Abstract
Trypanosoma brucei, the causative agent of African Sleeping sickness, is replete with unique biochemistry, including unusual features of gene transcription. The parasite also contains over 4500 non-annotated genes, representing novel biochemistry yet to be explored. Using tandem affinity purification (TAP)-tagged TbTFIIB, we identified and subsequently confirmed, one of the non-annotated Trypanosoma brucei proteins, Tb11.02.4300, as a TbTFIIB-interacting protein. The 49 kDa protein is nuclear and essential for parasite variability as determined by RNA interference studies; hence, the nomenclature T. brucei Essential Nuclear Factor (TbENF). TbENF is shown to interact with DNA in a sequence-independent fashion under the conditions examined. Furthermore, TbENF bears motifs associated with many eukaryotic transcription factors, such as a glutamine-rich region and a leucine zipper, yet TbENF is specific to trypanosomatids making it a potentially attractive therapeutic target. Taken together, our results suggest a role for TbENF in trypanosome gene transcription.
Keywords: Trypanosoma brucei, TFIIB-interacting protein, transcription, DNA-binding protein
1.1. Introduction
Parasitic protozoan trypanosomatids are the causative agent of world-wide diseases, including Chagas’ Disease, Leishmaniasis, and African Sleeping Sickness, afflicting millions[1]. Trypanosoma brucei, transmitted through the tsetse fly, causes human African Sleeping Sickness. The parasite also infects livestock, causing significant negative economic impact in sub-Saharan Africa. Treatments for the disease are limited, often toxic, and resistance can be problematic. New therapeutics are needed.
Trypanosomes have many unusual biochemical features which may serve as possible therapeutic targets, including transcription. RNA Polymerase (RNAP1) II-dependent gene transcription and the regulation of gene expression differ significantly from the process in higher eukaryotes. Protein-coding genes are transcribed polycistronically and are processed into mature individual mRNA through the trans-addition of a short capped Spliced Leader (SL) RNA and the addition of a poly(A) tail at the 5’ and 3’ ends of the transcript, respectively [2, 3]. The regulation of gene expression is hypothesized to occur mainly post-transcriptionally and, in some cases, has been found to occur at the level of translation [4–6]. Recent work also indicates that at least some transcriptional control is exercised at the level of chromatin remodeling in the parasites [2, 7].
Canonical RNAPII promoter elements for protein coding genes appear lacking. Indeed, the small nuclear SL RNA gene contains the only known RNAPII-dependent gene promoter in trypanosomes [8]. The transcriptional initiation of protein coding genes is not well understood, although evidence is accumulating that transcription initiates at regions of the genome where clusters of open reading frames switch from one strand to another (strand-switch regions) [9, 10].
Several of the basal transcription factors are either present in divergent forms or appear absent. For example, the trypanosomatid general transcription factor TFIIB is divergent in its sequence [11, 12] and the crystal structure reveals the presence of extra helices and loops speculated to participate in trypanosome-specific protein interactions [13]. The composition of TbTFIIH was examined and found to contain essential subunits unique to the parasite, while lacking the cyclin-activating kinase [14, 15]. The basal factors TFIIF and TFIIE appear either absent or are not readily discernable in the genome [16]. An extremely divergent form of the mediator head module was identified in trypanosomes and shown to be essential in small nuclear gene transcription [17]. The trypanosome small nuclear RNA-Activating Protein complex (SNAPc), involved in small nuclear RNA gene transcription, harbors one unique subunit and lacks others compared to its homologs in higher eukaryotes [18, 19]. T. brucei TATA binding protein (TBP)/TbTBP-related factor 4 (TRF4) is unique as several key DNA-interacting amino acids are not conserved in the trypanosome homologs [20]. Finally, the carboxyl terminal domain (CTD) of the largest subunit of TbRNAPII lacks the canonical heptad repeats of YSPTSPS, which in other eukaryotes orchestrate and coordinate the factors involved in the various stages of transcription [21, 22]. The observed divergence of the factors characterized thus far suggests transcription in trypanosomatids, in part, does not mirror the process in other eukaryotes. The unique properties of the characterized parasite proteins suggest novel, uncharacterized proteins may play roles in trypanosomatid transcription.
Currently, more than 4500 T. brucei gene products are non-annotated (http://tritrypdb.org/tritrypdb); among these are potential candidate proteins for roles in transcription. This pool of non-annotated proteins may also represent a host of targets for much needed therapeutics to treat the diseases inflicted by these obligate parasites. To seek out novel transcription-related proteins in trypanosomatids, we utilized a Tandem Affinity Purification (TAP)-tagged TbTFIIB and identified interacting proteins through mass spectrometry. Among the interacting proteins is one that harbors several hallmarks of known transcription factors, yet is specific to trypanosomatids. The protein, Essential Nuclear Factor 49kDa (ENF), strongly interacts with TbTFIIB, is essential for parasite viability, and binds tightly to DNA.
2. Materials and Methods
2.1 TAP-Tagged TbTFIIB generation and purification
To generate a carboxyl-terminal tandem affinity purification (TAP)-tagged TbTFIIB, the TFIIB gene (GenBank ID: 7083113) was amplified from T. brucei genomic DNA and inserted into the pLEWIII plasmid using the BamHI and HindIII sites. All primer sequences may be found in the Supplemental Tables 1 and 2. The N-terminal TAP tag was obtained from pJM26 (a kind gift from the Bellofatto laboratory, [18]) through BamHI digestion and insertion into the pLEWIII derivative containing the TbTFIIB gene. The resulting construct was verified by DNA sequencing and named pTAP-TbTFIIB. Ten µg of NotI-linearized pTAP-TbTFIIB was transfected into procyclic T. brucei cell line 29–13 [23], following the electroporation protocol outlined in [24]. TAP-TbTFIIB expression was induced through the addition of 500ng/mL tetracycline to the media for 24 hours.
TAP-TbTFIIB was purified from 2 L of parasites grown to 1.5×107 cells/mL based on the protocols of [18] and [25]. Nuclear extract was first prepared [26], nucleic acids were removed through ammonium sulfate precipitation, and the resulting proteins resuspended in immunoglobulin G (IgG) binding buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40, 1mM DTT, 10% glycerol, containing PMSF, pepstatin, and leupeptin). The proteins were applied to an IgG sepharose column, and after extensive washing, the column was equilibrated into tobacco etch virus (TEV) cleavage buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT, 5% glycerol). Protein was eluted from the column following digestion by 100 units of TEV protease at 4 °C for 2 hrs. The eluted proteins diluted into calmodulin binding buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10% glycerol, and 1 mM DTT), were applied to calmodulin resin (Sigma). After extensive washing, the proteins were eluted with calmodulin elution buffer (calmodulin binding buffer containing 5 mM EGTA). Eluted proteins were concentrated, separated by 10% SDS-PAGE, and analyzed by MALDI-TOF and Q-TOF mass spectrometry at the Center for Advanced Proteomics Research (UMDNJ-New Jersey Medical School, Newark, NJ).
2.2 Glutathione-S-Transferase (GST)-tagged TbENF generation and purification
To generate recombinant amino-terminal GST-tagged TbENF, the TbENF gene was amplified from T. brucei genomic DNA and inserted into pGEX-6P-1 (GE Healthcare) at the XhoI site. The resulting construct was verified by DNA sequencing and named pGST-TbENF. Recombinant GST-TbENF was expressed in E. coli BL21 cells and purified to approximate homogeneity using glutathione agarose according to the manufacturer’s guidelines. PreScission protease (GE Healthcare) was used to remove the GST tag and the resulting recombinant TbENF protein was used to generate custom polyclonal antibodies in rabbits (Lampire Biologicals, Pipersville, PA).
2.3 Verification of the TbENF-TbTFIIB interaction
To verify the interaction between TbTFIIB and TbENF, recombinant GST-TbENF or GST and T. brucei 427 whole cell extract, prepared as described in [27], were used. Ten µg of recombinant GST-TbENF purified to homogeneity was bound to 35 µL of glutathione sepharose and mixed with 700 µg of whole cell extract. The reaction, 300 µL total volume in pulldown buffer (150 mM sucrose, 20 mM potassium glutamate, 10 mM HEPES-KOH, pH 7.9, 2.5 mM MgCl2, 1 mM EDTA, and 2.5 mM DTT containing protease inhibitors), was incubated with rotation at 4 °C for 30 min. The beads were then washed extensively with 150 mM sucrose, 10 mM HEPES-KOH, pH 7.9, 2.5 mM MgCl2, 1 mM EDTA, 0.2% NP-40, 2.5 mM DTT, and either KCl ranging from 0.3–0.9 M or potassium glutamate ranging from 0.2–0.4 M. After the stringent wash, the interacting proteins were analyzed by Western blot analysis using TbTFIIB, TbTBP, and TbRNAPII antibodies (kind gifts from the Bellofatto laboratory). In every case, a control reaction carried out under identical conditions using GST was performed in parallel with the GST-TbENF pulldown reaction. Pulldown reactions in which the whole cell extract was treated with DNase were carried out under identical conditions using a 0.7M KCl wash except that the extract was pretreated with 2 Units of DNase at room temperature for 15 minutes. The DNase was determined to be active at room temperature in whole cell extract using plasmid DNA.
2.4 TbENF RNA interference studies and examination of transcript levels
To target TbENF by RNA interference, a 309 bp region corresponding to nucleotides 22–329 of the open reading frame was amplified from T. brucei genomic DNA and inserted into p2T7–177 [28] to yield pTbENF-RNAi. TbENF-RNAi was transfected into cell line 29–13 through electroporation and clonal cell lines generated by limiting dilution. Production of double-stranded RNA was induced by the addition of 1 µg/mL of tetracycline to the SDM-79 media daily. Parasite growth was monitored over 8 days. Daily, 8×106 cells were removed from culture, resuspended in Laemmli buffer [29], and TbENF levels monitored by Western blot using custom rabbit polyclonal antibody against TbENF (serum was used at dilution of 1:800) and goat anti-rabbit-IgG conjugated with alkaline phosphatase used at 1:20,000.
To measure transcript levels, RNA was isolated from 1 × 108 parasites using Trizol reagent. The RNA samples were DNase-treated and 0.5µg RNA was used in a reverse transcription reaction (Finnzyme Phusion RT-PCR kit used according to the manufacturer’s instructions) using gene-specific primers for Tb11.02.4300, the spliced leader RNA intron region, and U6 snRNA (Supplemental Table 3). The resultant cDNA was used as template in PCR reactions (20 cycles) and the products visualized on 8% polyacrylamide gels stained with SYBR green. The relative amounts of cDNA used in each reaction are provided in Figure 5C. The primers to examine Tb11.02.4300 did not correspond to the region used to target Tb11.02.4300 for knockdown.
Figure 5. RNA interference studies reveal TbEFN49 is essential.
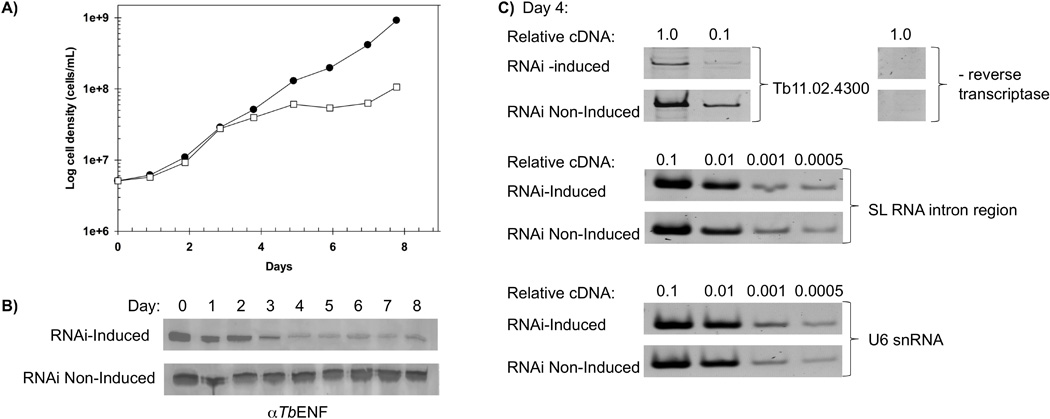
(A) Growth curves for clonal cell lines in which RNAi against TbENF was tetracycline induced (open squares) or non-induced (closed circles) in SDM-79 media. Parasite density, determined daily, is presented on a log scale. (B) Western blot analysis of TbENF levels in the RNAi-induced and non-induced cell lines. Each lane of the 10% SDS-PAGE gels contained 8×106 parasites. (C) Semiquantitative PCR to assess TbENF, Spliced Leader, and U6 RNA transcript levels on Day 4 of the RNA interference experiment.
2.5 TbENF-DNA interaction studies
Biotinylated probes corresponding to regions of the SL promoter (GenBank ID: X00633.1), T. brucei U6 gene promoter (GenBank ID: X13017.1), Tb11.02.4300 open reading frame (GenBank ID: 71755606), bla open reading frame (GenBank ID: HQ284188.1), and a region of lambda phage DNA (GenBank ID: 9626243) were generated using 5’ biotinylated primers (Eurofins MWG Operon, Huntsville, AL.). Nuclear extracts were prepared as described in [18]. The pulldown protocol was modified from [30] and [31]. Five hundred ng of biotinylated DNA was bound to 100 µg of M-280 Streptavidin Dynabeads (Invitrogen) in 1X PBS. The beads were then washed, equilibrated into, and blocked with Buffer 1 containing 5 mg/mL BSA (150 mM sucrose, 20 mM HEPES-KOH pH 7.9, 20 mM potassium glutamate, 20 mM KCl, 2.5%w/v PEG, 0.2 mM EDTA, 0.5 mM EDTA, 3 mM MgCl2, 4 mM DTT and protease inhibitors). After the block step, beads were equilibrated into Buffer 1 and 100 µg of nuclear extract was added to the beads. The binding was allowed to proceed for 5 min on ice. Following the binding, the magnetic beads were washed with either Buffer 1 containing TWEEN-20 (0.1–0.2 %) or KCl (0.5–1.0 M) for three 5 min washes, followed by one 5 min wash in Buffer 1. Samples were then resuspended in 30 µL of 2X Laemmli sample buffer and the associated proteins analyzed by Western blot.
2.6 Localization of TbENF by sucrose cushion
Trypanosome nuclei and cytoplasm were fractionated from 3×109 wild-type 427 procyclic trypanosomes by centrifugation on a sucrose cushion. The cells were harvested, washed, resuspended in hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 2.5 mM DTT, and protease inhibitors), and swelled on ice for 10 min. NP-40 was added to a final concentration of 0.2 % and the cells were homogenized using a dounce. The resulting extract was placed on a 0.8 M sucrose cushion (hyptonic buffer containing 0.8 M sucrose) and centrifuged in a swinging bucket rotor at 8,000 × g for 10 min. The resultant fractions were probed by Western blot analysis using custom polycolonal antibodies against TbTBP, TbENF, and TbTFIIB and a commercial antibody against human elongation factor 2 (Santa Cruz Biotechnology, Inc).
3. Results and Discussion
3.1 TAP-TbTFIIB Interacting Proteins: The Identification of TbENF, a Trypanosomatid-Specific Protein
The T. brucei basal transcription factor TFIIB was TAP-tagged and used to identify interacting proteins. TAP-TbTFIIB was overexpressed in procyclic parasites, purified, and mass spectrometry was used to identify the following potential interacting proteins from the TAP-TbTFIIB purfication: microtubule-associated protein (GenBank ID: 70832917), heat shock protein 70 (GenBank ID: 123633), conserved hypothetical protein Tb11.02.4300 (GenBank ID: 70834104), β-tubulin (GenBank ID: 115504281), TAP-TbTFIIB and non-tagged TbTFIIB, as determined by the apparent mass on the gel, (GenBank ID: 70831131), and conserved hypothetical protein Tb927.6.4440 (GenBank ID: 62358814). The final concentrated elution containing the protein bands identified by mass spectrometry is shown in Figure 1.
Figure 1.
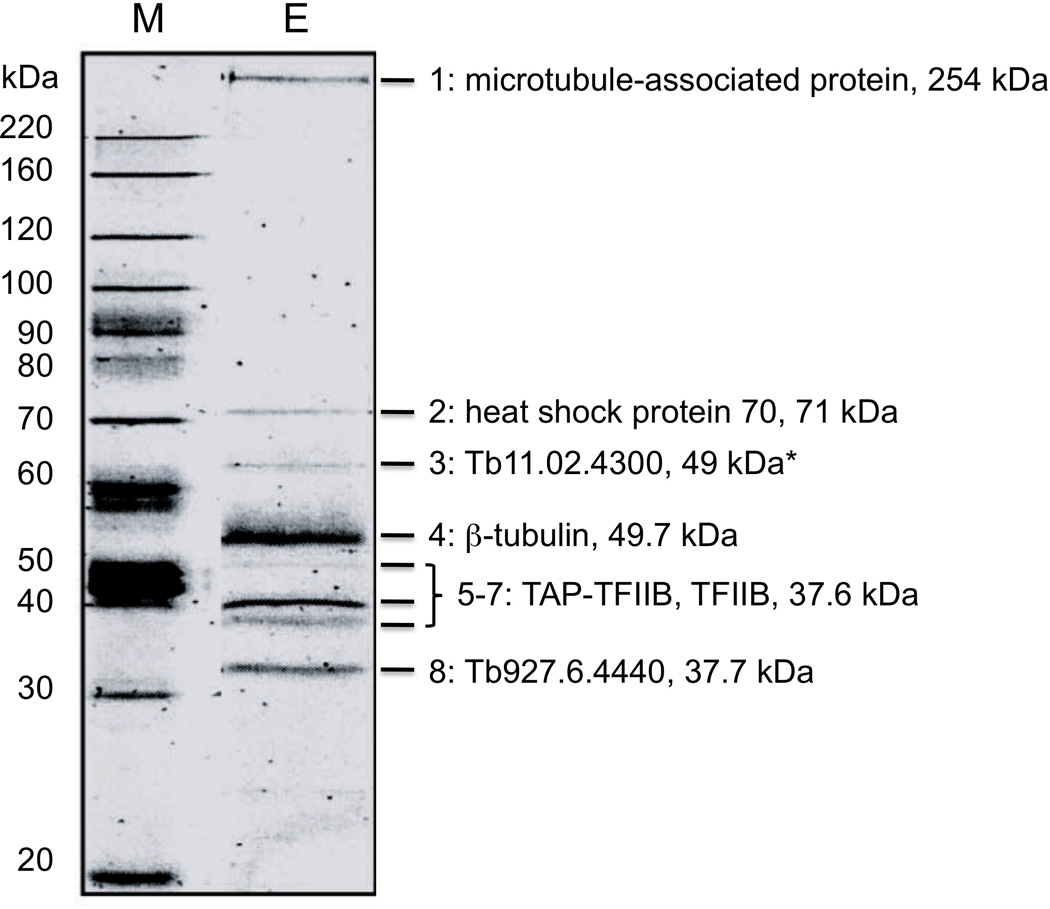
Potential TbTFIIB-interacting proteins from the TAP-TbTFIIB purification. The concentrated elution from the calmodulin resin was separated by electrophoresis in 10% polyacrylamide containing 0.1% sodium dodecyl sulfate. Proteins in bands 1–8 were identified through mass spectrometry and found to be 1) microtubule-associated protein (GenBank ID: 70832917), 2) heat shock protein 70 (GenBank ID: 123633), 3) conserved hypothetical protein Tb11.02.4300 (GenBank ID: 70834104), 4) β-tubulin (GenBank ID: 115504281), 5–7) TbTFIIB (GenBank ID: 70831131), and 8) conserved hypothetical protein Tb927.6.4440 (GenBank ID: 62358814). *Tb11.02.4300 consistently migrates 10 kDa higher than its molecular weight.
The conserved hypothetical protein Tb11.02.4300, henceforth referred to as Essential Nuclear Factor, 49 kDa (ENF), has interesting features at the sequence level that led us to explore this protein further (Figure 2). All trypanosomatid ENF homologs have predicted coiled-coil regions containing leucine zippers [32], motifs traditionally associated with dimerization and DNA binding in several known eukaryotic transcription factors, including the transcriptional activator GCN4 [33]. Furthermore, all ENF homologs are glutamine-rich. Several other known transcription factors, including transcription activators Sp1[34], Oct1, and Oct 2, [35] are glutamine-rich. We analyzed TbENF using Robetta [36], a program that predicts protein structure ab initio. The C-terminal domain of TbENF (residues 178–441) is predicted to have tertiary structure similar to that found in many known transcription factors. Several models were generated having confidence scores indicating a greater than 50% probability of the correct fold, including a winged-helix DNA binding domain found in such factors as HNF-3 [37], a homeodomain-like fold which is associated with DNA binding [38], and a leucine zipper domain. Finally, TbENF is reported to interact with both T. brucei RNAPII [39] and TFIIH [15]. In the later work, the authors suggest possible similarity between TbENF and the transcription factor MRG1 (Cbp/p300 interacting transactivator 2). Collectively, these results are highly suggestive of a role in transcription for TbENF.
Figure 2. Multiple sequence alignment of trypanosomatid ENF homologs.
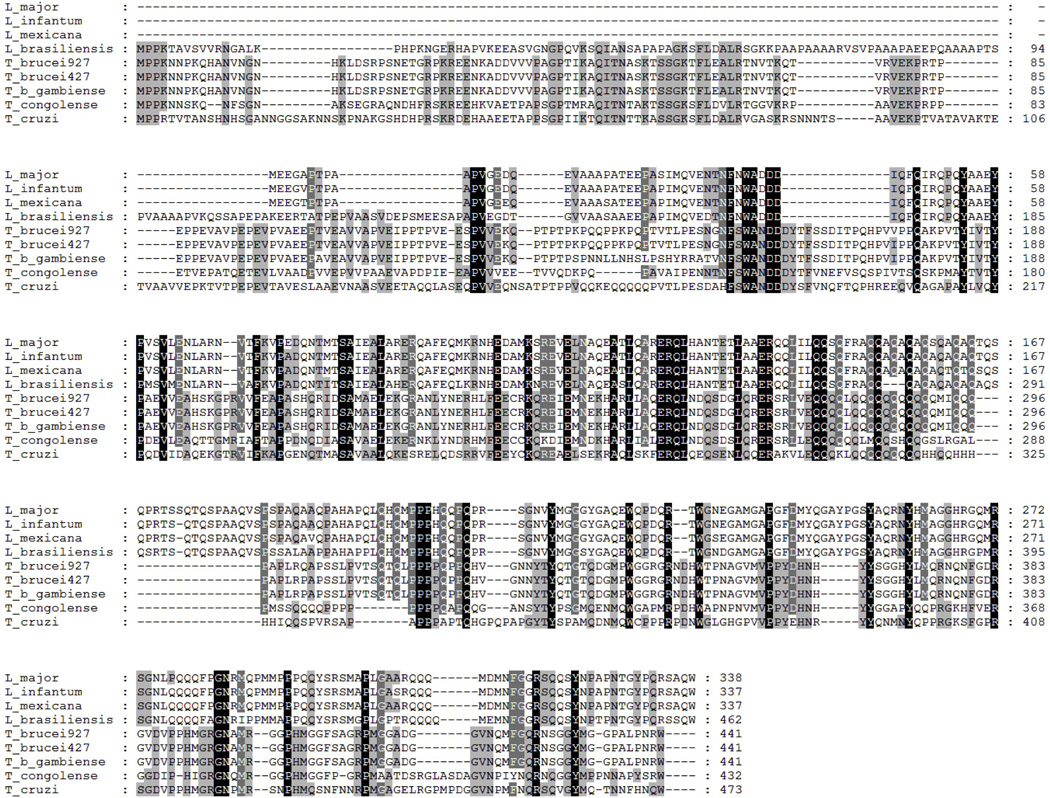
The sequences of Leishmaina major (GenBank ID:68124848), Leishmania infantum (GenBank ID:134067970), Leishmania Mexicana (GeneDB ID: LmxM.11.0820.1), Leishmania brasilienses (GenBank ID: 134060002), Trypanosoma brucei 927 (GenBank ID: 70834104), Trypanosoma brucei 427 (GeneDB ID: Tb427tmp.02.4300), Trypanosoma brucei gambiense (GeneDB ID: Tbg972.11.7260), Trypanosoma congolense (GeneDB ID: TcIL3000.11.7010), and Trypanosoma cruzi (GenBank ID: 708866170) ENF are shown. A black background with white lettering represents complete conservation of an amino acid, a gray background with white lettering indicates the amino acid is conserved in 80% of the sequences shown, and a gray background with black lettering indicates the amino acid is conserved in 60% of the sequences shown. The predicted coiled-coil region containing the leucine zipper is found approximately from residues 113–140 in the Leishmania species, excluding brasiliensis, and from residues 240–295 in the Trypanosoma species and Leishmania brasiliensis. Alignments were generated with ClustalX [50] and visualized with GeneDoc (http://www.nrbsc.org/gfx/genedoc/).
ENF appears unique to trypanosomatids. Through protein-protein BLAST [40], no ENF homologs were detected beyond trypanosomatids. Using PSI-BLAST (Position Specific Iterative-BLAST [41]), the hits above the threshold in the early iterations include myb-like DNA-binding domain-containing protein, several ankyrin repeat containing proteins (domains known to mediate protein interactions [42]), and many non-annotated proteins. The homology to these proteins appears to be based mainly upon the glutamine-rich region and the many prolines present in the ENF sequence. Thus, ENF has no closely related homologs beyond trypanosomatids.
While syntenic ENF homologs are discernable across the trypanosomatids, the homologs in Leishmania infantum, major, and mexicana (GeneDB ID: LinJ.11.0820, GeneDB ID: LmjF.11.0820, and GeneDB ID: LmxM.11.0820.1, respectively) are each approximately 100 amino acids shorter at the amino-terminal region than the other ENF homologs. Furthermore, the L. braziliensis protein is Ala rich in the N-terminus, distinguishing it from the Trypanosoma proteins. Together, these illustrate the diversity found in the amino-termini of ENF and suggest the conserved function of this protein may reside in the carboxyl-terminal domain. However, all ENF homologs share a leucine zipper motif and predicted coiled-coil structure.
3.2 Verification of the TbTFIIB-TbENF Interaction
We sought to confirm the interaction between TbTFIIB and TbENF through GST-pulldown experiments. Purified, recombinant GST-TbENF was incubated with procyclic T. brucei whole cell extract, GST-TbENF was captured through glutathione agarose beads, and any resultant protein interactions challenged through stringent salt washes using up to 0.9 M KCl. Western blot analysis revealed a strong interaction with TbTFIIB, even when challenged with 0.9 M KCl (Figure 3A). Minimal to no TbFIIB was detected in the control reaction with GST alone. To control for the possibility that the apparent interaction between TbENF and TbTFIIB is DNA-dependent, T. brucei whole cell extract was treated with DNase prior to the pulldown experiment and an interaction was still observed (Figure 3B). An interaction between TbENF and TbRNAPII or TbTBP was also examined (Figure 3A). Even at relatively low salt concentrations (0.2 M) interaction with these proteins was not observed. This result suggests that TbENF is not simply a protein involved in many non-specific protein-protein interactions. The fact that TbRNAPII was reported to interact with ENF but is not seen here may reflect interference by the GST-tag with certain protein interactions [39]. Curiously, the crystal structure of TbTFIIB reveals the trypanosome factor has additional structure relative to TFIIB from higher eukaryotes [13]. It is speculated that these extra regions allow for interactions with trypanosomatid-specific factors. Perhaps the TbTFIIB-TbENF interaction is mediated by one of these trypanosome-specific regions of TbTFIIB.
Figure 3. Verification of the TbENF-TbTFIIB interaction using GST-TbENF.
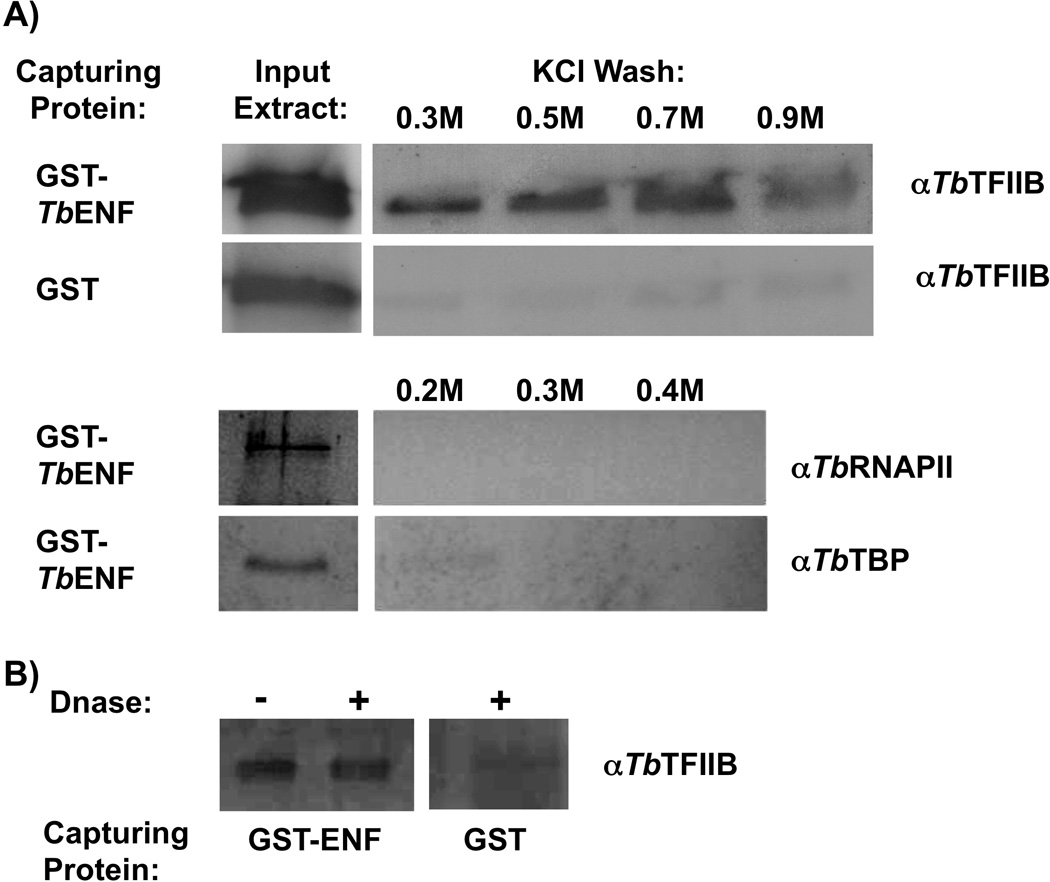
(A) GST-TbENF interacting proteins were captured through incubation with procyclic T. brucei whole cell extract and the interactions were challenged with up to 0.9 M KCl washes (three 5 minute washes at room temperature). Interacting proteins were queried through Western blot. Using antibodies against TbTFIIB, TbRNAPII, and TbTBP, it was found that GST-TbENF interacts tightly with TbTFIIB (top panels), but not with TbRNAPII or TbTBP (bottom panels) under these conditions. (B) As a control, the GST-TbENF TbTFIIB interaction studies were carried out with T. brucei whole cell extract treated with DNase.
3.3 TbENF is a Nuclear Protein
Given the TbTFIIB-TbENF interaction, we anticipated TbENF to be a nuclear protein. Through a sucrose cushion experiment, trypanosomes were fractionated into nuclear and cytoplasmic fractions. Western blot analysis using polyclonal TbENF antibody revealed that TbENF is in the nuclear fraction, along with the nuclear control proteins TbTFIIB and TbTBP (Figure 4). The commercial antibody against human elongation factor 2 (HsEF-2) recognizes T. brucei EF-2 and has previously been used as a cytoplasmic control [43]. EF-2 was only detected in the cytoplasmic fraction. TbENF, TbTBP, and TbTFIIB, were not detected in the cytoplasmic fraction. Thus, as anticipated for a TFIIB-interacting protein, TbENF is a nuclear protein.
Figure 4. TbENF is a nuclear protein.
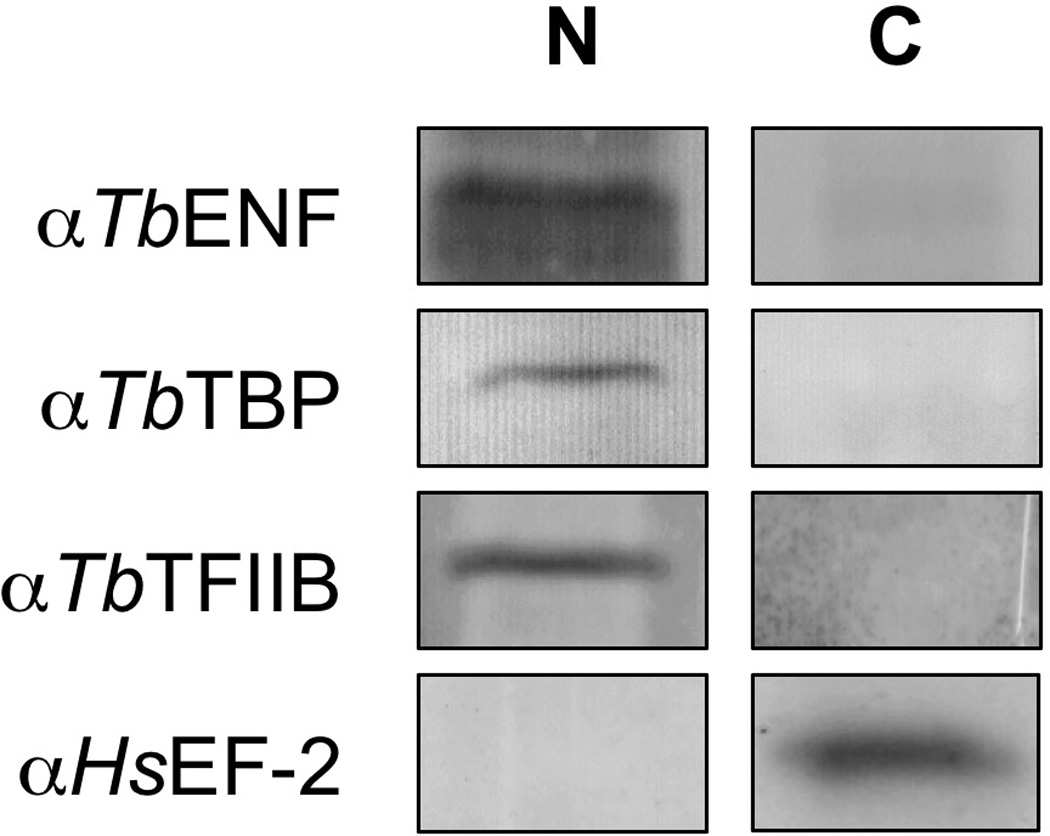
The nuclei (N) and cytoplasm (C) of procyclic T. brucei were fractionated through a 0.8 M sucrose cushion. TbENF was found to fractionate with TbTBP and TbTFIIB. Elongation factor 2 (EF-2) served as the cytoplasmic control, with the protein only observed in the cytoplasmic fraction.
3.4 TbENF is an Essential Protein
We next used RNA interference to begin to examine the role of TbENF in vivo. Trypanofan (http://trypanofan.path.cam.ac.uk/trypanofan/main/) was used to identify a unique region of the Tb11.02.4300 gene to target by RNAi. This region, corresponding to the amino-terminus of the open reading frame, was cloned into p2T7–177 [28]. The construct was then transfected into procyclic T. brucei cell line 29–13 [44], clonal cell lines were established by limiting dilution, and the production of dsRNA induced through tetracycline addition to the media. A difference in the growth was noted between the TbENF RNAi-induced and non-induced cells beginning at approximately day 4 (Figure 5A). Correspondingly, the levels of TbENF were markedly reduced in the RNAi-induced cells by day 4 as determined by Western blot analysis, but remained unaltered in the RNAi non-induced cells (Figure 5B). By day 5 of the RNAi induction, parasite growth stopped, indicating an essential role for TbENF.
While our studies were carried out in the procyclic form of the parasite, a recent high-throughput RNA interference screen in Trypanosoma brucei identified genes with essential functions throughout the lifecycle [45]. This work reveals that TbENF also is essential in the bloodstream forms, as well as for differentiation from the bloodstream form of the parasite to the procyclic form. Together, these findings reveal TbENF is essential for parasite viability throughout the lifecycle.
RNA was isolated from the T. brucei ENF RNAi-induced and non-induced cells on days 3 and 4 of the experiment, as the RNAi-induced cells were still viable but the protein levels were reduced. Semi-quantitative PCR was used to compare transcript levels between the two cell lines. As a positive control, ENF transcript levels were shown to be reduced in the RNAi-induced cell line (Figure 5C). Spliced Leader and U6 transcript levels also were assessed and neither appears altered at this stage of the RNAi experiment. While ENF may not play a role in SL RNA gene transcription, the experiment is not definitive as there is still a small amount of ENF protein present on these days (Figure 5B) as well as ENF transcript (Figure 5C). Furthermore, the cells do not display a growth phenotype on Day 3 and are just beginning to deviate from the RNAi non-induced growth phenotype on Day 4. Transcript levels in Day 5 cells were not examined because many cells were no longer viable.
3.5 TbENF Interacts with DNA
The presence of a conserved leucine zipper in the ENF homologs suggests the protein interacts with DNA. Given the interaction of TbENF with TbTFIIB, we tested the interaction between TbENF and the only known RNAPII-dependent gene promoter in trypanosomes, the SL RNA gene promoter region. Biotinylated DNA corresponding to the promoter region of the SL gene was mixed with nuclear extract and the DNA and interacting proteins captured through streptavidin beads. The interactions were challenged with stringent washes of either high salt (up to 0.9 M KCl) or high concentrations of the non-ionic detergent TWEEN-20 (up to 0.2%). Interaction between TbENF and the SL RNA gene promoter DNA was observed by Western blot (Figure 6). However, we noticed that TbENF also interacted very strongly with all other DNA probes tested; including a region from the trypanosome RNAPIII-dependent U6 promoter, internal regions of the open reading frames of the ENF gene (Tb11.02.4300) and the bla gene, and a region of lambda phage DNA. These interactions all withstood challenges from high salt and detergent. Little to no detectable binding of TbENF to the streptavidin beads was observed. Furthermore, we probed for TbTFIIB binding to the bla DNA probe under the conditions tested for TbENF. We were unable to detect TbTFIIB indicating that under the conditions used not all proteins are binding to DNA non-specifically (bottom panel, Figure 6). Additionally, this result demonstrates that the interaction between DNA and TbENF is not mediated through TbTFIIB.
Figure 6. TbENF interacts with double-stranded DNA.

(A) Biotinylated double-stranded DNA corresponding to the SL RNA gene promoter, U6 gene promoter, the ENF open reading frame (Tb11.02.4300), a region of the bla gene, or a region from lambda phage DNA was incubated with T. brucei nuclear extract. The DNA and interacting proteins were captured through streptavidin beads and the interactions were challenged with either high salt (KCl) or non-ionic detergent (TWEEN 20). An interaction with TbENF was detected through Western blot analysis. The no DNA control consisted of beads only. (B) As a control, the bla DNA pulldown sample was probed for an interaction with TbTFIIB.
To determine whether the tested DNA contains a conserved promoter element, an alignment and phylogenetic tree reveal the DNA probes used are not similar (Supplemental Figure 1). Further, the DNA probes were analyzed for motifs not detectable through an alignment using MEME [46]. No significantly scoring common motif was detected in the DNA sequences used. Thus, TbENF binds double stranded DNA and, under the conditions tested, in a sequence-independent fashion. Other factors are reported to bind DNA in a non-specific fashion, including the Adenovirus DNA binding protein (DBP) which is involved in a myriad of functions including transcription regulation [47]. DBP binds both dsDNA and ssDNA in a sequence-independent fashion. The Arc repressor protein has also been shown to associate with nucleic acids and polyanions in a non-specific manner [48, 49]. This work was carried out in the context of protein folding, but the property is hypothesized to play a role in Arc dimerization and DNA interaction. With regard to TbENF, it is possible that additional factors required for specificity were not present or functional under the in vitro conditions used for this experiment. Nevertheless, this experiment demonstrates that TbENF binds to DNA. The nature and specificity of the interaction is under further study.
4. Conclusions
Through this work, we have been able to identify one of the many non-annotated Trypanosoma brucei proteins, Tb11.02.4300, as a TbTFIIB-interacting protein. The large number of non-annotated proteins in T. brucei represents a wealth of novel science and biochemical processes unique to the parasites. They also may represent a reservoir of potential therapeutic targets; particularly so for TbENF, which is essential in all stages of the Trypanosoma brucei life cycle. The protein is curious at the sequence level, bearing motifs of known transcription factors, yet still unique to the parasite. The interaction of TbENF with known transcription factors and its association with nucleic acid points to a role for TbENF in transcription. Work continues in our laboratory to elucidate the function of this novel, trypanosomatid-specific protein.
Supplementary Material
A) A multiple sequence alignment of the DNA probes used in the TbENF-DNA interaction studies Regions from the T.brucei Spliced Leader RNA promoter (GenBank ID: X00633.1), T.brucei U6 promoter (GenBank ID: X13017.1), Tb11.02.4300 open reading frame (GenBank ID: 71755606), lambda phage DNA (GenBank ID: 9626243), and bla gene (GenBank ID: HQ284188.1) are shown. A black background with white lettering represents complete conservation of a nucleotide, a gray background with lettering indicates the nucleotide is conserved in 80% of the sequences shown, and a gray background with black lettering indicates the nucleotide is conserved in 60% of the sequences shown. B) Phylogenetic tree of the DNA probes sequences shown in A.
Acknowledgments
We thank the NIH (R15/AREA Award 1R15AI078396-01A1 to J.B.P.) and Villanova University (ORSP Summer Research Fellowship) for funding. We also thank Drs. Barry Selinsky and Dennis Wykoff for critical reading of the manuscript.
Role of Funding Sources
This work was funded by the NIH (R15/AREA award 1R15AI078396-01A1 to JBP) and Villanova University (ORSP Summer Research Fellowship). None of these organizations had involvement in study design, data collection, data interpretation, or in preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: RNAP, RNA Polymerase; SL, spliced leader; ENF, Essential Nuclear Factor; TAP, Tandem Affinity Purification; RNAi, RNA interference.
References
- 1.Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, et al. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martinez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol. 2010;2010:525241. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Nardelli SC, Avila AR, Freund A, Motta MC, Manhaes L, de Jesus TC, et al. Small-subunit rRNA processome proteins are translationally regulated during differentiation of Trypanosoma cruzi. Eukaryot Cell. 2007;6:337–345. doi: 10.1128/EC.00279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 2006;34:5312–5324. doi: 10.1093/nar/gkl598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gale M, Jr, Carter V, Parsons M. Translational control mediates the developmental regulation of the Trypanosoma brucei Nrk protein kinase. J Biol Chem. 1994;269:31659–31665. [PubMed] [Google Scholar]
- 7.Stanne TM, Kushwaha M, Wand M, Taylor JE, Rudenko G. TbISWI Regulates Multiple Polymerase I (Pol I)-Transcribed Loci and Is Present at Pol II Transcription Boundaries in Trypanosoma brucei. Eukaryot Cell. 2011;10:964–976. doi: 10.1128/EC.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito RM, Elgort MG, Campbell DA. A conserved upstream element is essential for transcription of the Leishmania tarentolae mini-exon gene. EMBO J. 1994;13:5460–5469. doi: 10.1002/j.1460-2075.1994.tb06881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009;10:152. doi: 10.1186/1471-2164-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palenchar JB, Liu W, Palenchar PM, Bellofatto V. A divergent transcription factor TFIIB in trypanosomes is required for RNA polymerase II-dependent spliced leader RNA transcription and cell viability. Eukaryot Cell. 2006;5:293–300. doi: 10.1128/EC.5.2.293-300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimanski B, Brandenburg J, Nguyen TN, Caimano MJ, Gunzl A. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2006;34:1676–1684. doi: 10.1093/nar/gkl090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibrahim BS, Kanneganti N, Rieckhof GE, Das A, Laurents DV, Palenchar JB, et al. Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei. Proc Natl Acad Sci U S A. 2009;106:13242–13247. doi: 10.1073/pnas.0904309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Jung HS, Gunzl A. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. 2009;37:3811–3820. doi: 10.1093/nar/gkp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecordier L, Devaux S, Uzureau P, Dierick JF, Walgraffe D, Poelvoorde P, et al. Characterization of a TFIIH homologue from Trypanosoma brucei. Mol Microbiol. 2007;64:1164–1181. doi: 10.1111/j.1365-2958.2007.05725.x. [DOI] [PubMed] [Google Scholar]
- 16.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Cai G, Panigrahi AK, Dunham-Ems S, Nguyen TN, Radolf JD, et al. A TFIIH-associated mediator head is a basal factor of small nuclear spliced leader RNA gene transcription in early-diverged trypanosomes. Mol Cell Biol. 2010;30:5502–5513. doi: 10.1128/MCB.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol Cell Biol. 2005;25:7314–7322. doi: 10.1128/MCB.25.16.7314-7322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimanski B, Nguyen TN, Gunzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan JP, Arhin GK, Ullu E, Tschudi C. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol Cell Biol. 2004;24:9610–9618. doi: 10.1128/MCB.24.21.9610-9618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 22.Evers R, Hammer A, Kock J, Jess W, Borst P, Memet S, et al. Trypanosoma brucei contains two RNA polymerase II largest subunit genes with an altered C-terminal domain. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- 23.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 24.McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 25.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 26.Huie JL, He P, Bellofatto V. In vitro transcription of the Leptomonas seymouri SL RNA and U2 snRNA genes using homologous cell extracts. Mol Biochem Parasitol. 1997;90:183–192. doi: 10.1016/s0166-6851(97)00146-1. [DOI] [PubMed] [Google Scholar]
- 27.Laufer G, Schaaf G, Bollgonn S, Gunzl A. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol Cell Biol. 1999;19:5466–5473. doi: 10.1128/mcb.19.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Schimanski B, Laufer G, Gontcharova L, Gunzl A. The Trypanosoma brucei spliced leader RNA and rRNA gene promoters have interchangeable TbSNAP50-binding elements. Nucleic Acids Res. 2004;32:700–709. doi: 10.1093/nar/gkh231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng WG, Zhu Y, Montero A, Wu KK. Quantitative analysis of binding of transcription factor complex to biotinylated DNA probe by a streptavidin-agarose pulldown assay. Anal Biochem. 2003;323:12–18. doi: 10.1016/j.ab.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Bornberg-Bauer E, Rivals E, Vingron M. Computational approaches to identify leucine zippers. Nucleic Acids Res. 1998;26:2740–2746. doi: 10.1093/nar/26.11.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 34.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, Clouston WM, Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai E, Clark KL, Burley SK, Darnell JE., Jr Hepatocyte nuclear factor 3/fork head or "winged helix" proteins: a family of transcription factors of diverse biologic function. Proc Natl Acad Sci U S A. 1993;90:10421–10423. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 39.Das A, Li H, Liu T, Bellofatto V. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol Biochem Parasitol. 2006;150:201–210. doi: 10.1016/j.molbiopara.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utter CJ, Garcia SA, Milone J, Bellofatto V. Poly(A)-Specific Ribonuclease (PARN-1) Function in Stage-Specific mRNA Turnover in Trypanosoma brucei. Eukaryot Cell. 2011;10:1230–1240. doi: 10.1128/EC.05097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirtz E, Hoek M, Cross GA. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 1998;26:4626–4634. doi: 10.1093/nar/26.20.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alsford S, Turner D, Obado S, Sanchez-Flores A, Glover L, Berriman M, et al. High throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011 doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 47.Chang LS, Shenk T. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–2109. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rentzeperis D, Jonsson T, Sauer RT. Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat Struct Biol. 1999;6:569–573. doi: 10.1038/9353. [DOI] [PubMed] [Google Scholar]
- 49.Marcovitz A, Levy Y. Arc-repressor dimerization on DNA: folding rate enhancement by colocalization. Biophys J. 2009;96:4212–4220. doi: 10.1016/j.bpj.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) A multiple sequence alignment of the DNA probes used in the TbENF-DNA interaction studies Regions from the T.brucei Spliced Leader RNA promoter (GenBank ID: X00633.1), T.brucei U6 promoter (GenBank ID: X13017.1), Tb11.02.4300 open reading frame (GenBank ID: 71755606), lambda phage DNA (GenBank ID: 9626243), and bla gene (GenBank ID: HQ284188.1) are shown. A black background with white lettering represents complete conservation of a nucleotide, a gray background with lettering indicates the nucleotide is conserved in 80% of the sequences shown, and a gray background with black lettering indicates the nucleotide is conserved in 60% of the sequences shown. B) Phylogenetic tree of the DNA probes sequences shown in A.


