Abstract
Objective
Several confirmed genetic susceptibility loci for lupus have been described. To date, no clear evidence for genetic epistasis is established in lupus. We test for gene-gene interactions in a number of known lupus susceptibility loci.
Methods
Eighteen SNPs tagging independent and confirmed lupus susceptibility loci were genotyped in a set of 4,248 lupus patients and 3,818 normal healthy controls of European descent. Epistasis was tested using a 2-step approach utilizing both parametric and non-parametric methods. The false discovery rate (FDR) method was used to correct for multiple testing.
Results
We detected and confirmed gene-gene interactions between the HLA region and CTLA4, IRF5, and ITGAM, and between PDCD1 and IL21 in lupus patients. The most significant interaction detected by parametric analysis was between rs3131379 in the HLA region and rs231775 in CTLA4 (Interaction odds ratio=1.19, z-score= 3.95, P= 7.8×10−5 (FDR≤0.05), PMDR= 5.9×10−45). Importantly, our data suggest that in lupus patients the presence of the HLA lupus-risk alleles in rs1270942 and rs3131379 increases the odds of also carrying the lupus-risk allele in IRF5 (rs2070197) by 17% and 16%, respectively (P= 0.0028 and 0.0047).
Conclusion
We provide evidence for gene-gene epistasis in systemic lupus erythematosus. These findings support a role for genetic interaction contributing to the complexity of lupus heritability.
Introduction
Recent candidate gene and genome-wide association studies (GWAS) led to the discovery and validation of multiple susceptibility loci for systemic lupus erythematosus (1). However, the heritability of lupus cannot be completely explained by the susceptibility loci already discovered. We suggest that the missing heritability in lupus can be explained by three potential mechanisms: A heritable epigenetic component, common and rare disease susceptibility variants yet to be discovered, and gene-gene interactions involving known and perhaps yet to be discovered genetic variants for disease susceptibility. There are very limited and controversial data regarding gene-gene interaction (epistasis) in lupus (2–3). Consequently, it is widely accepted that the known lupus susceptibility loci operate additively rather than epistatically to increase the risk for lupus.
Herein, we sought to examine gene-gene interactions in some of the previously established and confirmed susceptibility loci for lupus, using a large set of lupus patients and controls. We discovered and confirmed 6 novel gene-gene interactions for lupus, using both parametric and non-parametric statistical methodologies.
Methods
Study participants and genotyping
A total of 4,248 lupus patients and 3,818 normal healthy controls of European descent were included in this study. Eighteen SNPs representing previously confirmed and independent autosomal lupus susceptibility loci were genotyped (Table 1). A summary for the allelic association results in these loci using the patients and controls included in this study is shown in Supplementary Table 1. We genotyped 2 tag SNPs in the HLA region. These 2 SNPs were selected as they were recently shown to have independent genetic effects using logistic regression analysis of a large number of lupus-associated SNPs in the HLA region (4). Likewise, 3 tag SNPs representing independent genetic susceptibility effects in IRF5 were genotyped (5). All lupus patients fulfilled the ACR lupus classification criteria (6–7). Genotyping was performed using Illumina Custom Bead system on the iSCAN instrument as part of a large lupus candidate gene association study to reduce cost of genotyping and maximize sample size. We genotyped 347 ancestry informative markers (AIMs) in our samples (8–11). Individuals with a genotype success rate of <90% (361 samples) were excluded from the analysis. The remaining samples were then evaluated for duplicates or related individuals and one individual from each pair was removed (117 samples) if the proportion of alleles shared identical by descent (IBD) > 0.4. Samples were assessed for mismatches between their reported gender and their genetic data and 112 samples were removed from the analysis as they did not meet the following criteria: an assigned male was required to have chromosomal X heterozygosity ≤10% and be heterozygous at rs2557524 and an assigned female was required to have chromosomal X heterozygosity >10% and be homozygous at rs2557524. The SNP rs2557524 is mapped on a region on chromosome X and Y that is identical except for this 1 base. Because of this 1 base difference males generate a heterozygous genotype (due to the presence of both X and Y chromosomes) and females generate a homozygous genotype (due to the presence of only X chromosomes).
Table 1.
Previously reported lupus susceptibility loci analyzed for gene-gene interaction in this study.
| Gene/region | Chromosome | Associated SNP | Risk allele | Odds ratio* | Reference |
|---|---|---|---|---|---|
| BANK1 | 4q24 | rs10516487 | G | 1.38 | (26) |
| C8orf13-BLK | 8p22–23 | rs13277113 | A | 1.39 | (27) |
| CTLA4 | 2q33 | rs231775 | G | 1.23 | (28) |
| FCGR2A | 1q23 | rs1801274 | C | 1.35 | (29) |
| HLA region1 | 6p21.33 | rs3131379 | A | 2.36 | (30) |
| HLA region2 | 6p21.32 | rs1270942 | G | 2.35 | (30) |
| IL-21 | 4q26 | rs907715 | G | 1.29 | (31) |
| IRF5 | 7q32 | rs2070197 | C | 1.85** | (5) |
| IRF5 | 7q32 | rs729302 | A | 1.39** | (5) |
| IRF5 | 7q32 | rs10954213 | A | 1.25** | (5) |
| ITGAM | 16p11.2 | rs1143679 | A | 1.78 | (32) |
| KIAA1542 | 11p15.5 | rs4963128 | C | 1.28 | (30) |
| MBL | 10q11 | rs1800450 | A | 1.41 | (33) |
| PDCD1 | 2q37.3 | rs11568821 | A | 2.85 | (34) |
| PTPN22 | 1p13 | rs2476601 | A | 1.53 | (35) |
| PXK | 3p14.3 | rs6445975 | C | 1.25 | (30) |
| STAT4 | 2q32.2 | rs7574865 | T | 1.55 | (36) |
| TNFSF4 | 1q25 | rs2205960 | T | 1.28 | (37) |
SLE patients versus healthy controls as reported in previous studies referenced above
Transmitted/untransmitted ratio reported as this was based on trio and family studies (5).
Next, samples with increased heterozygosity (>5 standard deviation around the mean) were removed from the analysis (5 samples). Finally, 42 genetic outliers were removed from further analysis as determined by principal components analysis. An additional 2 outlier samples identified by admixture proportions calculated using ADMIXMAP were also removed. After applying the quality control measures detailed above, samples included in our analysis consisted of 3,936 European-derived lupus patients (3,592 females, 344 males), and 3,491 European-derived normal healthy controls (2,340 females, 1,151 males).
Detection of gene-gene interaction
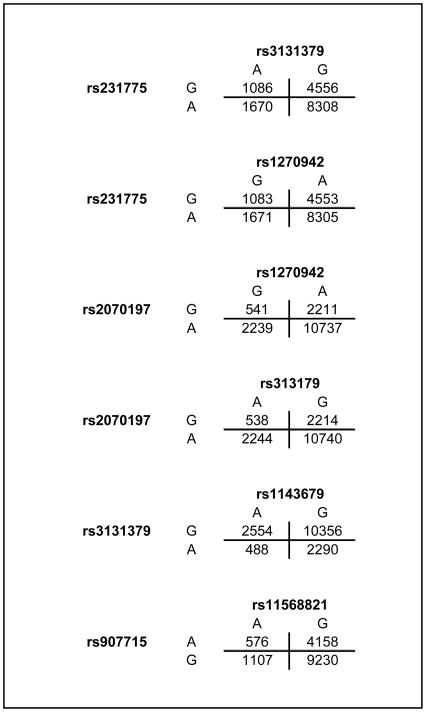
Testing for gene-gene interaction was performed sequentially using two independent statistical approaches. First, a parametric analysis for epistasis was applied as implemented in PLINK (12). Epistatic interactions detected using PLINK were validated using allelic 2×2 tables among lupus patients to calculate interaction odds ratios and identify the specific alleles in each SNP pair that contributed to the interaction detected. Allelic 2×2 tables (Figure 1) were obtained from 3×3 genotypic tables (Supplementary Figure 1) for each interaction tested. The allelic 2×2 tables are based on 4N allele counts, where N is the total number of individuals, with each individual contributing a total of 4 independent alleles. Z-scores were calculated as the natural logarithm of the odds ratio divided by the square root of the variance, and associated P values were assigned from the z-scores for each interaction. Chi-square statistics for pair-wise interaction were calculated as were chi-square derived P values. Second, a pair-wise non-parametric epistasis test was applied utilizing multifactor dimensionality reduction analysis (MDR) (13–14).
Figure 1.
Allelic 2×2 tables in lupus patients used to calculate interaction odds ratios and identify the specific alleles in each SNP pair that contributed to the interaction detected.
The false discover rate (FDR) method as described by Benjamini and Hochberg was used to correct for multiple comparisons (15–16).
Results
To test for gene-gene interactions within the known lupus susceptibility loci examined, we performed a 2-step epistasis analysis using a parametric approach followed by a non-parametric analysis. This 2-step approach has the strength of examining and confirming epistatic interactions using 2 independent statistical methods. This is necessary as the best methodology to detect gene-gene interaction remains controversial.
We first used a case-only pair-wise epistasis analysis implemented in PLINK. The case-only analysis was selected as it was shown to be a more powerful test for epistasis compared to case-control analysis (17–18). Interactions with FDR of ≤ 0.05 were considered established, and those with FDR >0.05 and ≤ 0.25 were considered suggestive interactions that require confirmation. A high FDR was used in the initial screening for suggestive interactions to avoid excluding true gene-gene interactions from confirmatory analyses.
We discovered six gene-gene interactions using parametric analysis (Table 2). The two most significant interactions were between CTLA4 and the two SNPs representing two independent genetic effects within the HLA region (FDR≤ 0.05). The detected epistasis signal between the risk alleles in CTLA4 and rs3131379 (HLA region 1) and CTLA4 and rs1270942 (HLA region 2) showed an interaction odds ratio of 1.19 and 1.18 (z-score= 3.95, P= 7.8×10−5, and z-score= 3.88, P= 1.0×10−4, respectively). These data indicate that in lupus patients, the presence of the lupus-risk allele in CTLA4 increases the odds of carrying the risk allele in either of the HLA lupus associated loci by ~20% and vice versa (Figure 1). Four additional suggestive gene-gene interactions (FDR≤ 0.25) were found between the HLA and IRF5, the HLA and ITGAM, and IL21 and PDCD1 (Table 2). The presence of the risk allele in the two HLA lupus-associated loci examined (rs1270942 and rs3131379) increases the odds of carrying the lupus-risk allele in IRF5 (rs2070197) by 17% and 16%, respectively, and vice versa (P= 0.0028 and 0.0047). Interestingly, our data suggest that the presence of the risk allele in ITGAM increases the odds of carrying the protective allele in rs3131379 (HLA) by 16% (P= 0.0075).
Table 2.
Gene-gene interaction results in 18 known independent lupus susceptibility loci, using logistic regression analysis implemented in PLINK. Only interactions with FDR ≤0.25 are shown.
| Gene | Polymorphism | Risk Allele | Interacting Alleles | Interaction Odds Ratio | Z-score | Z-score P | Chi2 | Chi2P |
|---|---|---|---|---|---|---|---|---|
| CTLA4 | rs231775 (A/G) | G | ||||||
| HLA | rs3131379 (A/G) | A | GXA | 1.19 | 3.95 | 7.8×10−5 | 15.19 | 9.7×10−5 |
|
| ||||||||
| CTLA4 | rs231775 (A/G) | G | ||||||
| HLA | rs1270942 (A/G) | G | GXG | 1.18 | 3.88 | 1.0×10−4 | 14.87 | 1.0×10−4 |
|
| ||||||||
| IRF5 | rs2070197 (A/G) | G | ||||||
| HLA | rs1270942 (A/G) | G | GXG | 1.17 | 2.99 | 0.0028 | 8.93 | 0.0028 |
|
| ||||||||
| IRF5 | rs2070197 (A/G) | G | ||||||
| HLA | rs3131379 (A/G) | A | GXA | 1.16 | 2.83 | 0.0047 | 7.98 | 0.0047 |
|
| ||||||||
| HLA | rs3131379 (A/G) | A | ||||||
| ITGAM | rs1143679 (A/G) | A | GXA | 1.16 | 2.67 | 0.0075 | 6.93 | 0.0085 |
|
| ||||||||
| IL21 | rs907715 (A/G) | G | ||||||
| PDCD1 | rs11568821 (A/G) | A | AXA | 1.16 | 2.64 | 0.0084 | 6.80 | 0.0091 |
Z-scores were calculated as the natural logarithm of the odds ratio divided by the square root of the variance
Next, and in order to confirm the two gene-gene interactions that we established using parametric tests, and to test if the other 4 suggestive gene-gene interactions can be established, we applied the multifactor dimensionality reduction test (MDR) to the interactions initially discovered using parametric analysis. MDR is a non-parametric test for non-linear epistasis. A pair-wise MDR analysis was applied to test the specific interactions discovered in stage 1. It should be noted, however, that results obtained using the MDR non-parametric analysis reflect a joint effect consisting of the main genetic association effect in the loci examined and the interaction effect. These results are presented in Table 3 (See Supplementary Table 2 and Supplementary Figure 2 for details).
Table 3.
Multifactor dimensionality reduction (MDR) analysis for pair-wise interactions detected using parametric analysis in lupus patients and controls.
| Interaction | Cross Validation Consistency | Balanced Accuracy | Chi2 | P Value |
|---|---|---|---|---|
| CTLA4 (rs231775) × HLA (rs3131379) | 10/10 | 0.5737 | 208.57 | 5.9×10-45 |
| CTLA4 (rs231775) × HLA (rs1270942) | 10/10 | 0.5744 | 212.76 | 7.4×10-46 |
| HLA (rs1270942) × IRF5 (rs2070197) | 10/10 | 0.5949 | 270.60 | 2.3×10-58 |
| HLA (rs3131379) × IRF5 (rs2070197) | 10/10 | 0.5946 | 268.81 | 5.6×10-58 |
| HLA (rs3131379) × ITGAM (rs1143679) | 10/10 | 0.5985 | 287.71 | 4.6×10-62 |
| PDCD1 (rs11568821) × IL21 (rs907715) | 10/10 | 0.5235 | 17.44 | 5.7×10-4 |
Df of 3 was used to calculate P values
Cross validation consistency reflects the number of times MDR found the same model as it divided up the data into different segments. Balanced accuracy is defined as (sensitivity+specificity)/2 where sensitivity = true positives/(true positives +false negatives) and specificity = true negatives/(false positives+true negatives). This gives an accuracy estimate that is not biased by the larger class (38).
Discussion
The very existence and nature of genetic epistasis in lupus has been elusive. We combined the strengths of two independent approaches to test for genetic epistasis in lupus, and identified several novel gene-gene interactions using a large European-derived sample. The most significant interaction we identified was between the HLA region and CTLA4. Indeed, two independent lupus-associated SNPs within the HLA region (rs3131379 and rs1270942) showed evidence for significant interaction with rs231775 in CTLA4 (Tables 2 and 3). The HLA-CTLA4 interaction in lupus underscores antigen presentation and T cell stimulation as an important process involved in the pathogenesis of lupus. This interaction is biologically logical as CTLA4 is upregulated on T cells following T cell activation by antigen presenting cells (19). Following T cell activation via the binding of MHC:antigen complex to the T cell receptor (signal 1), the binding of CD80/CD86 on antigen presenting cells to CD28 on the surface of T cells (signal 2) ensures T cell activation and IL-2 production (19). CTLA4 competes with CD28 to bind CD80/CD86 and provides a negative signal that suppresses T cell activation. This process is thought to be important to control T cell activation and prevent autoimmunity.
A role for antigen presenting cells in lupus is highlighted again with the HLA/ITGAM gene-gene interaction, though this interaction is between the risk and protective alleles in these 2 loci. ITGAM (integrin, alpha M) encodes for CD11b, the alpha chain in the integrin molecule CD11b/CD18 (MAC-1, CR3). It is expressed on the surface of antigen presenting cells and neutrophils, and plays a role in cell-cell adhesions, leukocyte extravasation, and in complement-mediated phagocytosis of C3bi opsonized antigens (20–21).
We also showed evidence for gene-gene interaction between the 2 independent lupus-associated SNPs within the HLA region with rs2070197 in IRF5. This interaction emphasizes the role of the interferon pathway in the pathogenesis of lupus.
The other gene-gene interaction we identified was between rs907715 in IL21 and rs11568821 in PDCD1. This interaction is very interesting as it highlights a role for follicular helper T cells (TFH) in lupus. High PDCD1 expression and IL-21 production is a hallmark of TFH cells (22). TFH cells promote germinal center formation, plasma cell differentiation, and antibody isotype switching (23). PDCD1 deficiency results in impaired germinal center B cell survival and diminished production of long-lived plasma cells (24). Indeed, the production of IL-21 is reduced in TFH cells from Pdcd1−/− mice (24). IL-21 deficiency results in impaired germinal center formation, plasma cell differentiation, and isotype class switching (23), emphasizing a central role for IL-21 in TFH function. Of interest, a higher fraction of circulating TFH cells was detected in the peripheral blood from patients with lupus compared to normal controls (25).
In summary, we provided strong evidence that the presence of one risk allele can influence the presence or absence of other risk alleles in lupus patients across different loci. We have identified novel gene-gene epistatic interactions in lupus. Gene-gene interactions might help explain at least part of the “missing heritability” in complex diseases. Our findings argue against a simple “additive” genetic model in autoimmunity, and highlight antigen presentation and T cells activation, the interferon pathway, and follicular helper T cells as important contributors to the pathogenesis of lupus.
Supplementary Material
Supplementary Figure 1: Genotypic 3×3 tables in lupus patients used to generate the allelic 2×2 tables presented in Figure 1.
Supplementary Figure 2: The optimal two-locus models as detremined by MDR for CTLA4/HLA region 1 (A), CTLA4/HLA region 2 (B), IRF5/HLA region 2 (C), IRF5/HLA region 1 (D), HLA region 1/ITGAM (E), PDCD1/IL21 (F). (0= no risk alleles, 1= 1 risk allele, 2= 2 risk alleles, 9= undetermined). The numbers within each small square represent number of cases (left) and controls (right). For each square, dark-shading indicates high risk of disease, wheras light shading represents low risk of disease.
Supplementary Table 1: Genetic association analysis for each locus included in the gene-gene interaction analysis using cases and controls included in this study.
Supplementary Table 2: Detailed cross-validation and whole dataset statistics derived from MDR analysis. MDR analysis was performed for the gene-gene interactions discovered using parametric analysis.
Acknowledgments
This work was made possible by the NIH R03AI076729, P20RR020143, P20RR015577, P30AR053483, R01AR042460, R37AI024717, R01AI031584, N01AR62277, P50AR048940, P01AI083194, RC1AR058554, U19AI082714, HHSN266200500026C, P30RR031152, P01AR049084, R01AR043274, R01AI063274, K24AR002138, P602AR30692, UL1RR025741, R01DE018209, R01AR043727, UL1RR025005, R01AR043814, K08AI083790, P30DK42086, L30AI071651, UL1RR024999, R01AR044804, M01RR000079, R21AI070304, Arthritis National Research Foundation, American College of Rheumatology/Research and Education Foundation, Lupus Research Institute, Kirkland Scholar awards, Alliance for Lupus Research, US Department of Veterans Affairs, US Department of Defense PR094002, European Science Foundation to the BIOLUPUS network (07-RNP-083), the Swedish Research Council, and the Instituto de Salud Carlos III ( PS09/00129) Cofinanced partly through FEDER funds of the European Union and the grant PI0012 from the Consejería de Salud de Andalucía.
We are thankful to Dr. Peter Gregersen for providing DNA control samples for our study.
The BIOLUPUS network is composed of: Johan Frostegård, MD, PhD (Huddinge, Sweden), Lennart Truedsson, MD, PhD (Lund, Sweden), Enrique de Ramón, MD PhD (Málaga, Spain), José M. Sabio, MD, PhD (Granada, Spain), María F. González-Escribano, PhD (Sevilla, Spain), Norberto Ortego-Centeno (Granada, Spain), José Luis CAllejas MD (Granada, Spain), Julio Sánchez-Román, MD (Sevilla, Spain), Sandra D’Alfonso, PhD (Novara, Italy), Sergio Migliarese MD (Napoli, Italy), Gian-Domenico Sebastiani MD (Rome, Italy), Mauro Galeazzi MD (Siena, Italy), Torsten Witte, MD, PhD (Hannover, Germany), Bernard R. Lauwerys, MD, PhD (Louvain, Belgium), Emoke Endreffy, PhD (Szeged, Hungary), László Kovács, MD, PhD (Szeged, Hungary), Carlos Vasconcelos, MD, PhD (Porto, Portugal), Berta Martins da Silva, PhD (Porto, Portugal).
Footnotes
Financial conflict of interest: None of the authors has any financial conflict of interest to report.
References
- 1.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6(3):461–79. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellquist A, Jarvinen TM, Koskenmies S, Zucchelli M, Orsmark-Pietras C, Berglind L, et al. Evidence for genetic association and interaction between the TYK2 and IRF5 genes in systemic lupus erythematosus. J Rheumatol. 2009;36(8):1631–8. doi: 10.3899/jrheum.081160. [DOI] [PubMed] [Google Scholar]
- 3.Suarez-Gestal M, Calaza M, Gonzalez A. Lack of interaction between systemic lupus erythematosus-associated polymorphisms in TYK2 and IRF5. J Rheumatol. 2010;37(3):676–7. doi: 10.3899/jrheum.090823. author reply 8. [DOI] [PubMed] [Google Scholar]
- 4.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104(16):6758–63. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 7.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 8.Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118(3–4):382–92. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 9.Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80(6):1014–23. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez E, Webb RD, Rasmussen A, Kelly JA, Riba L, Kaufman KM, et al. Genetically determined amerindian ancestry correlates with increased frequency of risk alleles for systemic lupus erythematosus. Arthritis Rheum. 2010;62(12):3722–9. doi: 10.1002/art.27753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-dimensionality reduction revealshigh-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JH. Detecting, characterizing, and interpreting nonlinear gene-gene interactions using multifactor dimensionality reduction. Adv Genet. 2010;72:101–16. doi: 10.1016/B978-0-12-380862-2.00005-9. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 16.Lage-Castellanos A, Martinez-Montes E, Hernandez-Cabrera JA, Galan L. False discovery rate and permutation test: an evaluation in ERP data analysis. Stat Med. 2010;29(1):63–74. doi: 10.1002/sim.3784. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10(2):167–70. [PubMed] [Google Scholar]
- 18.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janeway CA, Travers P, Walport M, Shlomchik MJ. T cell-mediated immunity. In: Janeway CA, Travers P, Walport M, Shlomchik MJ, editors. Immunobiology:The immune system in health and disease. 6. New York: Garland Sciences; 2005. pp. 319–65. [Google Scholar]
- 20.Li Y, Zhang L. The fourth blade within the beta-propeller is involved specifically in C3bi recognition by integrin alpha M beta 2. J Biol Chem. 2003;278(36):34395–402. doi: 10.1074/jbc.M304190200. [DOI] [PubMed] [Google Scholar]
- 21.Fagerholm SC, Varis M, Stefanidakis M, Hilden TJ, Gahmberg CG. alpha-Chain phosphorylation of the human leukocyte CD11b/CD18 (Mac-1) integrin is pivotal for integrin activation to bind ICAMs and leukocyte extravasation. Blood. 2006;108(10):3379–86. doi: 10.1182/blood-2006-03-013557. [DOI] [PubMed] [Google Scholar]
- 22.DiPlacido LD, Craft J. Emerging from the shadows: follicular helper T cells in autoimmunity. Arthritis Rheum. 2010;62(1):6–8. doi: 10.1002/art.25045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHeyzer-Williams LJ, Pelletier N, Mark L, Fazilleau N, McHeyzer-Williams MG. Follicular helper T cells as cognate regulators of B cell immunity. Curr Opin Immunol. 2009;21(3):266–73. doi: 10.1016/j.coi.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11(6):535–42. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62(1):234–44. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- 26.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nature genetics. 2008;40(2):211–6. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 27.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. The New England journal of medicine. 2008;358(9):900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 28.Barreto M, Santos E, Ferreira R, Fesel C, Fontes MF, Pereira C, et al. Evidence for CTLA4 as a susceptibility gene for systemic lupus erythematosus. Eur J Hum Genet. 2004;12(8):620–6. doi: 10.1038/sj.ejhg.5201214. [DOI] [PubMed] [Google Scholar]
- 29.Magnusson V, Johanneson B, Lima G, Odeberg J, Alarcon-Segovia D, Alarcon-Riquelme ME. Both risk alleles for FcgammaRIIA and FcgammaRIIIA are susceptibility factors for SLE: a unifying hypothesis. Genes and immunity. 2004;5(2):130–7. doi: 10.1038/sj.gene.6364052. [DOI] [PubMed] [Google Scholar]
- 30.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature genetics. 2008;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, et al. Genetic association of IL-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis. 2007 doi: 10.1136/ard.2007.075424. [DOI] [PubMed] [Google Scholar]
- 32.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM)is associated with systemic lupus erythematosus. Nature genetics. 2008;40(2):152–4. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 33.Lee YH, Witte T, Momot T, Schmidt RE, Kaufman KM, Harley JB, et al. The mannose-binding lectin gene polymorphisms and systemic lupus erythematosus: two case-control studies and a meta-analysis. Arthritis and rheumatism. 2005;52(12):3966–74. doi: 10.1002/art.21484. [DOI] [PubMed] [Google Scholar]
- 34.Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature genetics. 2002;32(4):666–9. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 35.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases--a meta-analysis. Rheumatology (Oxford, England) 2007;46(1):49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 36.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. The New England journal of medicine. 2007;357(10):977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nature genetics. 2008;40(1):83–9. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007;31(4):306–15. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Genotypic 3×3 tables in lupus patients used to generate the allelic 2×2 tables presented in Figure 1.
Supplementary Figure 2: The optimal two-locus models as detremined by MDR for CTLA4/HLA region 1 (A), CTLA4/HLA region 2 (B), IRF5/HLA region 2 (C), IRF5/HLA region 1 (D), HLA region 1/ITGAM (E), PDCD1/IL21 (F). (0= no risk alleles, 1= 1 risk allele, 2= 2 risk alleles, 9= undetermined). The numbers within each small square represent number of cases (left) and controls (right). For each square, dark-shading indicates high risk of disease, wheras light shading represents low risk of disease.
Supplementary Table 1: Genetic association analysis for each locus included in the gene-gene interaction analysis using cases and controls included in this study.
Supplementary Table 2: Detailed cross-validation and whole dataset statistics derived from MDR analysis. MDR analysis was performed for the gene-gene interactions discovered using parametric analysis.