Abstract
Background
Serum prostate specific antigen (PSA) concentrations lack the specificity to differentiate prostate cancer from benign prostate hyperplasia (BPH), resulting in unnecessary biopsies. We identified 5 autoantibody signatures to specific cancer targets which might be able to differentiate prostate cancer from BPH in patients with increased serum PSA.
Methods
To identify autoantibody signatures as biomarkers, a native antigen reverse capture microarray platform was used. Briefly, well-characterized monoclonal antibodies were arrayed onto nanoparticle slides to capture native antigens from prostate cancer cells. Prostate cancer patient serum samples (n=41) and BPH patient samples (collected starting at the time of initial diagnosis) with a mean follow-up of 6.56 y without the diagnosis of cancer (n=39) were obtained. One hundred micrograms of IgGs were purified and labeled with a Cy3 dye and incubated on the arrays. The arrays were scanned for fluorescence and the intensity was quantified. Receiver operating characteristic curves were produced and the area under the curve (AUC) was determined.
Results
Using our microarray platform, we identified autoantibody signatures capable of distinguishing between prostate cancer and BPH. The top 5 autoantibody signatures were TARDBP, TLN1, PARK7, LEDGF/PSIP1, and CALD1. Combining these signatures resulted in an AUC of 0.95 (sensitivity of 95% at 80% specificity) compared to AUC of 0.5 for serum concentration PSA (sensitivity of 12.2% at 80% specificity).
Conclusion
Our preliminary results showed that we were able to identify specific autoantibody signatures that can differentiate prostate cancer from BPH, and may result in the reduction of unnecessary biopsies in patients with increased serum PSA.
Keywords: autoantibodies, biomarkers, immunomics, prostate cancer, prostate specific antigen, protein microarray
Introduction
The American Cancer Society estimates that in 2011, 240,890 men will be diagnosed with prostate cancer (1). Serum prostate specific antigen (PSA) as a biomarker has long been the gold standard for detecting prostate cancer in males. In a normal healthy male, serum PSA concentrations are usually well below 4ng/ml while men with prostate cancer generally present with higher concentrations of PSA (2). Serum PSA concentrations, however, can fluctuate in a patient depending on his diet and exercise and PSA concentrations can be increased in patients with other prostatic diseases such as benign prostate hyperplasia (BPH), which often lead to unnecessary biopsies (2). Therefore, there is a need to develop better biomarkers which can specifically differentiate prostate cancer patients from patients with BPH (2,3).
Diagnosing cancers based on serum profiling is a particularly attractive concept. One potential strategy to identify cancer biomarkers is to take advantage of the body’s own immune system. Cancer sera contain antibodies that react with a unique group of autologous cellular antigens called tumor-associated antigens (TAAs). Proteins not present in normal cells may elicit a host immune response, which affords a dramatic amplification of signal in the form of antibodies relative to the amount of the corresponding antigen (4,5). In addition, it has been postulated that the production of autoantibodies to TAAs constitutes an integral component of the anti-tumor immune response in cancer patients. And since the immune response is generated locally, even small amounts of antigens, so few that they might be undetectable by any other means, may be identified and amplified by the immune response, especially during early stages of cancer formation (4,5). Therefore, autoantibody profiling may be a useful approach for identifying cancer biomarkers.
In order to identify autoantibody signatures that might differentiate between prostate cancer and BPH, a customized version of our previously reported reverse capture protein microarray technology (6-8) was developed. From experiments using an initial set of over 500 cancer related antigens, as well as from literature searches and the Cancer Immunome Database (www2.licr.org/CancerImmunomeDB), a customized array containing 27 unique antigens was tested. Specifically, 27 well characterized monoclonal antibodies to targeted cancer antigens were arrayed onto a nanoparticle slide. These antibodies capture the corresponding antigens that are found in prostate cancer cell lysates. The immobilized antigens would then act as bait to capture the appropriate autoantibodies from a patient. Through comparative analysis of the autoantibody signatures from prostate cancer patients and signatures from patients with BPH, autoantibody signatures capable of differentiating prostate cancer from BPH were identified. Combining the top 5 of the 27 autoantibody signatures resulted in greater sensitivity and specificity compared to serum concentration PSA when differentiating between prostate cancer and BPH in our cohort. The development of these signatures into a diagnostic tool may eliminate unnecessary biopsies for those patients with increased concentrations of PSA who have a benign prostatic disease and not prostate cancer.
Materials and Methods
Patient samples
Serum samples were collected from patients with BPH and patients with prostate cancer prior to any treatments and according to an IRB approved protocol. After obtaining signed patient consent, 5 ml samples were collected in Serum Separator Tubes (Sherwood Medical, St. Louis, MO) and processed within 8 h. Samples were centrifuged at 1500 rpm for 10 min, the serum layer collected, and stored at 50 ul aliquots at −80°C until use. Samples were chosen so that the overall serum PSA concentrations were similar for both the BPH and cancer patients. After identifying the serum samples, 39 BPH patients with a mean follow-up of 6.56 y with no evidence of cancer and 41 prostate cancer patients were used for autoantibody profiling. Although these patients were followed, we used sera collected at the time of diagnosis. Tables 1 and 2 list the clinical characteristics of the patients.
Table 1.
Prostate Cancer Patient Clinical Characteristics.
Listed in the table are the patient ID number, age, PSA level, Gleason score, and follow-up time for each patient sample. The mean value for each characteristic is displayed at the bottom of the table. 41 prostate cancer samples were used.
| Sample | Age (years) | PSA | Gleason Score | Follow Up (years) |
|---|---|---|---|---|
| PC19 | 61 | 4.2 | 3+3 = 6 | 7 |
| PC20 | 58 | 4.7 | 3+3 = 6 | 7 |
| PC21 | 58 | 4.2 | 3+4 = 7 | 7 |
| PC32 | 57 | 5 | 3+3 = 6 | 7 |
| PC33 | 52 | 3.8 | 3+4 = 7 | 7 |
| PC34 | 56 | 4.3 | 3+3 = 6 | 7 |
| PC42 | 57 | 4.7 | 3+3 = 6 | 7 |
| PC48 | 56 | 2.4 | 3+3 = 6 | 7 |
| PC50 | 56 | 5.5 | 3+3 = 6 | 7 |
| PC54 | 53 | 2.8 | 3+3 = 6 | 7 |
| PC59 | 49 | 4.2 | 4+3 = 7 | 7 |
| PC63 | 56 | 4.8 | 4+3 = 7 | 7 |
| PC75 | 58 | 4.3 | 3+4 = 7 | 7 |
| PC78 | 51 | 2.3 | 3+3 = 6 | 7 |
| PC79 | 57 | 4.9 | 3+4 = 7 | 7 |
| PC81 | 55 | 4.6 | 3+3 = 6 | 7 |
| PC82 | 46 | 3.9 | 3+3 = 6 | 7 |
| PC85 | 56 | 2.8 | 3+3 = 6 | 7 |
| PC87 | 61 | 5 | 3+3 = 6 | 7 |
| PC88 | 56 | 3.8 | 3+3 = 6 | 6 |
| PC89 | 69 | 5.3 | 4+3 = 7 | 6 |
| PC90 | 52 | 4.8 | 3+3 = 6 | 7 |
| PC91 | 48 | 5.6 | 3+4 = 7 | 7 |
| PC92 | 54 | 0.9 | 3+3 = 6 | 6 |
| PC94 | 53 | 4.3 | 3+4 = 7 | 7 |
| PC96 | 53 | 4 | 4+5 = 9 | 7 |
| PC101 | 62 | 5.2 | 3+4 = 7 | 6 |
| PC104 | 49 | 3.6 | 3+3 = 6 | 6 |
| PC107 | 56 | 3.3 | 3+3 = 6 | 6 |
| PC115 | 60 | 5.3 | 3+4 = 7 | 6 |
| PC116 | 69 | 4.8 | 3+4 = 7 | 6 |
| PC117 | 46 | 4.3 | 3+3 = 6 | 6 |
| PC122 | 47 | 3.1 | 3+3 = 6 | 6 |
| PC131 | 66 | 4.8 | 3+3 = 6 | 6 |
| PC134 | 56 | 5.4 | 4+3 = 7 | 6 |
| PC135 | 65 | 4.1 | 3+4 = 7 | 6 |
| PC139 | 55 | 5.2 | 3+3 = 6 | 6 |
| PC142 | 58 | 3.7 | 3+4 = 7 | 6 |
| PC149 | 68 | 4.2 | 3+3 = 6 | 6 |
| PC151 | 55 | 4.2 | 3+4 = 7 | 6 |
| PC155 | 59 | 4 | 5+4 = 9 | 6 |
| Average | 56.3 | 4.2 | 6.53 | 6.56 |
Table 2.
BPH Patient Clinical Characteristics.
Displayed are the patient ID number, age, PSA level, and follow-up year for each BPH patient sample. The mean value for each characteristic is displayed at the bottom of the table. 39 BPH samples were used.
| Sample | Age (years) | PSA | Follow Up (years) |
|---|---|---|---|
| BPH2 | 64 | 2.8 | 8 |
| BPH9 | 53 | 3.6 | 8 |
| BPH10 | 55 | 7 | 8 |
| BPH11 | 74 | 5 | 7 |
| BPH12 | 77 | 1.2 | 8 |
| BPH13 | 56 | 4.5 | 7 |
| BPH14 | 67 | 3.6 | 8 |
| BPH15 | 71 | 3.6 | 8 |
| BPH17 | 57 | 5.8 | 8 |
| BPH20 | 65 | 4.8 | 8 |
| BPH25 | 87 | 3 | 7 |
| BPH26 | 56 | 5.7 | 7 |
| BPH29 | 67 | 3.2 | 7 |
| BPH31 | 68 | 4.8 | 6 |
| BPH33 | 59 | 5.3 | 7 |
| BPH35 | 72 | 6.3 | 5 |
| BPH47 | 70 | 5 | 5 |
| BPH48 | 64 | 5.6 | 5 |
| BPH49 | 79 | 4.1 | 5 |
| BPH59 | 68 | 4.7 | 6 |
| BPH61 | 76 | 3.9 | 5 |
| BPH65 | 74 | 3 | 6 |
| BPH69 | 68 | 1.9 | 6 |
| BPH70 | 63 | 1.9 | 4 |
| BPH72 | 64 | 1.1 | 7 |
| BPH73 | 60 | 4.5 | 6 |
| BPH75 | 64 | 3.1 | 7 |
| BPH76 | 79 | 4.1 | 6 |
| BPH85 | 61 | 1.8 | 7 |
| BPH88 | 59 | 5.9 | 7 |
| BPH89 | 60 | 1.9 | 7 |
| BPH91 | 87 | 6.8 | 7 |
| BPH98 | 69 | 7 | 6 |
| BPH99 | 69 | 6.7 | 6 |
| BPH102 | 63 | 3.1 | 7 |
| BPH112 | 48 | 2 | 7 |
| BPH121 | 58 | 4 | 5 |
| BPH123 | 54 | 4.1 | 6 |
| BPH129 | 74 | 3.1 | 6 |
| Average | 66 | 4.1 | 6.56 |
Cell culture
Native antigens were obtained from human prostate cancer cell lines. Androgen-responsive LNCaP cells and androgen-independent PC3 cells were purchased from the American Type Culture Collection in Rockville, MD. However, these cells have not been reauthenticated after purchase. Cells were cultured in RPMI-1640 with L-glutamine (Invitrogen, Carlsbad, CA), 10% fetal bovine serum, and 100 IU/ml penicillin and 100 mg/ml streptomycin. Whole cell extracts were obtained by scraping cells from cell culture plates and extracting the proteins with Protein Extraction/Labeling Buffer (Clontech Laboratories, Mountain View, CA) according to the manufacturer’s instructions. After inverting the suspension for 10 min at room temperature, the insoluble fraction was removed by 30 min centrifugation at 10,000 × g at 4°C. Protein concentrations were determined by using a BCA Protein Assay Reagent kit according to the manufacturer’s instructions (Thermo Fisher Scientific Inc, Rockford, IL).
Serum IgG isolation and purification
IgGs were isolated from 50 ul of patient sera using Melon Gel IgG Purification kits (Thermo Fisher Scientific Inc., Rockford, IL) as described by the manufacturer. Sample purity was determined by running each purified sample on an 8-16% Tris-HCL Criterion Precast Gel (Bio-Rad Laboratories, Hercules, CA). If a sample produced bands other than those expected for the heavy and light chain of IgG, the sample was re-purified.
Following isolation, the concentration of IgG in each sample was determined by using a BCA Protein Assay Reagent kit to ensure that a consistent amount of antibodies were dye-labeled and applied to each microarray. One hundred micrograms of purified IgG in 100 microliters solution was dye-labeled with green fluorescing Cy3 maleimide mono-reacting dye (Thermo Fisher Scientific Inc., Rockford, IL) as previously described. Excess dye was removed by Protein Desalting Spin Columns (Thermo Fisher Scientific Inc., Rockford, IL) as previously described (8).
Reverse capture microarray protocol
Twenty-seven-plex reverse capture microarrays were constructed using gold nanoparticle glass slides with monoclonal antibodies to 27 antigens. These monoclonal antibodies were chosen to antigens that were identified from our previous work (6-8) as well as from literature searches and the Cancer Immunome Database (www2.licr.org/CancerImmunomeDB). Each array on the nanoparticle glass slide was first fitted with gaskets which separated the 16 individual arrays on a slide. Two hundred microliters of I-block buffer (Inanovate, Inc., Raleigh, NC) was added to each array, and the entire slide was gently rocked for 30 min. The blocking solution was removed and 6.25 ul of a 1 ug/ul mix of LNCaP/PC-3 cell lysate was combined with 93.75 ul of I-wash buffer (Inanovate Inc., Raleigh, NC) and added to each array on the slide. After two hours of incubation with gentle rocking at room temperature, each array was thoroughly washed using a plate-washer filled with a 10% I-wash buffer solution. Following the wash step, Cy3 dye-labeled IgGs were added to each array on the nanoparticle slide to a predetermined layout. A schematic of the array protocol is shown in Figure 1. Each sample was tested using two different sample concentrations. The first concentration was 4 ul of 1 ug/ul of Cy3 dye-labeled patient IgG mixed with 96 ul of I-wash buffer. The second concentration was 2 ul of 1 ug/ul of Cy3 dye-labeled patient IgG in 98 ul of I-wash buffer. After incubating for one hour with gentle rocking at room temperature, the slides were washed using a plate-washer filled with a 10% I-wash buffer solution. The slides were then spun dry for 20 min at room temperature by centrifuging at 1000 rpm.
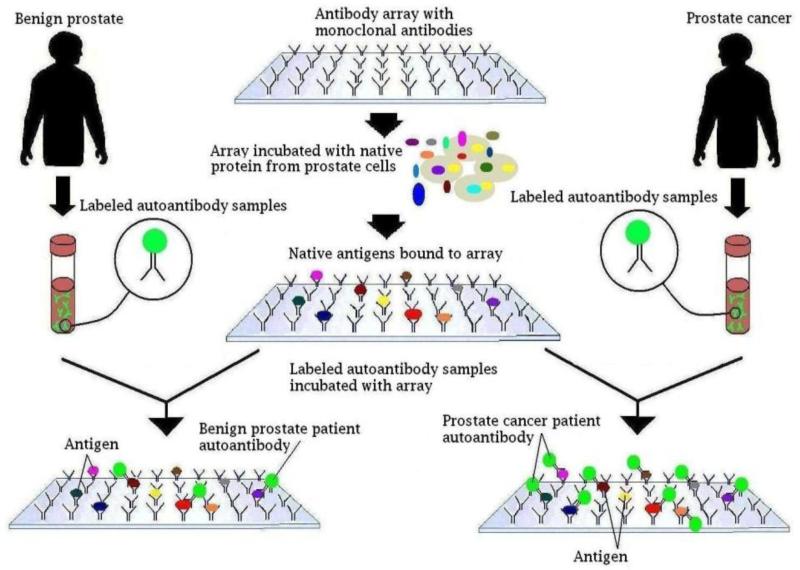
Figure 1.
Array protocol scheme. The reverse capture autoantibody microarray platform is based on the ELISA dual-antibody sandwich immunoassay. Monoclonal antibodies are used to immobilize native antigens from prostate cancer cells. These monoclonal antibodies have been quality controlled and well-characterized by Western blot, and have been demonstrated to bind only to their specific antigens. Using the native antigens as bait, Cy3 dye labeled autoantibody from patients with either prostate cancer or BPH is used as probes and the reactivity to target antigens is then determined.
Image scanning and data collection
A PerkinElmer ScanArray 4000XL scanner and ScanArray Express software (PerkinElmer Inc., Waltham, MA) were used to scan each array for fluorescence and to generate Tiff images. The Tiff images were then uploaded into GenePix Pro 6.0 (Molecular Devices, Sunnyvale, CA) where the data was collected and organized.
Receiver operator characteristic curve and area under the curve
Statistical Analysis Software Ver. 9.1 (SAS Institute Inc., Cary, NC) was used to generate receiver operator characteristic (ROC) curves, which were then used to determine the area under the curve (AUC) values for autoantibody reactivity to each antigen. The curve was based on the fluorescence values for autoantibody reactivity to each specific antigen from all of the patients, cancer and BPH. After arranging the values from highest to lowest for a particular autoantibody reactivity, the intensity of each fluorescence value was plotted on a sensitivity vs. 1-specificity graph.
Results
Preferential reactivity of autoantibodies from sera of patients with prostate cancer versus patients with BPH
We tested the feasibility of autoantibody profiling as a potential strategy to distinguish age-matched prostate cancer patients from BPH patients with similar serum PSA concentrations. The mean serum PSA for patients with prostate cancer is 4.2 ng/ml with a Gleason score of 6 or 7 (see Table 1), and the mean serum PSA for patients with BPH is 4.1 ng/ml (see Table 2). To establish our control group, the BPH samples were histologically confirmed and had a mean follow-up time of 6.56 y to rule-out a diagnosis of cancer. As illustrated in Figure 2, results show that there is clearly preferential autoantibody reactivity with the immobilized antigens between patients with prostate cancer. Since the antigens were immobilized with known monoclonal antibodies on the array, the antigens recognized by the autoantibodies were easily identified.
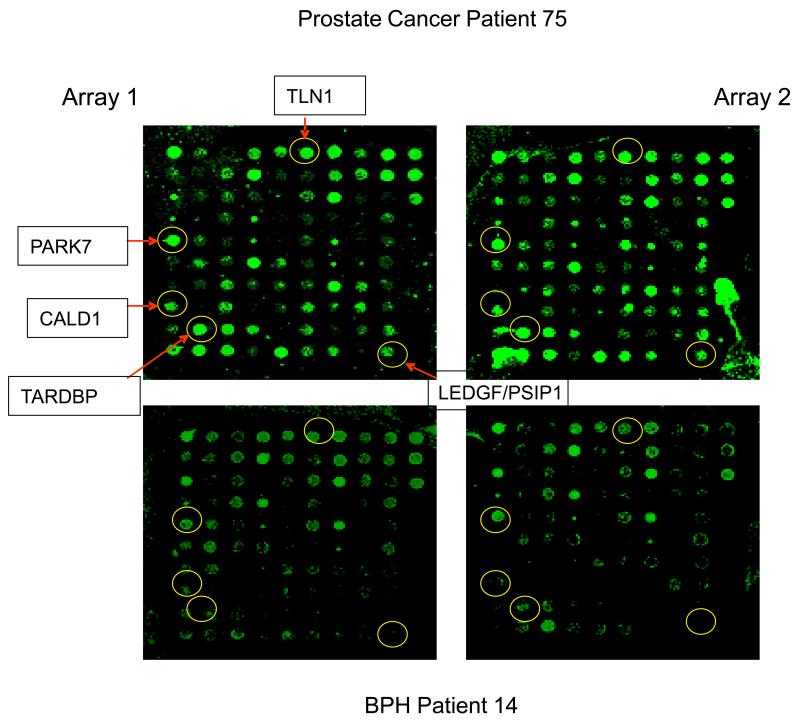
Figure 2.
This figure highlights the finding that patients with prostate cancer and BPH consistently express distinct, disease-related autoantibody reactivity to the target antigens. The top two images are from the same prostate cancer patient in duplicate runs, and the bottom two images are from the same BPH patient in duplicate runs. Circled spots show greater fluorescence, or autoantibody reactivity to the antigen, in the cancer sample than in the BPH sample.
Figure 2 also illustrates the reproducibility of our customized reverse capture microarray. The images of this figure show the autoantibody reactivity of the same patient in duplicate runs. The images clearly show the similarity of the reactivity in the duplicate runs, i.e., Array 1 vs. Array 2 for the same patient.
Receiver operator characteristic (ROC) curve and area under the curve (AUC)
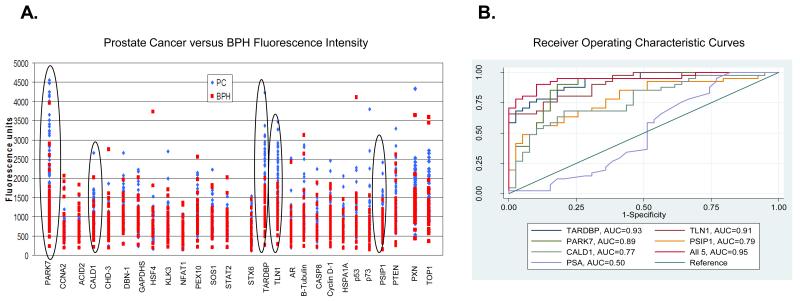
A scatter plot of the fluorescence intensity units is also shown for all 27 autoantibody reactivity (Fig. 3A). Out of the 27 autoantibody reactivities, 17 were found to be statistically significant (p ≤ 0.05) in separating prostate cancer from BPH. However, the 5 circled signatures demonstrate the best separation of prostate cancer from BPH samples (Fig. 3A). In addition, a ROC curve for autoantibody reactivity was constructed for each of the antigens using individual fluorescence intensity values for each of the cancer cases and BPH controls. Table 3 shows the AUC values for autoantibody reactivity to the antigens, ranking from the highest AUC to the lowest AUC. In addition, the ROC curves for the top 5 autoantibody signatures, as determined by their AUC, are shown in Figure 3B. The top 5 autoantibody signatures were identified to be TARDBP, TLN1, PARK7, LEDGF/PSIP1, and CALD1. When combined, these autoantibody signatures produced an AUC value of 95% compared with serum concentration PSA at the time of sample collection with an AUC value of 50% (Table 3 and Fig. 3B). In addition, the ROC curve also shows that when combined, the top 5 autoantibody signatures had a sensitivity of 95% at 80% specificity, and that serum PSA had a sensitivity of 12.2% at 80% specificity (Fig. 3B). Finally, when we compared the AUC of the top 5 autoantibody signatures combined versus the AUC of the top 4, 3, and 2 autoantibody signatures combined, the AUC decreased from 0.95 (all 5 included) to 0.93 (top 2 or 3 included).
Figure 3.
(A): A scatter plot of the fluorescence intensity units is shown for all autoantibody reactivity. Fluorescence units for the BPH are shown in red, and the fluorescence units for the prostate cancer are shown in blue. The 5 circled signatures highlight the best separation of prostate cancer from BPH samples. (B): A receiver operating characteristic (ROC) curve was constructed for each of the immobilized antigens using individual fluorescence intensity values for each case and BPH control. The top 5 autoantibodies to the immobilized antigens are shown in the figure. Also included in the figure is the area under the curve (AUC) for each of the 5 antigens. ROC curve for serum PSA is also shown. The PSA values for our cases and BPH controls reflect the levels of the samples (ng/ml) at the time of collection. PSA has an AUC of 0.50 for our cohort.
Table 3.
This table displays the AUC values for all antibodies tested on the 27-plex microarray platform. They are ranked according to AUC values for concentration 1, which is 4 ul of 1 ug/ul of Cy3 dye-labeled patient IgG mixed with 96 ul of I-wash buffer. AUC for PSA is calculated from the sample’s serum PSA level at the time of blood drawn.
| Concentration 1 | ||
|---|---|---|
| Ab Name | AUC | rank |
| TARDBP | 0.927 | 1 |
| TLN1 | 0.911 | 2 |
| PARK7 | 0.886 | 3 |
| LEDGF/PSIP1 | 0.789 | 4 |
| CALD1 | 0.77 | 5 |
| p73 | 0.76 | 6 |
| PTEN | 0.675 | 7 |
| PXN | 0.667 | 8 |
| PEX10 | 0.667 | 9 |
| KLK3 | 0.606 | 10 |
| DBN1 | 0.6 | 11 |
| NFAT1 | 0.592 | 12 |
| B Tubulin | 0.591 | 13 |
| SOS1 | 0.583 | 14 |
| HSF4 | 0.581 | 15 |
| TOP1 | 0.567 | 16 |
| HSPA1A/B | 0.567 | 17 |
| ACID2 | 0.556 | 18 |
| STAT2 | 0.54 | 19 |
| p53 | 0.512 | 20 |
| CHD 3 | 0.506 | 21 |
| CASP8 | 0.498 | 22 |
| STX6 | 0.498 | 23 |
| AR | 0.478 | 24 |
| GAPDHS | 0.476 | 25 |
| Cyclin D1 | 0.465 | 26 |
| CCNA2 | 0.438 | 27 |
| Serum level PSA (ng/ml) | 0.50 | |
Coefficient of variance and platform characteristics
To further evaluate the robustness of the assay, CV data within an array (between duplicate spots) as well as between duplicate arrays (well to well) across samples was calculated. CVs averaged 16% from spot to spot and 15% from well to well (Tables 4 and 5).
Table 4.
Coefficient of Variance for Prostate Cancer Arrays.
Shown is the antigen-autoantibody reactivity with the CV from spot-to-spot (within an array). Also shown are the CV values for the antigen-autoantibody reactivity in well-to-well (in duplicate arrays). At the bottom of the table, the mean CV value for all antigens is shown.
| Array 1 | Array 2 | Array 1 and Array 2 | |
|---|---|---|---|
| Spot # | Spot to Spot CV | Spot to Spot CV | Well to Well CV |
| 1 Control | 12% | 16% | 9% |
| 2 Control | 18% | 20% | 17% |
| 3 Control | 11% | 17% | 13% |
| 4 Control | 13% | 18% | 14% |
| 5 PARK7 | 11% | 8% | 12% |
| 6 CCNA2 | 13% | 13% | 18% |
| 7 ACID2 | 13% | 17% | 19% |
| 8 CALD1 | 14% | 16% | 17% |
| 9 CHD-3 | 19% | 14% | 16% |
| 10 DBN1 | 18% | 19% | 21% |
| 11 GAPDHS | 18% | 17% | 17% |
| 12 HSF4 | 17% | 16% | 17% |
| 13 KLK3 | 15% | 16% | 18% |
| 14 NFAT1 | 13% | 16% | 16% |
| 15 PEX10 | 14% | 14% | 19% |
| 16 SOS1 | 18% | 20% | 24% |
| 17 STAT2 | 14% | 14% | 21% |
| 18 STX6 | 17% | 26% | 27% |
| 19 TARDBP | 14% | 14% | 11% |
| 20 TLN1 | 10% | 9% | 12% |
| 21 AR | 20% | 16% | 21% |
| 22 b-Tubulin | 8% | 12% | 15% |
| 23 CASP8 | 12% | 12% | 18% |
| 24 Cyclin D1 | 10% | 15% | 21% |
| 25 HSPA1A/B | 14% | 14% | 20% |
| 26 p53 | 12% | 18% | 19% |
| 27 p73 | 14% | 13% | 19% |
| 28 LEDGF/PSIP1 |
16% | 15% | 22% |
| 29 PTEN | 18% | 20% | 16% |
| 30 PXN | 14% | 17% | 19% |
| 31 TOP1 | 9% | 10% | 16% |
| Averages | 14% | 16% | 17% |
Table 5.
Coefficient of Variance Data for BPH Arrays.
Shown is the antigen-autoantibody reactivity with the CV from spot-to-spot (within an array). Also shown are the CV values for the antigen-autoantibody reactivity in well-to-well (in duplicate arrays). At the bottom of the table, the mean CV value for all antigens is shown.
| Array 1 | Array 2 | Array 1 and Array 2 | |
|---|---|---|---|
| Spot # | Spot to Spot CV | Spot to Spot CV | Well to Well CV |
| 1 Control | 8% | 9% | 6% |
| 2 Control | 19% | 23% | 13% |
| 3 Control | 14% | 14% | 12% |
| 4 Control | 15% | 21% | 13% |
| 5 PARK7 | 15% | 22% | 14% |
| 6 CCNA2 | 14% | 14% | 15% |
| 7 ACID2 | 15% | 14% | 14% |
| 8 CALD1 | 13% | 16% | 17% |
| 9 CHD-3 | 16% | 20% | 11% |
| 10 DBN1 | 20% | 18% | 14% |
| 11 GAPDHS | 16% | 21% | 12% |
| 12 HSF4 | 17% | 21% | 16% |
| 13 KLK3 | 14% | 17% | 13% |
| 14 NFAT1 | 16% | 18% | 15% |
| 15 PEX10 | 14% | 15% | 14% |
| 16 SOS1 | 13% | 16% | 14% |
| 17 STAT2 | 11% | 19% | 16% |
| 18 STX6 | 20% | 25% | 21% |
| 19 TARDBP | 21% | 21% | 13% |
| 20 TLN1 | 15% | 17% | 12% |
| 21 AR | 16% | 18% | 13% |
| 22 b-Tubulin | 10% | 13% | 10% |
| 23 CASP8 | 14% | 16% | 13% |
| 24 Cyclin D1 | 12% | 16% | 12% |
| 25 HSPA1A/B | 15% | 18% | 12% |
| 26 p53 | 13% | 19% | 16% |
| 27 p73 | 16% | 21% | 17% |
| 28 LEDGF/PSIP1 |
15% | 19% | 18% |
| 29 PTEN | 30% | 28% | 16% |
| 30 PXN | 23% | 24% | 16% |
| 31 TOP1 | 13% | 14% | 10% |
| Averages | 16% | 18% | 14% |
Discussion
Serum prostate specific antigen (PSA) has remained the leading prostate cancer biomarker for the past 25 y. However due to its low specificity (50%), the number of false positives and false negatives has proven problematic, resulting in unnecessary, expensive biopsies for benign disorders such as BPH. Our own research (6-8) and the work of others (9) suggest that autoantibody profiling may offer an appealing alternative to PSA. Thus, we have continued to develop our approach to autoantibody profiling in an attempt to identify a set of signatures that may be able to distinguish between prostate cancer from BPH in patients with increased serum PSA concentrations.
For our current work, we tested 27 antigens. These antigens were identified from our prior work on autoantibody profiling (6-8), from published literature searches, and from the Cancer Immunome Database (www2.licr.org/CancerImmunomeDB). Unlike our previous studies where a two color dye-swap method was used, our current study uses a single Cy3 dye. This allowed us to analyze the results using ROC curves without concern for the potential errors and biases of two different dyes.
The robustness of our assay is shown in Figure 2 as well as by the CVs (Tables 4 and 5). Autoantibody signatures were consistent between arrays, with CVs averaging 15%, as well as within arrays (spot to spot), with CVs averaging 16%. In addition, similar autoantibody reactivity was found at two separate sample concentrations (results not shown).
Using this customized reverse capture microarray platform, we have identified a set of autoantibodies which has greater sensitivity and specificity than serum PSA concentrations in patients with prostate cancer vs. BPH. Figure 2 illustrates examples of preferential autoantibody reactivity in patients with prostate cancer over patients with BPH.
Along with cyclin D1, p53, p57, CASP8, and AR, which were all previously reported to have preferential reactivity in patients with prostate cancer (6,7), we identified a set of five autoantibody signatures that demonstrated superior performance than serum PSA concentrations (Table 3 and Fig. 3). These 5 autoantibody signatures were TARDBP, TLN1, PARK7, LEDGF/PSIP1, and CALD1. In addition, when combined, the 5 autoantibody signatures achieved a sensitivity of 95% at 80% specificity; whereas serum PSA concentrations achieved a sensitivity of only 12.2% at 80% specificity (Fig. 3B). Finally, when we compared the AUC of the top 5 autoantibody signatures combined versus the AUC of the top 4, 3, and 2 autoantibody signatures combined, the AUC decreased from 0.95 (all 5 included) to 0.93 (top 2 or 3 included), suggesting that each of the top 5 autoantibody signatures contributes toward some concentration of discrimination between prostate cancer and BPH.
The TARDBP gene produces the protein TAR DNA-binding protein 43. The protein functions to regulate transcription and splicing via direct DNA and RNA binding (10), and inhibition of TARDBP has been shown to result in apoptosis (11). With an AUC of 92.7%, with sensitivity of 85% at 80% specificity, autoantibodies to TARDBP may play a role in distinguishing prostate cancer from BPH in patients with increased serum PSA.
TLN1 produces talin-1, and has been reported to be associated with prostate cancer. Talin-1 was previously identified by Taylor et al as a tumor-associated antigen in prostate cancer patients (12). Furthermore, talin-1 has been identified to play a role in increasing prostate cancer cell migration, and assisting in the process of metastasis (13). Our results showed that autoantibodies to talin-1 had an AUC of 91.1%, with sensitivity of 80.5% at 80% specificity, suggesting that it is also an appealing biomarker for prostate cancer.
Our third top autoantibody signature is to PARK7 and its protein product DJ-1. DJ-1 is thought to act as a chaperone protein and a transcription regulator for androgen receptor-dependent transcriptions (14). In a study by Ronquist et al (15), they probed proteins derived from prostasomes using Western blotting of 2-D SDS-PAGE and sera from prostate cancer patients. Proteins that were immuno-reactive with patient’s sera were further identified by mass spectrometry, and one such proteins was identified as PARK7 (15). Our result is consistent with their preliminary findings, as autoantibody reactivity to PARK7 generated an AUC of 88.6%, with sensitivity of 90% at 80% specificity.
LEDGF/p75 from PSIP1 is a transcription coactivator with potential protective properties from stress-induced apoptosis (16). LEDGF/p75 expression was detected in 93% of prostate tumors but not in normal prostate tissue (16). Using ELISA, autoantibodies to LEDGF/p75 was previously identified in over 18% of prostate cancer patients and was absent from BPH patients (16). Our results showed that autoantibodies to LEDGF/p75 resulted in an AUC of 78.9% with 58% sensitivity at 80% specificity, and LEDGF/p75 is our fourth top autoantibody signature.
The fifth top autoantibody signature is the gene product for CALD1, which produces the protein caldesmon. Caldesmon is an actomyosin binding protein involved in many cell types and processes. Specifically, it has been shown that caldesmon is highly involved in early tumor vascularization in several tissue types, including prostate cancer (17). It has also been demonstrated that caldesmon regulates podosome and invadopodium activity, two molecules believed to play a role in facilitating cancer cells to spread (18). Our results showed that autoantibodies to CALD1 resulted in an AUC of 77% with 63% sensitivity at 80% specificity.
When combined, these 5 autoantibody signatures achieved a superior performance than serum concentration PSA. As recent studies have shown (19-21), there is a need to move from a single biomarker to a panel of markers in order to improve the sensitivity and specificity of these tests. Our results showed that these 5 autoantibody signatures may be strong candidates for a panel of markers that would distinguish prostate cancer from BPH in patients with increased serum PSA.
However, this study has some limitations. One such limitation is with our reverse capture microarray platform. This platform uses native antigens, and although the capture monoclonal antibodies are quality controlled and well-characterized by Western blot to bind only to their specific antigens, we cannot rule out the possibility that there are antigen complexes captured by the platform. Therefore, we are currently validating the antigens recognized by the autoantibodies through a series of experiments (e.g., knock-out experiments in the cells). Although such validation studies are underway, they are beyond the scope of this paper. Another weakness of our study is with our sample size. However, the strength of our study is that we limited our BPH controls to samples that had adequate follow-up and assuring that there is no prostate cancer that might co-exist in the BPH group. Thus, our BPH controls had a mean follow-up of 6.56 y. Another limitation is that we did not include any age-matched normal healthy controls. But our initial objective was to test the hypothesis that there are autoantibody signatures that might distinguish between prostate cancers from BPH in patients with increased serum PSA concentrations, and thus eliminated the use of normal healthy controls from this study. However, preliminary studies did not demonstrate any consistent or significant concentrations of autoantibody reactivity with the antigens when probed with random normal healthy sera (data not shown). Finally, we will need to demonstrate the utility of these autoantibody signatures in a larger and independent cohort of patients and controls to validate these results. Such work is ongoing. As such, our results should be considered only preliminary. Our findings, however, have validated using utility of some of the autoantibody-antigen reactivities that have been reported by us and others. Future work will include enlarging the sample size and to generate some quantitative measurements for the autoantibody signatures. In addition, we will need to address whether well-differentiated (Gleason score of 2 to 4) prostate cancer or patients with higher serum PSA concentration (>10 ng/ml) will have similar or different and unique autoantibody signatures. If successful, a panel of autoantibody signature may complement the current PSA assay, perhaps as a secondary test, and may prevent unnecessary biopsies in patients with BPH.
Highlights.
-
➢
PSA lacks the ability to differentiate between prostate cancer and benign prostatic hyperplasia
-
➢
In this study, we identified autoantibody signatures unique to prostate cancer patients
-
➢
Our top 5 autoantibody signatures, when combined, achieved a sensitivity of 95% at 80% specificity
-
➢
Our findings may result in a blood test to reduce unnecessary biopsies in patients with elevated PSA
Acknowledgments
This work was supported in part by grants CA143232 (J.N.), DK063665 (B.L.), DK078566 (B.L.) from the NIH, and a grant from Inanovate, Inc (D.U.). The authors thank Esther H. Orr for drawing Figure 1. DJO and DAD performed this work as part of their undergraduate research co-op program at Northeastern University, Boston, MA. DJO was the recipient of the “Best Abstract” award from the Clinical Diagnostic and Immunology Division, the Clinical Translational Science Division, and the Proteomics Division at the 2011 Annual Meeting of the American Association for Clinical Chemistry for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: Dennis O’Rourke, Daniel DiJohnson, Robert Caiazzo, Jr., Brian Liu, James Nelson, and David Ure have filed a patent on the biomarkers and work disclosed in this manuscript. In addition, James Nelson and David Ure are employees of Inanovate, Inc.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011. The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Brawer MK. Prostate-specific antigen: current status. CA Cancer J Clin. 1999;49:264–81. doi: 10.3322/canjclin.49.5.264. [DOI] [PubMed] [Google Scholar]
- 3.Gretzer MB, Partin AW. PSA markers in prostate cancer detection. Urol Clin North Am. 2003;30:677–86. doi: 10.1016/s0094-0143(03)00057-0. [DOI] [PubMed] [Google Scholar]
- 4.Finn OJ. Immune response as a biomarker for cancer detection and a lot more. N Engl J Med. 2005;353:1288–90. doi: 10.1056/NEJMe058157. [DOI] [PubMed] [Google Scholar]
- 5.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–33. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin S, Qiu W, Ehrlich JR, Ferdinand AS, Richie JP, O’Leary MP, et al. Development of a “reverse capture” autoantibody microarray for studies of antigen-autoantibody profiling. Proteomics. 2006;6:3199–209. doi: 10.1002/pmic.200500673. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich JR, Caiazzo RJ, Jr., Qiu W, Tassinari OW, O’Leary MP, Richie JP, Liu BCS. A native antigen “reverse capture” microarray platform for autoantibody profiling of prostate cancer sera. Proteomics Clin Appl. 2007;1:476–85. doi: 10.1002/prca.200700012. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich JR, Qin S, Liu BCS. The “reverse capture” autoantibody microarray: a native antigen-based platform for autoantibody profiling. Nat Protoc. 2006;1:452–60. doi: 10.1038/nprot.2006.66. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–35. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 10.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–84. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA. 2008;105:3785–9. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor BS, Pal M, Yu J, Laxman B, Kalyana-Sundaram S, Zhao R, et al. Humoral response profiling reveals pathways to prostate cancer progression. Mol Cell Proteomics. 2008;7:600–11. doi: 10.1074/mcp.M700263-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin 1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–95. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niki T, Takahashi-Niki K, Taira T, Iguchi-Ariga SMM, Ariga H. DJBP: a novel DJ-1 binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex. Mol Cancer Res. 2003;1:247–61. [PubMed] [Google Scholar]
- 15.Ronquist KG, Carlsson L, Ronquist G, Nilsson S, Larsson A. Prostasome-derived proteins capable of eliciting an immune response in prostate cancer patients. Int J Cancer. 2006;119:847–53. doi: 10.1002/ijc.21895. [DOI] [PubMed] [Google Scholar]
- 16.Daniels T, Zhang J, Gutierrez I, Elliot ML, Yamada B, Heeb MJ, et al. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 17.Zheng PP, van der Weiden M, Kros JM. Differential expression of HeLa-type caldesmon in tumour neovascularization: a new marker of angiogenic endothelial cells. J Pathol. 2005;205:408–14. doi: 10.1002/path.1700. [DOI] [PubMed] [Google Scholar]
- 18.Yoshio T, Morita T, Kimura Y, Tsujii M, Hayashi N, Sobue K. Caldesmon suppresses cancer cell invasion by regulating podosome/invadopodium formation. FEBS Lett. 2007;581:3777–82. doi: 10.1016/j.febslet.2007.06.073. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev. 2003;12:136–143. [PubMed] [Google Scholar]
- 20.Zhang JY. Tumor-associated antigen arrays to enhance antibody detection for cancer diagnosis. Cancer Detect Prev. 2004;28:114–8. doi: 10.1016/j.cdp.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Casiano CA, Mediavilla-Varela M, Tan EM. Tumor-associated antigen arrays for the serological diagnosis of cancer. Mol Cell Proteomics. 2006;5:1745–59. doi: 10.1074/mcp.R600010-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]