was translocated across the thylakoid membrane, resulting in the appearance of translocation intermediate-2 (Ti-2) that exhibited a bitopic topology with the N-terminus facing the stroma and the C-terminus in the lumen (the periplasm equivalent). Here, we identify for the first time similar translocation intermediates in Escherichia coli and provide evidence that formation of Ti-2 but not Ti-1 is dependent upon a functional signal peptide, an intact Tat translocase, and correct folding of the substrate. Furthermore, we have exploited the Ti-2 intermediate to create MAD-TRAP (membrane-anchored display for Tat-based recognition of associating proteins), a new method for isolating ligand-binding proteins from combinatorial libraries that are displayed as Ti-2 intermediates on the periplasmic face of the E. coli inner membrane (IM). By combining the quality control mechanism of the Tat pathway with bacterial membrane display, MAD-TRAP permits simultaneous engineering of in vivo folding efficiency and antigen-binding activity of proteins such as single-chain variable fragment (scFv) antibodies in as few as one or two rounds of mutagenesis and screening.
Results
Anchoring Tat substrates to the IM
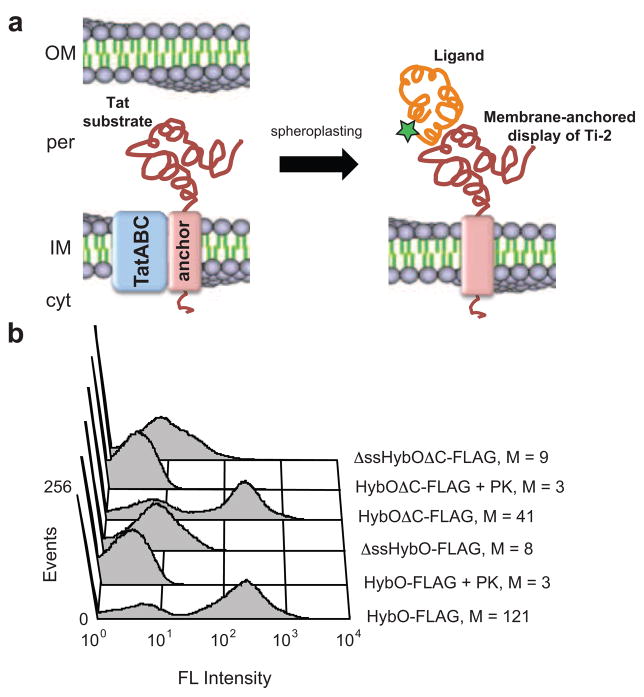
We set out to develop a method for anchoring Tat-exported proteins to the periplasmic side of the IM of E. coli. Such a strategy would allow facile detection and functional interrogation of these proteins using a two-step strategy that involves permeabilizing E. coli cells followed by immunolabeling (Fig. 1a). Because Tat proteins are subject to folding quality control 8; 11, we hypothesized that this procedure would have an in-built fitness filter such that only correctly folded proteins would be displayed on the IM. To enable Tat-mediated membrane anchoring, we first investigated a class of endogenous E. coli Tat substrates that possesses C-terminal transmembrane α-helices (TMs) and are thus C-tail anchored integral membrane proteins 20. One example is HybO, a nonessential Tat substrate that assembles with HybC to form a hydrogenase respiratory complex. Previous studies demonstrated that a 22-residue TM at the extreme C-terminus of HybO was sufficient to anchor this subunit to the periplasmic side of the IM 20. Moreover, addition of the HybO C-tail to soluble proteins rendered these proteins membrane-bound. To determine if C-tail anchored HybO could be immunodetected on the periplasmic side of the IM, wildtype (wt) E. coli cells were induced to express HybO with an N-terminal FLAG tag (inserted just upstream of the C-terminal TM) and then incubated with EDTA and lysozyme to disrupt the outer membrane (OM) and cell wall. The resulting spheroplasts were mixed with a FITC-conjugated anti-FLAG antibody, and the cell fluorescence was determined by flow cytometry (FC). Spheroplasts expressing HybO-FLAG were highly fluorescent whereas those that had been treated with proteinase K (PK) prior to labeling or those expressing a version of HybO without a signal peptide were 40- and 15-times less fluorescent, respectively (Fig. 1b). The fluorescent signal from cells expressing HybO-FLAG was dependent on spheroplasting, as labeling of untreated cells resulted in only background fluorescence. However, to our surprise, spheroplasts expressing a variant of HybO that lacked the C-tail anchor were highly fluorescent (Fig. 1b). Treatment with PK eliminated this fluorescence, as did expression of HybO without an export signal (Fig. 1b). The observation that HybO remained attached to the IM without a C-tail anchoring motif but not without a functional Tat export signal suggested that the N-terminal signal peptide served as a membrane anchor.
Figure 1. IM-anchored display of Tat substrates.
(a) Correctly folded Tat substrates are transported from the cytoplasm (cyt) to the periplasm (per) of E. coli, but remain N-terminally anchored to the inner membrane (IM). After removing the outer membrane (OM) and periplasm, the protein can be detected by immunolabeling and/or probed for interactions with other proteins. (b) FC analysis of spheroplasts expressing HybO constructs with C-terminal FLAG epitope tags. Constructs lacking a native Tat signal peptide (ΔssHybO) and/or a C-tail membrane anchor (HybOΔC) were included as controls. Specified samples were treated with proteinase K (PK). FLAG tags were detected using a FITC-conjugated anti-FLAG antibody. Median fluorescence values (M) are shown for each construct.
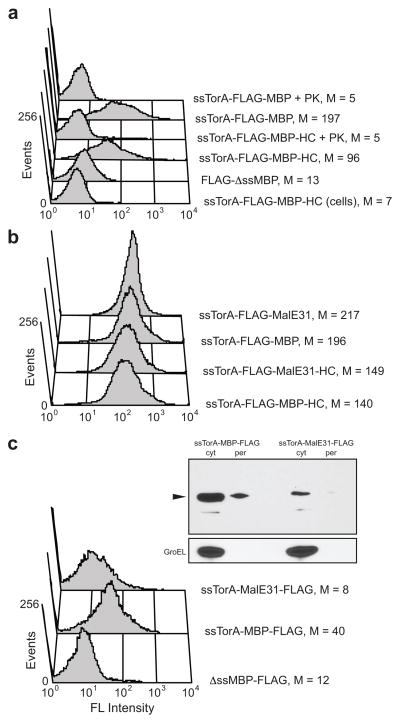
To determine whether this could be extended to other proteins, we next attempted to anchor E. coli maltose binding protein (MBP) to the IM. MBP is a soluble protein that can be rerouted to the Tat pathway by replacing its native Sec-dependent signal with a Tat-dependent signal (e.g., ssTorA) 21. We modified mature MBP with an N-terminal ssTorA signal immediately followed by a FLAG epitope tag and a C-terminal HybO C-tail (HC) anchor motif. Spheroplasts expressing the ssTorA-FLAG-MBP-HC chimera were 14- and 9-times more fluorescent than unpermeabilized cells expressing the same construct or spheroplasts expressing the chimera without ssTorA, respectively (Fig. 2a). PK treatment of spheroplasts was sufficient to eliminate the signal. Similar to HybO, immunodetection of MBP did not require the C-tail anchor (Fig. 2a). In fact, spheroplasts expressing ssTorA-FLAG-MBP were nearly twice as fluorescent as their ssTorA-FLAG-MBP-HC-expressing counterparts. Taken together, these data indicate that plasmid-expressed Tat substrates become anchored in the IM by their N-terminal signal peptide.
Figure 2. IM-anchored display of MBP.
(a) FC analysis of MBP constructs with and without a C-terminal HybO C-tail (HC). FLAG tags were placed between the ssTorA signal peptide and MBP. Specified samples were treated with proteinase K (PK). Constructs lacking a signal peptide (ΔssMBP) and cells that were not spheroplasted were included as controls. (b) FC analysis of misfolding MBP variant. An aggregation-prone MBP variant (MalE31) and wt MBP were expressed with and without the C-terminal HC. FLAG tags were between the ssTorA signal peptide and MBP. (c) FC and Western blot analysis of MBP constructs after repositioning of the FLAG tag to the C-terminus of wt MBP and MalE31. For Western blots, periplasmic (per) and cytoplasmic (cyt) fractions from an equivalent number of cells were probed with an anti-FLAG antibody and arrow indicates expected size of fusion protein. Detection of GroEL with an anti-GroEL antibody was used to verify quality of fractionations. For (a)–(c), FLAG tags were detected with a FITC-conjugated anti-FLAG antibody, and median fluorescence values (M) are shown.
To determine if IM display of Tat substrates was regulated by the folding quality control feature of the bacterial Tat system 8; 11, we modified the MBP domain in ssTorA-FLAG-MBP with two amino acid substitutions, namely Gly32Asp and Ile33Pro. The resulting MBP variant, called MalE31, is highly aggregation prone and thus blocked for export via the Tat pathway as we showed previously 11. To our surprise, however, the fluorescence of spheroplasts expressing ssTorA-FLAG-MalE31 was indistinguishable from that of spheroplasts expressing ssTorA-FLAG-MBP (Fig. 2b), irrespective of whether a C-tail anchor was present or not. To determine if the position of the FLAG epitope was important, we generated constructs in which the FLAG epitope was positioned C-terminally (i.e., ssTorA-MBP-FLAG). Interestingly, when the FLAG epitope was repositioned at the C-terminus of MBP, only the correctly folded wt MBP could be immunodetected on spheroplasts (Fig. 2c). The inability to detect the C-terminal FLAG epitope on the misfolded MalE31 domain suggested that this domain was blocked for Tat export. In support of this notion, both ssTorA-MBP-FLAG and ssTorA-MalE31-FLAG were detected in the cytoplasm whereas only ssTorA-MBP-FLAG appeared in the periplasm (Fig. 2c).
Based on these results, we speculated that the labeling strategy was detecting two distinct translocation intermediates. These intermediates, called Ti-1 and Ti-2, were previously observed in the plant thylakoidal Tat system 19 but have not been reported for the bacterial Tat system. Formation of Ti-1 and Ti-2 requires development of membrane spanning segments, one of which is likely provided by the hydrophobic domain present in the Tat signal peptide 19. Placement of the FLAG tag immediately after the signal peptide appears to position this epitope in a periplasmically-oriented hydrophilic region between the two presumed membrane segments (Fig. 3a). Since formation of Ti-1 is “unassisted” and does not depend on the Arg-Arg motif or functional Tat machinery 19, this epitope is accessible regardless of whether the substrate protein is ultimately translocated to the periplasm. This explains why the export-competent wt MBP and the export-incompetent MalE31 could both be immunolabeled when the FLAG tag was positioned N-terminally. On the other hand, a C-terminal FLAG epitope is sequestered in the cytoplasm and only becomes accessible to immunolabeling if the protein is competent for Tat export. Indeed, when the FLAG tag was located at the C-terminus, only correctly folded wt MBP but not misfolded MalE31 was efficiently labeled. Interestingly, even though both Ti-1 and Ti-2 are naturally transient intermediates, HybO and MBP remained anchored to the IM for more than 24 h after spheroplasting. Thus, we suspect that the high substrate expression levels used here saturated both the translocation machinery and signal peptidase I such that these intermediates were long-lived.
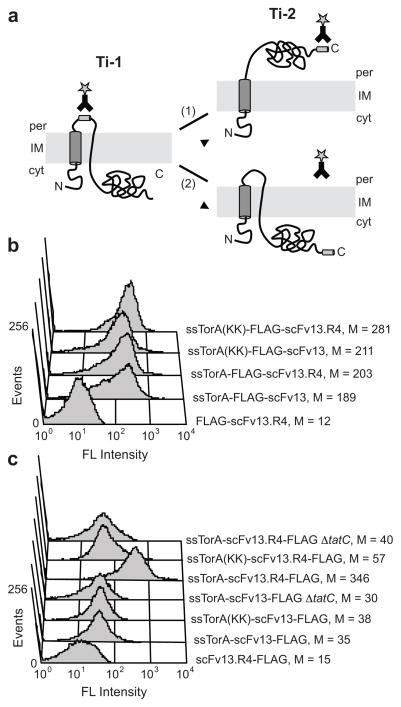
Figure 3. Separate detection of Ti-1 and Ti-2.
(a) Schematic of Tat translocation intermediates. Formation of Ti-1 can be detected when an epitope tag is inserted between the signal peptide and the N-terminus of the protein. If the protein is incorrectly folded (1), it cannot form Ti-2 and a C-terminal epitope tag is not accessible for immunolabeling. If the protein is properly folded (2), it is transported to the periplasm to form Ti-2, and a C-terminal epitope tag can be detected on the periplasmic face of the inner membrane. (b) FC analysis to detect Ti-1 for a poorly folded scFv (scFv13) and a well folded scFv (scFv13.R4). FLAG tags were inserted between the N-terminal Tat signal peptide and the scFvs. (c) FC analysis to detect Ti-2 for scFv13 and scFv13.R4. FLAG tags were placed at the C-terminus of scFvs. ssTorA(KK) indicates mutation of the Arg-Arg motif to Lys-Lys in the Tat signal peptide; ΔtatC indicates cells that lacked the TatC protein. FLAG tags were detected with a FITC-conjugated anti-FLAG antibody, and ssTorA-scFv13 lacking an epitope tag was included as a control. Median fluorescence values (M) are shown.
Separate immunodetection of Ti-1 and Ti-2
To further illustrate separate labeling of Ti-1 and Ti-2 and to eliminate any biases that may be introduced by using proteins such as HybO and MBP that are naturally exported in bacteria, we created a series of chimeras based on scFv13, a human antibody fragment specific for β-galactosidase (β-gal) 22. Our previous studies demonstrated that wt scFv13, which folds poorly in the cytoplasm of E. coli, was incapable of Tat export 23. However, folding-enhanced variants of scFv13 (e.g., scFv13.R4) have been isolated 22, and these are efficiently localized to the periplasm by the Tat export machinery 23. In agreement with our model (Fig. 3a), both the poorly folded wt scFv13 and the folding-enhanced scFv13.R4 could be detected on the IM when the FLAG tag was positioned between the signal peptide and the scFv domain (Fig. 3b). Consistent with the model for Ti-1 formation 19, labeling proceeded even in the presence of a defective signal peptide containing twin Lys residues or in a host lacking the tatC gene (Fig. 3b), which encodes one of the essential components of the Tat translocase 1. Only removal of the Tat signal peptide was sufficient to eliminate labeling (Fig. 3b). When the FLAG tag was moved to the C-terminus of each scFv, labeling of ssTorA-scFv13.R4-FLAG resulted in a nearly 10-fold increase in fluorescence compared to the background level of fluorescence from the export-incompetent ssTorA-scFv13-FLAG (Fig. 3c). Ti-2 formation by scFv13.R4 was dependent on the Tat signal peptide (Fig. 3c). Unlike Ti-1, Ti-2 detection also depended on the conserved twin Arg residues in the signal peptide and the tatC gene (Fig. 3c). Thus, Ti-2 formation depended on both functional Tat targeting and on correct substrate folding, whereas Ti-1 formation was insensitive to these parameters.
MAD-TRAP detection of ligand binding
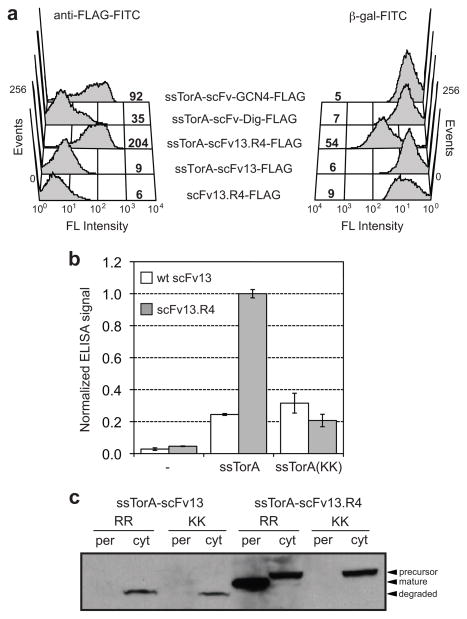
Collectively, the data above form the basis of a new technology that we termed MAD-TRAP. To evaluate the utility of MAD-TRAP for detecting correctly folded and functional antibody fragments displayed on the IM, we examined the ability of membrane tethered scFv13.R4 to bind β-gal. To test this, spheroplasts expressing ssTorA-scFv13.R4-FLAG were mixed with β-gal that had been conjugated to FITC (β-gal-FITC) and analyzed by FC. Strong fluorescence was associated with spheroplasts (Fig. 4a), indicating binding of β-gal-FITC by the membrane-anchored antibody fragment. In contrast, no antigen binding was observed for wt scFv13 or for a version of scFv13.R4 that lacked a signal peptide, confirming that correct folding and Tat targeting are required for IM display. To confirm binding specificity, we expressed two unrelated scFv sequences: scFv-Dig that is specific for the cardiac glycoside digoxin and scFv-GCN4 that is specific for the bZIP domain of the yeast transcription factor Gcn4. The scFv-Dig antibody misfolds in the cytoplasm of wt E. coli 8 and, as a result, was barely detected on the IM following immunolabeling with anti-FLAG-FITC antibodies (Fig. 4a). The misfolded scFv-Dig also did not bind to β-gal-FITC. In the case of scFv-GCN4, which was previously optimized for intracellular expression 24, expression on the IM was readily detected with anti-FLAG-FITC antibodies (Fig. 4a). However, correctly folded scFv-GCN4 that was displayed on the IM did not cross-react with β-gal-FITC, confirming the binding specificity of the assay. In parallel, we observed that antigen-coated ELISA plates (or beads) could be used to readily discriminate correctly folded, functional antibody fragments from those that are incorrectly folded or improperly targeted to the Tat pathway (Fig. 4b). Western blot analysis of subcellular fractions confirmed that only the folding-enhanced scFv13.R4 was exported from the cytoplasm and that export depended on a functional signal peptide (Fig. 4c). In the case of wt scFv13, the lack of export for this protein was coupled to cytoplasmic degradation, consistent with previous findings that export-incompetent Tat substrates are proteolytically cleared from cells 8.
Figure 4. Expression and activity of IM-anchored scFvs.
(a) FC analysis to detect Ti-2 formation and antigen-binding activity for the scFvs. The antigen used was FITC-labeled β-gal. Poorly folded scFvs (scFv13 and scFv-Dig) and scFvs specific for irrelevant antigens (scFv-Dig and scFv-GCN4) were included. scFv13.R4-FLAG lacking a Tat signal peptide was used as a negative control. FLAG tags were detected with a FITC-conjugated anti-FLAG antibody. Median fluorescence values are shown directly in histograms. (b) Detection of ligand binding by ELISA for spheroplasts displaying scFv13-FLAG and scFv13.R4-FLAG. Binding activity was measured using β-gal-coated ELISA plates. Bound scFvs were detected with an anti-FLAG antibody. ELISA signals were normalized to the signal for scFv13.R4. Hyphen (−) indicates constructs lacking a signal peptide; KK indicates an Arg-Arg to Lys-Lys substitution in ssTorA. Data represents the average of three replicates, and error bars represent standard error of the mean. (c) Western blot analysis of periplasmic and cytoplasmic fractions from cells expressing scFv13 and scFv13.R4 fused to either ssTorA or ssTorA(KK). Blot was probed with an anti-FLAG antibody. An equivalent number of cells was loaded in each lane.
Clonal enrichment using MAD-TRAP
Based on the above findings, we hypothesized that MAD-TRAP could be useful for screening combinatorial libraries of antibody fragments (Supplemental Fig. S1). To test this notion, we first performed enrichment experiments where mixtures of treated cells were labeled with β-gal-FITC followed by a single round of biopanning using β-gal-coated magnetic beads. When spheroplasts expressing ssTorA-scFv13.R4-FLAG were mixed 1:1 or 1:100 with spheroplasts cells expressing ssTorA-MBP-FLAG and panned on β-gal, 100% of the recovered clones were identified as ssTorA-scFv13.R4-FLAG (Fig. S2a). Enrichment was similarly achieved when ssTorA-scFv13.R4-FLAG was mixed 1:1 or 1:100 with ssTorA-scFv-GCN4-FLAG (Fig. S2b).
Next, we implemented a directed evolution strategy to engineer variants of wt scFv13 that exhibited improved expression and/or antigen binding (Fig. S1a). For this, the gene encoding wt scFv13 was mutagenized by error-prone PCR 25, and the PCR products were cloned between the ssTorA sequence and a FLAG epitope tag. The resulting plasmid DNA library was then transformed into E. coli, giving rise to 2.4×106 independent clones. DNA sequencing of 12 library clones selected at random revealed an average of ~0.5% nucleotide substitutions per gene. Cells expressing the ssTorA-scFv13-FLAG library were treated with EDTA-lysozyme and mixed with β-gal-coated magnetic beads. Bead-bound spheroplasts were isolated, and the genes encoding the antigen-binding scFvs were rescued by PCR amplification of the DNA from the isolated cells. This was aided by the fact that the conditions used for PCR amplification result in the quantitative release of cellular DNA from the cells that have partially hydrolyzed cell walls due to the EDTA-lysozyme treatment during labeling 26. It should also be noted that direct PCR amplification of scFv sequences rather than cell plating was used to recover positive hits, because the plating efficiency of isolated clones was low, as reported previously for EDTA-lysozyme treated E. coli cell libraries 26. After 30 rounds of PCR amplification, the isolated scFv sequences were cloned back into the original plasmid backbone and transformed into fresh E. coli. The resulting sublibrary was subjected to an additional round of biopanning exactly as above. FC screening of the library cells prior to enrichment as well as cells isolated from the first and second rounds of biopanning revealed a clear enrichment in both cell surface expression and antigen binding (Fig. S2c). Next, 30 clones from the second round were randomly chosen for rescreening using FC. Following β-gal-FITC labeling, fluorescence for 29 of these clones was confirmed to be significantly greater than the parental wt scFv13 clone. The most active of these, clone 1–4, was chosen for further characterization. Sequencing of this clone revealed only a single S55R substitution in complementarity determining region 2 (CDR2) of the heavy chain (V H; Table 1 and Fig. S3), which is also one of the 7 mutations isolated previously in scFv13.R4 22. To determine the effect of this substitution on in vivo folding and activity, we first compared the cytoplasmic expression of clone 1–4 versus its progenitor wt scFv13. Western blot analysis revealed that clone 1–4 was much more soluble in the cytoplasm than wt scFv13 when each was expressed from plasmid pET-21a(+) without an N-terminal Tat signal peptide (Fig. 5a). We next compared the binding activity of 1–4 and wt scFv13 following their purification from the cytoplasm. By ELISA, clone 1–4 exhibited a significant increase in binding activity compared to wt scFv13 (Fig. 5b). These results indicate that just a single amino acid substitution in clone 1–4 was sufficient to enhance both in vivo folding and affinity for its cognate antigen and suggest an unexpectedly short evolutionary distance between a stable, well folded scFv and its less stable, poorly expressed parental sequence. The importance of this residue was highlighted by the fact that substitution of most amino acids in position S55 resulted in complete loss of binding activity (Fig. S4), which in some but not all cases was due to poor solubility. However, an S55K mutant bound to β-gal at a level that rivaled the S55R mutant, indicating that the improvement in binding and solubility of clone 1–4 may be due to the introduction of a positive charge in this position.
Table 1.
Summary of mutations, equilibrium dissociation constants, and scFv yields for isolated clones
| scFv clone | Rounds of mutagenesis and screening | VH CDR and framework mutationsa | VL CDR and framework mutationsa | Kdb (nM) | Purified scFv yieldc (mg scFv/L culture) | ||
|---|---|---|---|---|---|---|---|
| CDR (#) | FR (#) | CDR (#) | FR (#) | ||||
| wt scFv13 | n/a | n/a | n/a | n/a | n/a | 200 ± 87 | 1.1 |
|
| |||||||
| scFv13.R4 | 4 | G10S (1) | G51D (2) | - | 128 ± 49 | 5.8 | |
| S52aG (2) | V48I (2) | ||||||
| S55R (2) | K75T (3) | ||||||
| A93V (3) | |||||||
|
| |||||||
| 1–4 | 1 | S55R (2) | - | - | - | nd | 3.2 |
|
| |||||||
| 2-1 | 2 | S55R (2) | - | S27aC (1) | S72F (3) | 91.5 ± 6.8 | 1.1 |
| G51D (2) | |||||||
|
| |||||||
| 2–3 | 2 | S55R (2) | L11P (1) | K42R (2) | - | nd | 3.1 |
| V48I (2) | G51D (2) | ||||||
| A93T (3) | V97D (3) | ||||||
Kabat numbering is used for VH and VL amino acids; see Fig. S3 for complete sequences. Value in parentheses refers to CDR or framework number. nd = not determined.
Equilibrium dissociation constants in the solution phase were determined by ELISA according to the protocol described by Martineau [ref. 41].
Yields of purified scFvs from a representative experiment.
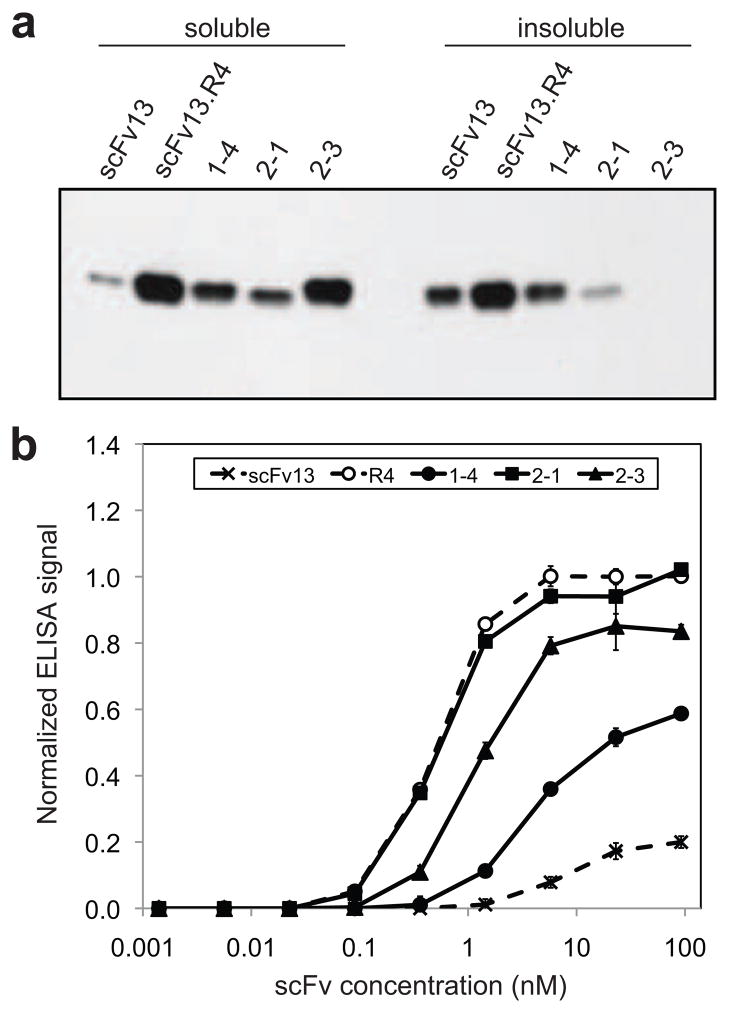
Figure 5. Expression and activity of scFv clones isolated using MAD-TRAP.
(a) Western blot analysis of soluble and insoluble fractions from cells expressing scFvs in the cytoplasm. Clone 1–4, 2-1, and 2–3 were isolated using MAD-TRAP. scFv13 was the starting sequence for the first-round library, and scFv13.R4 was isolated in a previous study after four rounds of directed evolution 22. Samples were normalized by total protein concentration in the soluble fraction, and blot was probed with an anti-6x-His antibody. (b) ELISA data for binding of isolated clones to βgal. scFvs were purified from cell lysate, and their binding to β-gal-coated ELISA plates was measured. Bound scFvs were detected with an anti-6x-His antibody. Data represent the average of six replicates and are normalized to the signal for scFv13.R4 at ~20 nM. Error bars represent standard error of the mean.
Affinity maturation of clone 1–4 using MAD-TRAP
To determine whether MAD-TRAP could be used to further improve the expression and/or binding activity of clone 1–4, we performed an additional round of mutagenesis and screening (Fig. S1b). An error-prone library of clone 1–4 was created as described above. However, to favor the isolation of higher affinity clones, we performed competitive biopanning in the presence of purified 1–4 protein that served as a competitor for β-gal binding. Following this procedure, 13 candidates were identified of which 10 were determined to be true positives by ELISA-based rescreening. The two most active clones from this group, namely clones 2-1 and 2–3, were characterized in more detail. A total of 3 and 6 additional mutations were acquired by clones 2-1 and 2–3, respectively (Table 1). The mutations in clone 2-1 were clustered in the light chain (VL) only and included the G51D mutation in CDR2 that is also present in scFv13.R4. Clone 2–3 also carried a VL G51D substitution as well as mutations in the VL CDRs and the VH frameworks. Interestingly, the VH framework mutations were identical to (e.g., V48I), similar to (e.g., A93T), or nearby to (e.g., L11P) substitutions in scFv13-R4. The effect of these mutations on folding and activity is clearly distinct. For instance, clone 2-1 showed no detectable increase in cytoplasmic expression and a small decrease in the amount of protein that partitioned to the insoluble fraction compared to its parent 1–4 (Fig. 5a). However, binding activity and affinity of this clone increased to a level on par with scFv13.R4 (Fig. 5b and Table 1). In the case of clone 2–3, the binding properties showed a more modest increase compared to parental clone 1–4 (Fig. 5b), but in vivo folding was dramatically improved compared to clone 1–4 as evidenced by a large increase in soluble expression and no detectable accumulation in the insoluble fraction (Fig. 5a). Gel permeation chromatography revealed that each purified scFv was predominantly monomeric (Fig. S5). Hence, we conclude that binding differences observed above were not due to the presence of inactive but soluble scFv aggregates. Collectively, these results show the Tat mechanism can be leveraged for the selection of binding proteins with significant improvements in both in vivo folding efficiency and antigen-binding activity.
Discussion
We have identified two long-lived Tat translocation intermediates, Ti-1 and Ti-2, that can be detected on the IM of permeabilized E. coli cells and are likely to be equivalent to Ti-1 and Ti-2 previously identified for the plant thylakoidal Tat system 18; 19. From a mechanistic standpoint, these results help us to dissect the transport process into several distinct steps that are characterized by separate translocation intermediates. For instance, detection of Ti-1 suggests that in the case of the Tat substrates tested here, the precursor assumes a loop-like structure involving the signal peptide and the early part of the mature region, leaving the N- and C- termini at the cytoplasmic face. Such an insertion mechanism is not unique to the Tat pathway as certain Sec pathway substrates such as OmpA have long been known to transiently adopt loop-like conformations 27. Interestingly, Ti-1 formation in E. coli can proceed with a non-functional Lys-Lys signal peptide or in the absence of proteinaceous transport machinery, similar to the situation in plant thylakoids. This suggests that initial membrane insertion and adoption of the transient loop topology for at least some E. coli Tat substrates occurs spontaneously. It is noteworthy that a similar spontaneous membrane insertion mechanism has been described for several thylakoidal membrane proteins that do not rely on the Tat (or Sec) protein export systems 28; 29. Following formation of Ti-1, proteins that are competent for export (i.e., have a functional Arg-Arg signal peptide and are correctly/completely folded) undergo transition to a bitopic topology during which the C-terminus is translocated across the membrane while the N-terminus remains in the cytoplasm. The fact that both folded and misfolded substrates formed the Ti-1 conformation, whereas only folded substrates formed Ti-2 suggests that any interrogation of substrate folding state likely occurs after insertion into the inner membrane, either preceding or coincident with the Ti-1 to Ti-2 transition. Moreover, since Ti-2 formation (but not Ti-1) is dependent on tatC, the Tat translocase may contribute to the quality control mechanism, as we speculated previously 8. In line with this hypothesis, the TatBC proteins were previously shown to interact with both folded and unfolded Tat substrates 13. However, site-specific cross-linking revealed a perturbed interaction between the signal peptide of the unfolded precursor and the TatBC receptor site, consistent with some degree of quality control by TatBC. Since TatB molecules oligomerize to form a transient cytoplasmic binding site for folded Tat substrates 30, Ti-1 intermediates could potentially access the TatB binding site prior to membrane translocation and formation of Ti-2. At present, however, the exact details of the quality control mechanism and the role played by the translocase in this process remain a mystery.
From an applied standpoint, we used the Ti-2 intermediate to create MAD-TRAP—a new protein engineering platform that enables simultaneous engineering of the solubility and antigen-binding properties of an scFv antibody. Because a Tat substrate must pass an in-built fitness filter to form Ti-2 and become displayed on the inner membrane, our MAD-TRAP technique effectively eliminates poorly folded scFv clones prior to panning for antigen binding. This should prove especially useful for the development of scFv antibodies that can be expressed in an intracellular compartment (i.e., intrabodies). Intrabodies have shown great potential as therapeutics for infectious diseases, neurodegenerative disorders and cancer 31; 32; 33; they are even being tested in cancer clinical trials 34. However, generation of intrabodies is challenging because formation of the disulfide bonds connecting the two β-sheets in each of the VH and VL domains is disfavored in the reducing environment of the cytoplasm. The absence of these bonds causes a large decrease in the ΔG of folding (~4–5 kcal/mol) 35 and accompanied loss of antigen-binding activity, susceptibility to proteolysis, and aggregation 36; 37. We anticipate that the MAD-TRAP strategy will simplify the generation of scFv variants that are well suited to function as intrabodies. Unlike most existing platforms, the incorporation of the Tat folding quality control allows screening for antigen binding and intracellular solubility in a single step. This point is best illustrated by clone 1–4, which was isolated after just a single round of mutagenesis and screening and exhibited marked improvements in both soluble expression and binding affinity. It is noteworthy that we previously developed a Tat-based selection for intrabodies called FLI-TRAP that relied on co-expression of antigens in the cytoplasm 38. MAD-TRAP similarly exploits the Tat folding quality control mechanism to screen antibody libraries but in a notable departure does not require intracellular expression of the antigen. Hence, MAD-TRAP represents a useful new tool for the antibody engineering toolbox that should permit isolation of intrabodies against more challenging targets such as post-translationally modified proteins (e.g., phosphoproteins) or integral membrane proteins for which in vitro panning strategies are possible.
Materials and Methods
Strains and growth conditions
Wildtype E. coli strain MC4100 and its isogenic ΔtatC derivative called BILK0 1 were used for membrane-anchored display of proteins. BL21(DE3) was used for cytoplasmic expression of proteins. Cultures were grown in LB medium supplemented with the appropriate antibiotic, and protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.5–1.0 mM) or arabinose (2% w/v) depending on the plasmid used. Antibiotics were supplemented at the following concentrations: ampicillin (100 μg/mL), chloramphenicol (20 μg/mL), and kanamycin (50 μg/mL).
Plasmid construction
Native HybO and HybO lacking the C-terminal tail were expressed from pBAD18-HybO-FLAG and pBAD18-HybOΔC-FLAG 38. Plasmid pBAD18-Δss-HybO-FLAG and pBAD18-Δss-HybOΔC-FLAG were constructed by amplifying HybO or HybOΔC, respectively, downstream from the signal sequence and including a SacI site in the forward primer and a FLAG tag and XbaI site in the reverse primer. To create artificial Tat-dependent proteins, an expression plasmid for each protein of interest was generated that contained an N-terminal signal peptide derived from E. coli trimethylamine N-oxide reductase (ssTorA). To generate these plasmids, pSALect-ssTorA-MBP-Bla 11 was digested with NdeI and BstBI to remove the MBP and TEM1 β-lactamase (Bla) genes, which resulted in the pSALect-ssTorA cassette plasmid. All genes to be ligated in the pSALect-ssTorA cassette were PCR amplified using primers specific for the target gene that contained an NdeI site in the forward primer and a BstBI site in the reverse primer. For constructs with an N-terminal FLAG epitope tag (DYKDDDDK), the FLAG sequence was included in the forward primer. Likewise, for constructs with a C-terminal FLAG tag, the FLAG sequence was included in the reverse primer. For constructs with a C-terminal HybO C-tail (HC; AGAIGLLGGVVGLVAGVSVMAV) 20, the HC was introduced using two overlapping reverse primers. For the different MBP and scFv13 constructs, template plasmids used for PCR amplification of the target genes were as follows: pSALect-ssTorA-MBP-Bla for the mature domain of MBP lacking its native Sec signal peptide, pHCME31 for the insoluble variant of MBP (G32D, I33P) 39 also lacking its Sec signal peptide, pPM163 and pPM163-R4 for insoluble scFv13 and soluble variant scFv13.R4, respectively 22, pSALect-scFv2610 for anti-digoxin scFv (laboratory stock), and pBAD18-Cm-ssTorA-scFv-GCN4(λ)-FLAG for the anti-GCN4 scFv 38. To generate derivatives that lacked the ssTorA signal peptide, pSALect-ssTorA-MBP-Bla was digested with NotI, which cuts upstream of the ssTorA sequence, and BstBI to remove the ssTorA-MBP-Bla insert. The different MBP and scFv13 constructs described above were then ligated in-frame between the NdeI and BstBI sites of digested pSALect-ssTorA-MBP-Bla. The ssTorA(KK) derivatives of scFv13 and scFv13.R4 were generated using the QuikChange II site-directed mutagenesis kit (Stratagene) to mutate the RR motif in the signal sequence to KK. To generate expression plasmids for MBP constructs with N-terminal FLAG tags in pBAD18-Cm, the pBAD18-Cm plasmid was first digested with SacI and XbaI. Wildtype and mutant MBP genes with and without the HC and with N-terminal FLAG tags were amplified from pSALect constructs with a SacI cut site upstream of ssTorA in the forward primer and with an XbaI site in the reverse primer. A similar approach was used to clone constructs with C-terminal FLAG tags between SacI and PstI of pBAD18-Kan. To express proteins of interest in the cytoplasm, target genes from biopanning experiments were subcloned into a pET21a(+) plasmid with a C-terminal 6x-His tag using NdeI and NotI sites flanking the target gene.
Spheroplast formation
Expression of proteins from pSALect-based plasmids was induced overnight at 37°C, and expression of proteins from pBAD18-based plasmids was generally induced for 4 h at 37°C. Following induction, 3 mL of each culture was pelleted in a 1.5 mL microcentrifuge tube. The pellets were washed with 100 μL of ice-cold fractionation buffer (FB; 0.75 M sucrose, 0.1 M Tris buffer, pH 8.0) and resuspended in 350 μL of ice-cold FB supplemented with lysozyme (1 mg/mL). While slowly vortexing, 700 μL of EDTA (1 mM, pH 8.0) was added dropwise, and tubes were incubated for 20 min at room temperature. After adding 50 μL of cold MgCl2 (0.5 M), tubes were incubated on ice for 10 min and then spun down (12000 rcf) for 10 min at 4°C. The supernatant was removed, and the spheroplasts were resuspended in 1 mL of phosphate-buffered saline (PBS, pH 7.4). Spheroplasts were kept on ice until used in subsequent assays.
Flow cytometry
Flow cytometric analysis was used to detect membrane-anchored translocation intermediates and evaluate binding of anchored proteins to a target antigen, namely β-gal. β-Gal was labeled with FITC for these experiments using the FluoroTag FITC conjugate kit (Sigma). β-gal (Biochemika) was dissolved at 5 mg/mL in sodium bicarbonate buffer (0.1 M, pH 9.0) and then labeled following protocols provided with the FluoroTag kit. A molar extinction coefficient of 24,1590 M−1cm−1 was assumed for β-gal 40. Following labeling, the molar ratio of fluorescent dye to β-gal (F/P ratio) was 2.3, and the final concentration of FITC-β-gal was 0.6 mg/mL. Spheroplasts were prepared as described above, and 50 μL of spheroplasts was mixed with 50 μL of PBS containing a FITC-conjugated anti-FLAG antibody (Sigma, 10 μg/mL final concentration) or with 50 μL of PBS supplemented with 2 μL of FITC-conjugated β-gal. Spheroplasts were incubated with the FITC conjugates for 45 min in the dark, washed with 400 μL of PBS, and resuspended in 500 μL of PBS. Flow cytometry was performed using a FACSCalibur flow cytometer (BD).
Subcellular fractionation and Western blot analysis
To prepare subcellular fractions for Western blot analysis, 3 mL of induced culture was pelleted and washed with subcellular fractionation buffer (SFB; 30 mM Tris-HCl, 1 mM EDTA, 0.6 M sucrose). Cells were resuspended in 1 mL SFB and then incubated for 20 min at room temperature. After adding 266 μL of 5 mM MgSO4, cells were incubated for 10 min on ice. Cells were spun down, and the supernatant was taken as the periplasmic fraction. The pellet was treated with BugBuster Master Mix (Novagen) for 5 min at room temperature. Following centrifugation at 16000 rcf at room temperature for 5 min, the second supernatant was taken as the cytoplasmic soluble fraction, and the pellet was the insoluble fraction. To prepare samples for analysis of cytoplasmic solubility, 5 mL of induced culture was pelleted and resuspended in 1 mL of BugBuster Master Mix. Samples were incubated for 20 min at room temperature and then spun down at 16000 rcf for 20 min at 4°C. The supernatant was taken as the soluble fraction. The pellet was washed with Tris-HCl (50 mM) with EDTA (1 mM) and resuspended in PBS with 2% SDS. After boiling for 10 min, the resuspended pellets were centrifuged for 10 min at 16000 rcf. The supernatant was taken as the insoluble fraction. Proteins were separated by 12% SDS-polyacrylamide gels (Bio-Rad), and Western blotting was performed according to standard protocols. Briefly, proteins were transferred onto polyvinylidene fluoride (PVDF) membranes, and membranes were probed with either anti-FLAG antibodies conjugated to HRP (Abcam) or anti-6x-His antibodies conjugated to HRP (Abcam).
Combinatorial library construction
A random mutagenesis library was generated from scFv13 using the Genemorph II random mutagenesis kit (Stratagene). Four PCR reactions were used, with 1 ng pPM163 (containing scFv13 template) in each reaction. The resulting PCR product was digested with NdeI and BstBI, purified by gel electrophoresis, and cloned into the pSALectΔBla cassette digested with the same enzymes. The library was transformed into electrocompetent DH5a cells, and the library size and error rate were determined to be 2.4×106 members and ~5 mutations per gene, respectively. The resulting plasmid library was then transformed into MC4100 cells for bead-based screening. After the first round of mutagenesis and screening, a second-generation library was constructed in the same manner using first-round clone 1–4 as a template. The library size and error rate for this library were determined to be 1.2×106 members and ~7 mutations per gene, respectively. Saturation mutagenesis of residue S55 in the VH domain of scFv13 was generated using the QuikChange II site-directed mutagenesis kit (Stratagene) with primers that included random bases at the VH S55 site to produce an NNK codon, where N is A, C, G, or T and K is G or T. After sequencing ~60 randomly selected transformants, clones with all but three amino acids at position S55 were isolated. The remaining three amino acids (E, H, and K) were cloned individually using the same site-directed mutagenesis kit.
Panning with magnetic beads
Dynabead M-280 tosylactivated beads (Invitrogen) were coated with β-gal for panning scFv libraries. 1 mL of beads (2×109 beads) was washed with 0.1 M sodium phosphate buffer (pH 7.4; Buffer 1) and then resuspended in 1 mL of Buffer 1 containing 0.5 mg/mL β-gal. Following overnight incubation at 37°C, beads were washed twice with PBS with 0.1% BSA (w/v) and 2 mM EDTA (pH 7.4; Buffer 2). Beads were resuspended in 1 mL of 0.2 M Tris with 0.1% BSA (pH 8.0; Buffer 3), incubated for 2 h at 37°C to deactivate free tosyl groups, and washed with Buffer 2. Beads were mixed with spheroplasts (prepared as described above) using a ratio of approximately 10:1 cells:beads, with 1 mL total volume in each tube. Binding reactions were incubated with rotation at 4°C for 11 h. After incubation, bead-bound spheroplasts were washed four times with Buffer 2 and resuspended in 25 μL of distilled water in each tube. The DNA for the scFvs displayed on the bead-bound spheroplasts was recovered using PCR and then cloned back into the pSALectΔBla cassette. The panning procedure was repeated to enrich for scFvs that bind to β-gal. During screening of the second-generation library, purified scFv clone 1–4 (0.05–50 nM) was included as a competitor to increase the stringency of the screening.
Protein purification
After expressing scFvs in the cytoplasm and obtaining soluble fractions as described above, 6x-His-tagged scFvs were purified using protocols provided with Ni-NTA protein purification spin columns (Qiagen). Soluble lysates were put over the columns, and the columns were washed four times with a buffer containing 50 mM NaH2PO4, 300 mM sodium chloride, and 20 mM imidazole (pH 8.0). scFvs were eluted using a buffer containing 50 mM NaH2PO4, 300 mM sodium chloride, and 500 mM imidazole (pH 8.0). After spin-column purification, eluted scFvs were further purified using a 100 kDa molecular weight cut-off column (Sartorius Stedim) to remove a high molecular weight impurity. Final purity of scFvs was confirmed by SDS-PAGE and Coomassie staining (Fig. S6). Representative scFv yields are provided in Table 1.
Enzyme-linked immunosorbant assay
Enzyme-linked immunosorbant assay (ELISA) was used to evaluate the binding of purified scFvs to β-gal. ELISA plates were coated overnight at 4°C with 50 μL/well of β-gal in PBS (10 μg/mL). Plates were then blocked at room temperature for 2 h with 2% non-fat milk in PBS. After washing plates using PBS supplemented with 0.1% Tween 20 (PBST), purified protein samples serially diluted in PBS with 50 μg/mL BSA (PBS-BSA) were added to the plates (50 μL/well). Plates were incubated for 1 h at room temperature and then washed with PBST. Horseradish peroxidase (HRP)-conjugated anti-6x-His antibody (Abcam) in PBS-BSA was added to the plates (50 μL/well). After 1 h of incubation at room temperature, plates were washed and then incubated with SigmaFast OPD HRP substrate (Sigma) for 20 min. The reaction was quenched with H2SO4, and the absorbance of the wells was measured at 490 nm. ELISAs with spheroplasts were performed in a similar manner; membrane-bound scFvs were detected with an HRP-conjugated anti-FLAG antibody (Abcam).
Dissociation constant determination by competitive ELISA
Equilibrium dissociation constants for wt scFv13, scFv13.R4, and scFv clone 2-1 were determined as described by Martineau 41. Briefly, ELISA plates were coated with β-gal (2 μg/mL) and blocked with 2% non-fat milk in PBS. For each scFv, dilutions of β-gal (0–4000 nM) were prepared and mixed with an equal volume of scFv solution. Final concentrations of scFv solutions (5 μg/mL for scFv13 wt, 5 μg/mL for scFv13.R4, 0.35 μg/mL for clone 2-1) were chosen to produce ELISA signals within the linear range for each scFv. The β-gal/scFv solutions were incubated for 1 h and added to the coated and blocked ELISA plates. After incubating the solutions in the plates for 1 h, the solutions were removed and the plates were washed with PBST. scFv bound to the plate (scFv that did not bind to β-gal in solution) was detected using an anti-6x-His primary antibody (Genscript) and an anti-rabbit secondary antibody conjugated to alkaline phosphatase. Plates were incubated with liquid p-nitrophenylphosphate (pNPP) alkaline phosphatase substrate (Sigma) for 4 h prior to reading plates at 405 nm. The dissociation constant Kd for each scFv was extracted from the absorbance data using KaleidaGraph (Synergy Software) to fit the Kd binding model to the data.
Gel filtration chromatography
scFv samples were purified as described above, and the concentration was adjusted to 20 μM. For each scFv, 250 μL was injected into a Superdex 200 10/300 GL column (GE Healthcare) attached to a DuoFlow FPLC system (Bio-Rad), and samples were run at a flow rate of 0.25 mL/min at 4°C in buffer containing 50 mM sodium phosphate (pH 7.5) and 150 mM sodium chloride. The migration of the scFvs was monitored by absorbance at 280 nm, and 250 μL fractions were collected. Gel filtration protein standards (Bio-Rad) were run through the column in a similar manner.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Career Award CBET-0449080, the New York State Office of Science, Technology and Academic Research Distinguished Faculty Award and the National Institutes of Health Small Business Innovation Research Award R41GM090585 (all to M.P.D.). This work was also supported by award number F32CA150622 from the National Cancer Institute (to A.J.K). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations
- Tat
twin-arginine translocation
- ssTorA
trimethylamine N-oxide reductase signal peptide
- Ti
translocation intermediate
- MAD-TRAP
membrane-anchored display for Tat-based recognition of associating proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–6. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 2.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–50. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Settles AM, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–70. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 4.Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 5.Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigue A, Chanal A, Beck K, Muller M, Wu LF. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J Biol Chem. 1999;274:13223–8. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 7.Santini CL, Ize B, Chanal A, Muller M, Giordano G, Wu LF. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–12. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLisa MP, Tullman D, Georgiou G. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci U S A. 2003;100:6115–20. doi: 10.1073/pnas.0937838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders C, Wethkamp N, Lill H. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol Microbiol. 2001;41:241–6. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- 10.Matos CF, Robinson C, Di Cola A. The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J. 2008;27:2055–63. doi: 10.1038/emboj.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15:449–58. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim HK, Mansell TJ, Linderman SW, Fisher AC, Dyson MR, DeLisa MP. Mining mammalian genomes for folding competent proteins using Tat-dependent genetic selection in Escherichia coli. Protein Sci. 2009;18:2537–49. doi: 10.1002/pro.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panahandeh S, Maurer C, Moser M, DeLisa MP, Muller M. Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J Biol Chem. 2008;283:33267–75. doi: 10.1074/jbc.M804225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter S, Bruser T. Targeting of unfolded PhoA to the Tat translocon of Escherichia coli. J Biol Chem. 2005;280:42723–30. doi: 10.1074/jbc.M509570200. [DOI] [PubMed] [Google Scholar]
- 15.Maurer C, Panahandeh S, Moser M, Muller M. Impairment of twin-arginine-dependent export by seemingly small alterations of substrate conformation. FEBS Lett. 2009;583:2849–53. doi: 10.1016/j.febslet.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 16.Cline K, McCaffery M. Evidence for a dynamic and transient pathway through the Tat protein transport machinery. EMBO J. 2007;26:3039–49. doi: 10.1038/sj.emboj.7601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter S, Lindenstrauss U, Lucke C, Bayliss R, Bruser T. Functional Tat transport of unstructured, small, hydrophilic proteins. J Biol Chem. 2007;282:33257–64. doi: 10.1074/jbc.M703303200. [DOI] [PubMed] [Google Scholar]
- 18.Berghofer J, Klosgen RB. Two distinct translocation intermediates can be distinguished during protein transport by the TAT (Deltaph) pathway across the thylakoid membrane. FEBS Lett. 1999;460:328–32. doi: 10.1016/s0014-5793(99)01365-4. [DOI] [PubMed] [Google Scholar]
- 19.Hou B, Frielingsdorf S, Klosgen RB. Unassisted membrane insertion as the initial step in DeltapH/Tat-dependent protein transport. J Mol Biol. 2006;355:957–67. doi: 10.1016/j.jmb.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol. 2003;49:1377–90. doi: 10.1046/j.1365-2958.2003.03642.x. [DOI] [PubMed] [Google Scholar]
- 21.Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J Bacteriol. 2003;185:2811–9. doi: 10.1128/JB.185.9.2811-2819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martineau P, Jones P, Winter G. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol. 1998;280:117–27. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- 23.Fisher AC, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol. 2009;385:299–311. doi: 10.1016/j.jmb.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.der Maur AA, Zahnd C, Fischer F, Spinelli S, Honegger A, Cambillau C, Escher D, Pluckthun A, Barberis A. Direct in vivo screening of intrabody libraries constructed on a highly stable single-chain framework. J Biol Chem. 2002;277:45075–85. doi: 10.1074/jbc.M205264200. [DOI] [PubMed] [Google Scholar]
- 25.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–53. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 26.Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc Natl Acad Sci U S A. 2004;101:9193–8. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn A, Kiefer D, Kohne C, Zhu HY, Tschantz WR, Dalbey RE. Evidence for a loop-like insertion mechanism of pro-OmpA into the inner membrane of Escherichia coli. Eur J Biochem. 1994;226:891–7. doi: 10.1111/j.1432-1033.1994.00891.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim SJ, Jansson S, Hoffman NE, Robinson C, Mant A. Distinct “assisted” and “spontaneous” mechanisms for the insertion of polytopic chlorophyll-binding proteins into the thylakoid membrane. J Biol Chem. 1999;274:4715–21. doi: 10.1074/jbc.274.8.4715. [DOI] [PubMed] [Google Scholar]
- 29.Michl D, Robinson C, Shackleton JB, Herrmann RG, Klosgen RB. Targeting of proteins to the thylakoids by bipartite presequences: CFoII is imported by a novel, third pathway. EMBO J. 1994;13:1310–7. doi: 10.1002/j.1460-2075.1994.tb06383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurer C, Panahandeh S, Jungkamp AC, Moser M, Muller M. TatB functions as an oligomeric binding site for folded Tat precursor proteins. Mol Biol Cell. 2010;21:4151–61. doi: 10.1091/mbc.E10-07-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams BR, Zhu Z. Intrabody-based approaches to cancer therapy: status and prospects. Curr Med Chem. 2006;13:1473–80. doi: 10.2174/092986706776872899. [DOI] [PubMed] [Google Scholar]
- 32.Miller TW, Messer A. Intrabody applications in neurological disorders: progress and future prospects. Mol Ther. 2005;12:394–401. doi: 10.1016/j.ymthe.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Marasco WA, LaVecchio J, Winkler A. Human anti-HIV-1 tat sFv intrabodies for gene therapy of advanced HIV-1-infection and AIDS. J Immunol Methods. 1999;231:223–38. doi: 10.1016/s0022-1759(99)00159-3. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez RD, Barnes MN, Gomez-Navarro J, Wang M, Strong TV, Arafat W, Arani RB, Johnson MR, Roberts BL, Siegal GP, Curiel DT. A cancer gene therapy approach utilizing an anti-erbB-2 single-chain antibody-encoding adenovirus (AD21): a phase I trial. Clin Cancer Res. 2000;6:3081–7. [PubMed] [Google Scholar]
- 35.Frisch C, Kolmar H, Schmidt A, Kleemann G, Reinhardt A, Pohl E, Uson I, Schneider TR, Fritz HJ. Contribution of the intramolecular disulfide bridge to the folding stability of REIv, the variable domain of a human immunoglobulin kappa light chain. Fold Des. 1996;1:431–40. doi: 10.1016/s1359-0278(96)00059-4. [DOI] [PubMed] [Google Scholar]
- 36.Cattaneo A, Biocca S. The selection of intracellular antibodies. Trends Biotechnol. 1999;17:115–21. doi: 10.1016/s0167-7799(98)01268-2. [DOI] [PubMed] [Google Scholar]
- 37.Proba K, Honegger A, Pluckthun A. A natural antibody missing a cysteine in VH: consequences for thermodynamic stability and folding. J Mol Biol. 1997;265:161–72. doi: 10.1006/jmbi.1996.0726. [DOI] [PubMed] [Google Scholar]
- 38.Waraho D, DeLisa MP. Versatile selection technology for intracellular protein-protein interactions mediated by a unique bacterial hitchhiker transport mechanism. Proc Natl Acad Sci U S A. 2009;106:3692–7. doi: 10.1073/pnas.0704048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betton J, Hofnung M. Folding of a mutant maltose-binding protein of Escherichia coli which forms inclusion bodies. J Biol Chem. 1996;271:8046–52. doi: 10.1074/jbc.271.14.8046. [DOI] [PubMed] [Google Scholar]
- 40.Hoyoux A, Jennes I, Dubois P, Genicot S, Dubail F, Francois JM, Baise E, Feller G, Gerday C. Cold-adapted beta-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl Environ Microbiol. 2001;67:1529–35. doi: 10.1128/AEM.67.4.1529-1535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martineau P. Affinity measurements by competition ELISA. Antibody Engineering. 2010;1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.