Abstract
Although there have been numerous reports describing the isolation of liver progenitor cells from adult liver, their exact origin has not been clearly defined; and the role played by mature hepatocytes as direct contributors to the hepatic progenitor cell pool has remained largely unknown. Here we report strong evidence that mature hepatocytes in culture have the capacity to dedifferentiate into a population of adult liver progenitors without genetic or epigenetic manipulations. By using highly-purified mature hepatocytes, which were obtained from untreated, healthy rat liver and labeled with fluorescent dye PKH2, we found that hepatocytes in culture gave rise to a population of PKH2-positive liver progenitor cells. These cells, Liver Derived Progenitor Cells or LDPCS, which share phenotypic similarities with oval cells, were previously reported to be capable of forming mature hepatocytes both in culture and in animals. Studies done at various time points during the course of dedifferentiation cultures revealed that hepatocytes rapidly transformed into liver progenitors within one week through a transient oval cell-like stage. This finding was supported by lineage-tracing studies involving double-transgenic AlbuminCreXRosa26 mice expressing β-galactosidase exclusively in hepatocytes. Cultures set up with hepatocytes obtained from these mice resulted in generation of β-galactosidase-positive liver progenitor cells demonstrating that they were a direct dedifferentiation product of mature hepatocytes. Additionally, these progenitors differentiated into hepatocytes in vivo when transplanted into rats that had undergone retrorsine pretreatment and partial hepatectomy.
Conclusion
Our studies provide strong evidence for the unexpected plasticity of mature hepatocytes to dedifferentiate into progenitor cells in culture; and this may potentially have a significant impact on the treatment of liver diseases requiring liver or hepatocyte transplantation.
Keywords: Liver, cell culture, stem cells, oval cells, transgenic mice
Introduction
Liver is a unique organ with an enormous capacity to regenerate after injury. Up to 75% of liver mass can be regenerated in the rat within one week after partial hepatectomy (1). Under normal conditions and in the absence of toxic injury, hepatocytes are predominantly responsible for this process (2). When circumstances do not allow proliferation of resident hepatocytes as in prior exposure of the liver to certain toxins or chemicals, a population of stem/progenitor cells is activated. These cells, typically referred to as oval cells, are thought to exist in small numbers in terminal biliary ductules (3-4). Although a bone marrow origin for oval cells has been suggested (5), more recent studies demonstrated that they were of hepatic origin (6, 7). Oval cells have the capacity to repair the injured liver, giving rise to both functional hepatocytes and cholangiocytes in vivo (8). However, their clinical use is prohibited, in part, because of the methods used to retrieve them in adequate numbers and the high frequency of malignant transformation observed in transplant studies (9, 10).
Multipotent stem cell populations such as embryonic stem cells and more recently induced pluripotent stem cells have also been shown to possess hepatic differentiation ability even though their clinical applicability remains controversial (11-13). Bone marrow-derived (hematopoietic or mesenchymal) stem cells also have the capacity to produce hepatocytes in vitro and in vivo as evidenced by human bone marrow transplant studies as well as animal studies (14, 15). However, the frequency of this event appears to be low in the in vivo setting (16, 17). Therefore, there continues to be a major need for a stem/progenitor cell population that is safe and practical for clinical applications and treatment of a variety of human liver diseases. This has resulted in intensification of efforts to identify and study liver-specific stem/progenitor cells in parallel to those involving ES and iPS cells. We, too, have recently reported isolation and characterization a unique population of adult liver progenitors called Liver Derived Progenitor Cells or LDPCs (18).
LDPCs are a novel population of bipotential liver progenitor cells whose isolation does not require any chemicals or toxins. In addition to a mixture of hematopoietic and hepatic markers, LDPCs express a number of stem cell markers including CD34, c-kit and Thy-1; and they are phenotypically very similar to oval cells. More recently, we have shown that LDPCs are capable of differentiating into functional hepatocytes both in vitro and in vivo (19). However, the origin of these cells, which has both significant scientific and clinical implications, has remained largely unanswered. Our results demonstrated that LDPCs were a direct dedifferentiation product of isolated mature hepatocytes, thus radically altering the concept of lineage relationship between liver progenitors and hepatocytes. The results have significant implications for a variety of stem cell applications.
Materials and Methods
Animals
Albumin CreX Rosa26 double transgenic mouse strain was generated by cross breeding Albumin-Cre and Rosa26 mouse strains both of which were purchased from Jackson Laboratories (Bar Harbor, Maine). Sprague–Dawley and Fischer344 rats used in the experiments were 6 to 8 weeks old and were purchased from Harlan Laboratories (Madison, WI). All animal experiments were performed within the context of institutionally approved animal protocols and the animals were treated humanely in compliance with University of Minnesota regulations.
LDPC Cultures
Rat LDPCs were generated from Sprague–Dawley and Fischer344 rat livers as previously described (Sahin et al., 2008) with the exception that the liver cell preps were highly enriched for hepatocytes prior to LDPC cultures by additional × 3 low-gravity-centrifugation steps (50 G × 15 seconds). The protocol for isolation of mouse LDPCs was a modified form for rat LDPCs. Briefly, mouse hepatocytes were cultured in a medium which consisted of Dulbecco’s modified Eagles medium (DMEM-LG) (Sigma, St Louis, MO) and F12 (Sigma) medium at ratio of 3:1, supplemented with 15% mouse serum (Equitech, Kerville, TX), 1 mg/ml of bovine serum albumin (Sigma), 100 uM of β-mercaptoethanol (Sigma), 5mM of nicotinamide (Sigma), and penicillin (100 μg/ml) /streptomycin (100 μg/ml) (Gibco, Carlsbad, CA). The cells were plated at a density of 2×103 cells/cm2 on 24-well plates coated with type I collagen (Becton Dickinson, New Jersey, USA) at a concentration of 10 μg/cm2. Unlike rat LDPCs, mouse LDPCs were generated without further media change.
Immunofluorescence (IF) Staining
The expression of each antigen was examined on cells from two independent experiments. The cells were fixed in 2% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100, washed in PBS, pre-blocked with donkey serum (Sigma), and subsequently stained with sheep anti-human serum albumin (1:1000, Abcam Inc, Cambridge, MA), mouse anti-cytokeratin 7, CK7, (1:100, Abcam), rabbit anti LMO2 (1:100, Abcam), mouse anti-hepatocyte nuclear factor-1α, HNF-1α (1:500, BD Biosciences), PE-Rat anti-mouse CD45 (1:500, BD Biosciences, Franklin Lakes, NJ), PE-mouse anti-rat CD45 (1:20, BioLegend, San Diego, CA), mouse anti-gamma glutamine transaminase, GGT, (1:100, Sigma), mouse anti-beta galactosidase (1:500, Roche, Germany), mouse anti-human/rat OV-6 (1:100, R&D Systems, Minneapolis, MN), rabbit anti-SALL4 (1:100, GeneTex, Irvine, California, USA), chicken anti-Vimentin (1:10,000, Abcam, Cambridge, MA), mouse anti-Liv-2 (1:100, MBL, Nagoya, Japan), rabbit anti-CK19 (1:100, GeneTex, Irvine, California, USA), mouse anti-HNF4a (1:100, Abcam, Cambridge, MA), mouse anti-CD44 (1:100, Cell Signaling, Danvers, MA) and mouse anti-HepPar1 (1:100, Abcam, Cambridge, MA). Appropriate secondary antibodies, Dylight 488 donkey anti-sheep IgG, Dylight 549 donkey anti-rabbit IgG, and Dylight 594 donkey anti-mouse IgG (1:200; Jackson ImmunoResearch Laboratories, Inc. West Grove, PA) were used. In the control experiments, cells were stained with secondary antibodies only. The nuclei were labeled with 4, 6-diamidino-2-phenylindole (DAPI, Roche). Quantification of IF staining was also performed for a few select markers. For this purpose, NIH Image J software was used to measure the mean gray value (MGV) for staining of cells through the culture period. For each cell, MGV of the area of interest (Sall4 and HNF4a: nuclei, HepPar1: whole cell) was calculated and background was subtracted. An average of the MGV was then calculated for each group. The values were then extracted from that of negative control groups which were only stained with secondary antibodies.
Histopathology and β-galactosidase Staining
Mouse liver tissues were fixed in 2% paraformaldehyde for 30 min, and then embedded in Tissue-Tek OCT (Sakura Finetek, Torrance, CA) compound. The sections were sliced at a thickness of six-microns. After air-drying, the sections were incubated in β-galactosidase reporter gene staining solution (β-galactosidase reporter gene staining kit, Sigma) at 37°C over night and then counterstained with nuclear fast-red staining (Sigma) for 5 min at room temperature. For immunofluorescent staining, the sections were fixed in cold acetone. Rabbit anti-β-galactosidase (1:20, Cedarlane) was mixed with either sheep anti-Albumin antibody (1:500) or mouse anti-CK7 (1:100). After washing, dylight 549 anti-mouse, dylight 549 anti-sheep or dylight 488 anti-rabbit antibodies at a dilution of 1:200 were used as secondary antibodies.
RNA Isolation and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated using miniRNA kit (Qiagen, Valencia, CA) according to instructions of the supplier and was subsequently subjected to RT-PCR. 1 μg of total RNA was reverse-transcribed in a 20-μl volume with the Superscript III Reverse Transcription Kit (Invitrogen, Carlsbad, CA) as per the manufacturer’s recommendations. After the reverse-transcription, the volume of the reaction was increased to 100 μl, and 2 μl was used in each of the PCR assays with the Red kit (Sigma) on a Biorad thermocycler (Biorad, Hercules, CA) according to the manufacturer’s instructions. The reactions were repeated a minimum of three times.
Western Blot Analysis
Mouse tissues were lysed in 2X total protein buffer containing 10mM Tris at pH of 7.6, 1% NP40, protease inhibitors (Roche Complete Mini EDTA-Free), and 2mM DTT. Then, the cell lysates were sonicated and quantified using Bio-Rad Protein Assay. 40 μg of protein was loaded into each well and separated by 4-15% SDS Mini-Protean TGX gel (Biorad). After transfer, the PVDF membrane was blocked with 5% non-fat dried milk and then cut into two pieces. The upper panel was incubated with rabbit anti β-galactosidase (Cedarlane Labs, Burlington, NC); the lower panel was incubated with mouse monoclonal anti-β-Actin (Sigma) at 4°C overnight. After washing, the peroxidase-conjugated secondary antibodies were added for 1 hour at room temperature. The detection was achieved using SuperSignal West Dura Extended Duration Substrate (Pierce, Rockford, IL).
PKH-2 Staining and Flow Cytometry
Hepatocytes obtained from Sprague-Dawley rats were stained with PKH2 for cytoplasmic labeling according to the manufacturer’s instructions (Sigma). After staining, hepatocytes were plated on 24-well plates or T-150 culture flasks at a density of 1×104 cells/cm2. The fluorescence images of the cells were obtained at pre-determined time points on a Leica DMIL fluorescence microscope using Leica application suite V3.1 software. On days 1 and 14, the cells on flasks were collected and subjected to flow cytometry for quantitative analysis of their fluorescence. For flow cytometric analysis, the unlabeled cells were used as negative controls. Mean fluorescent intensity was determined for each sample; and total fluorescence was calculated by multiplying mean fluorescent intensity and the total number of cells. The value of total fluorescence on Day 1 was given an arbitrary unit of 1. The total fluorescence on Day 14 was calculated in an identical manner and then compared with that of Day 1. Triplicates were used for statistical calculations. For flow cytometric analysis of hepatocyte purity, the cells were fixed and permeabilized by 0.2% Triton X-100. After blocking with donkey IgG, the cells were incubated with an anti Albumin-FITC antibody (1:20, Cedarlane) for 30 min at room temperature. The negative control was FITC-rabbbit IgG. FACS acquisition was performed using a FACSCalibur flow cytometer (BD Biosciences), and the data were analyzed using Flow Jo 6.4. A statistical analysis was done by Student’s t test to identify significant differences. P value less than 0.05 was considered significant. The flow cytometric analysis of LDPC purity was performed in an identical fashion except for the antibody which was FITC-conjugated rabbit anti-CD45 antibody (BD Biosciences).
Transplantation of LDPCs
Five female Fischer344 rats were treated with retrorsine (Sigma) prior to transplantation to impair the ability of recipient hepatocytes to proliferate. Two intraperitoneal injections were administered two weeks apart at a dose of 30 mg/kg. One month after the second dose of retrorsine, the recipient animals were anesthetized using isoflurane and subjected to laparatomy and 66% partial hepatectomy under sterile conditions. This was followed by injection into spleen of 1× 107 PHK26-labeled male LDPCs obtained from male Fischer344 rats. (PKH26 labeling was performed following manufacturer’s (Sigma) instructions resulting in labeling of >90% of the cells.) Two of the rats died from surgical complications the day after transplantation. The livers of the remaining three rats were examined two months later for evidence of engraftment. The animal experiments were done within the framework of institutionally approved protocols and the animals were treated and euthanized humanely.
Fluorescence In Situ Hybridization (FISH) for Y-Chromosome
Liver sections of 6μm were pre-stained with albumin antibody and fixed in 4% formaldehyde at 37°C for 10 minutes. Then following the manufacturer’s protocol, the sections were washed with 2xSSC for 2 minutes at 73°C and treated with 0.005% pepsin for 10 minutes at 37°C. After rinsing in 1xPBS with glycine, the slides were dehydrated in ethanol; and rat IDetect Chr-Y Probe (ID labs, London, ON, Canada) applied. After 2 minutes of denaturation at 69°C, the slides were incubated for hybridization at 37°C overnight. After hybridization, the slides were washed with 0.4xSSC with 0.3% lgepal (Sigma) for 2 minutes at 73°C, and 2xSSC with 0.1% lgepal for 1 minute room temperature. After staining with DAPI, the samples were examined under a fluorescence microscope and images were obtained.
Results
Rat Liver Preparations Subjected to Low-G Centrifugations Are Highly Enriched for Hepatocytes
In order to identify the origin of LDPCs, isolated hepatocytes were highly purified by low-G centifugations. We performed RT-PCR for markers that were specific for various cell types found in the liver including desmin for stellate cells (20, 21); von Willebrand Factor (vWF) for endothelial cells; fucose receptor for Kupffer cells (22); cytokeratin 7 (CK7) for biliary epithelial cells (23); and albumin for hepatocytes. The cell prep prior to low-G spin showed clear signals for all of the markers except for CK7 indicating that the initial cell population contained hepatocytes, endothelial, Kupffer and stellate cells. It appears that our standard centrifugation steps prior to low-G spins eliminated CK7 positive ductal cells, which were present in the whole liver preparation prior to any manipulation. After low-G spins, we were able to detect only albumin, and the signals for CK7, desmin, vWF, or fucose receptor mRNAs were undetectable (Fig. 1A) indicating a virtual absence of other major cell types found in the liver. The purity of the hepatocyte prep was also confirmed morphologically by albumin and HNF-1α staining (Fig. 1B). Additionally, flow cytometric analysis showed that hepatocytes used in the LDPC cultures were over 99% pure (Fig. 1C).
Figure 1.
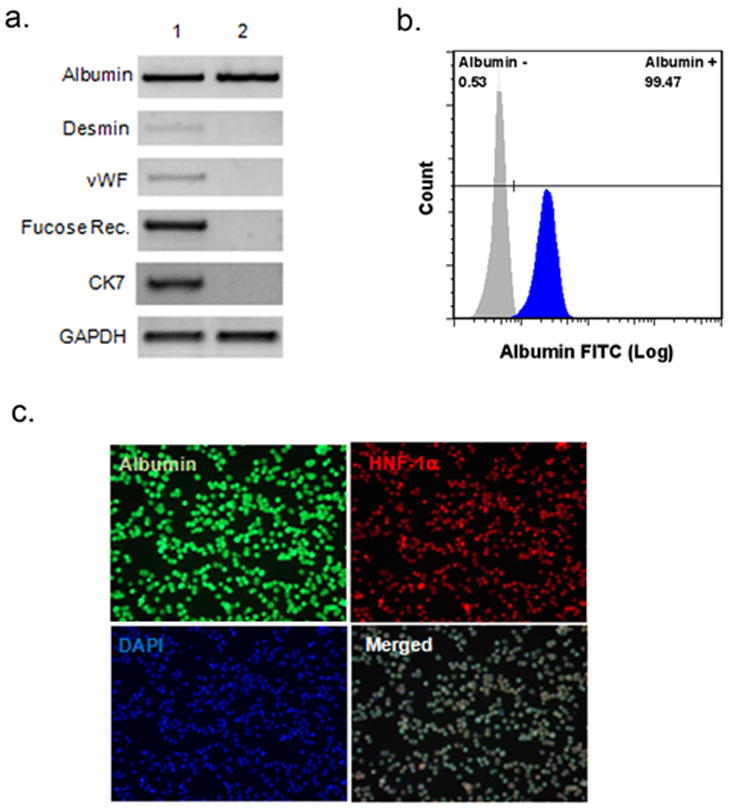
Purity of rat hepatocytes isolated by low-G centrifugation. (A) RT-PCR analysis of the cell purity. The fresh liver cells were subjected to RT-PCR before (lane 1) and after (lane 2) low-G spin for the hepatocyte marker albumin, stellate cell marker desmin, endothelial cell marker vWF, Kupffer cell marker fucose receptor, and biliary ductal cell marker CK7. After low-G spin, only hepatocyte marker albumin was detectable. (B) Flow cytometric analysis of hepatocyte purity using an anti-albumin antibody. Over 99% of the cells were albumin-positive. (C) Immunofluorescence staining of the of the same cell population. Albumin is stained with green, HNF-1α with red and cell nuclei with blue (DAPI) fluorescence. In the merged image, virtually all cells were triple positive consistent with a highly pure hepatocyte preparation (original magnification: 100x).
PKH2-labeled Rat Hepatocytes Give Rise to PKH2-labeled Liver Progenitor Cells
To track the fate of cultured hepatocytes, we initially labeled their cytoplasm with a fluorescent marker, PKH2 (Fig. 2A). The hepatocytes underwent drastic morphological changes including significant cell death in the first few days of culture. The remaining live cells either became flattened, forming cell clusters with many nuclei (polykaryons via possible endomitosis) or smaller as if they were undergoing apoptosis (cell shrinkage or condensation). Between days 5 and 7 of culture, LDPCs began to appear by either shrinkage of hepatocytes or by budding off from multinucleated cell clusters, a mechanism which is reminiscent of budding yeast (Fig. 2B). By day 14, LDPCs were the only cells left in culture with the exception of scattered fibroblast-like cells. The fluorescence images showed that virtually all LDPCs exhibited green fluorescence (i.e. PHK2 positive), which decreased over time. The results were consistent with the hypothesis that they were derived directly from PKH2-labeled hepatocytes and then underwent further cell divisions.
Figure 2.
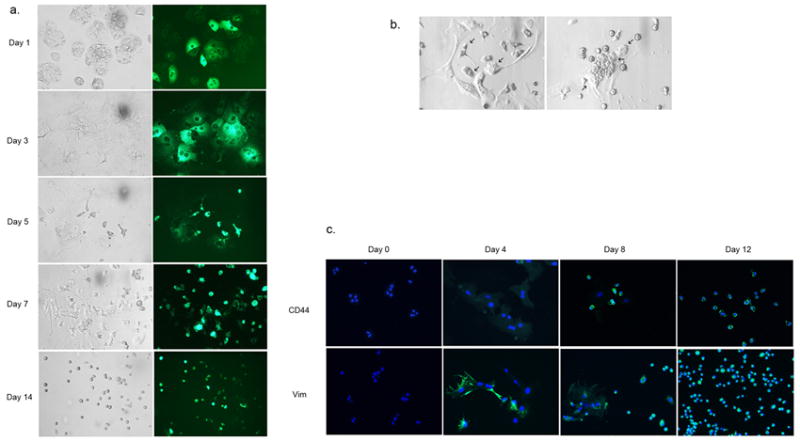
Dedifferentiation of rat hepatocytes into LDPCs. (A) Morphology of LDPCs cultures initiated with PKH2-stained (cytoplasmic) hepatocytes at various time points during the culture period. The panels on the left are the bright-field images of the cultures on the indicated days, the panels on the right are the corresponding fluorescence images. PKH2 positive LDPCs began to emerge from hepatocytes starting on day 5. By day 14, virtually all the cells in the culture were LDPCs (original magnification: 100x). (B) Potential mechanisms by which hepatocytes dedifferentiate into LDPCs. The panel on the left shows hepatocytes shrinking or undergoing condensation (arrows) to become LDPCs. The panel on the right demonstrates a single multinucleated dedifferentiating hepatocyte giving rise to LDPCs (cell membranes are forming around the cell nuclei) by what appears to be fragmentation of the cytoplasm and budding off (arrows, original magnification: 200x). (C) Expression of mesenchymal markers CD44 and vimentin during the culture period. These markers which were not present in hepatocytes on Day 0 became detectable around Day 4 and on Day 12, they were strongly positive in LDPCs suggesting that hepatocytes underwent an EMT during their transformation to LDPCs (original magnification: 100x).
The morphological changes in the LDPC cultures suggested transformation of hepatocytes (epithelial) into fibroblast-like cells (mesenchymal) before the appearance of LDPCs. Thus, we examined the expression of mesenchymal markers, CD44 and vimentin, in a time-dependent manner by the cells in culture. IF studies revealed that, while hepatocytes were negative for these mesenchymal markers on Day 0, the cells in the culture began to express both CD44 and vimentin around Day 4 and LDPCs were strongly positive for these markers on Day 12. This finding suggested that hepatocytes may be undergoing an Epithelial Mesenchymal Transition (EMT) before giving rise to LDPCs which appear to have a non-epithelial, mesenchymal phenotype.
To confirm our morphological findings and provide quantitative data, we examined the kinetics of LDPC cultures by performing cell count at certain time points during the culture period. This confirmed our previous observations showing that more than 80% of the plated hepatocytes died by Day 6 followed by rapid repopulation of the culture by LDPCs by Day 14 nearly restoring the original cell number (Sup.Fig.1A). Additionally, we performed a quantitative assessment of the total fluorescence of the cultured cells by flow cytometry as a further evidence for the origin of LDPCs. On days 1 and 14 of the LDPC cultures, we collected all the cells cultured within indentical flasks and measured their total fluorescence (Sup.Fig.1B). We found that non-hepatocyte cells constituted < 1% of all cells with a fluorescence intensity of 0.01 units (arbitrary units, total intensity of all cells on Day 1 was assigned a value of 1.0). The total fluorescence of LDPCs on day 14 averaged around 0.5 (average of three separate experiments), which was at least 50 times greater than the total fluorescence of non-hepatocyte cells on day 1. Considering that, PKH staining of a particular cells gets distributed among daughter cells and becomes undetectable after 6 to 8 cell divisions, it was highly improbable that a very small population of progenitors present in the initial cell prep could have expanded many times to give rise to LDPCs with significant fluorescence intensity. This result effectively ruled out the possibility that LDPCs could have originated from the initial non-hepatocyte cell population in the culture.
RT-PCR Analysis Confirms Rapid Transition of the Cells from Hepatocyte Phenotype to LDPC Phenotype During the Culture Period
Next, we wanted to substantiate our PKH staining results by documenting the phenotypic changes taking place during the transformation of hepatocytes into LDPCs. To that end, we performed RT-PCR and IF analyses of hepatocyte-specific and LDPC-specific markers at predetermined time points during the culture period. On days 0, 4, 8, and 12, the cultures were examined for expression of albumin, HNF-1α (hepatocyte-specific), CD45 and LMO2 (LDPC-specific). RT-PCR studies showed that in the beginning, the cells expressed albumin and HNF-1α and no identifiable CD45 and LMO2. By day 4, there was a rapid decline in hepatocyte specific markers and LDPC-specific markers became detectable at low levels. Subsequently, on days 8 and 12, hepatocyte markers became undetectable and LDPC markers were expressed at increasingly higher levels (Fig. 3A). IF studies revealed a similar pattern of marker expression, further confirming our RT-PCR data (Fig. 3B). In addition to these four markers, we examined the expression pattern of several other highly relevant hepatic genes during the culture period to better characterize the transformation process. We looked at the expression of mature hepatocyte markers HepPar1 and HNF-4α, immature hepatocyte marker Liv2 (24), biliary ductal /oval cell marker CK19 and liver progenitor/embryonic liver marker Sall4 (25) in a time-dependent manner. IF staining and quantitative analysis of the images revealed a pattern (Sup.Fig. 2 A and B) which was consistent with rapid transformation of mature hepatocytes into cells with liver progenitor phenotype thus supporting our findings shown in Fig.3. Both the RT-PCR and IF studies correlated well with the morphological changes that took place in the cultures including temporal appearance of LDPCs. Taken together, the rat studies strongly suggested that LDPCs originated from mature hepatocytes by direct dedifferentiation.
Figure 3.
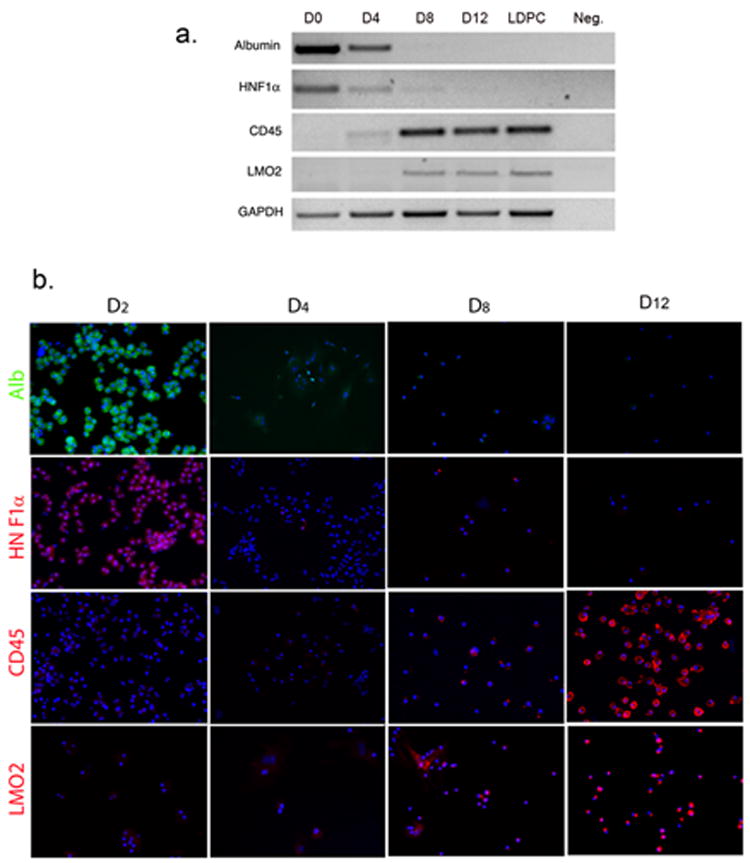
Analysis of hepatocyte-specific and LDPC-specific markers at various time points during the LDPC culture period. (A) RT-PCR for the hepatocyte markers; albumin and HNF-1α, and LDPCs markers; LMO2, CD45. On day 0, only hepatocyte markers were expressed and no signals for LDPC markers were detectable. Beginning around day 4, hepatocyte markers became weaker and virtually gone by day 8 while LDPC markers showed an opposite trend and became progressively stronger. Day 12 cultures and “pure” LDPCs obtained on day 14 by gentle EDTA treatment of the cultures showed no difference indicating that the transformation into LDPCs was completed (only the cell number increased after day 12). The lane marked Neg. shows the negative control reaction. (B) IF studies of the LDPC cultures for the same markers confirmed the RT-PCR results again showing rapid disappearance of hepatocyte markers by day 4 and progressively stronger expression of LDPC markers after day 8 (cell nuclei stained by DAPI in blue, original magnification, 100x). The observed patterns were consistent with the rapid transformation of hepatocytes into LDPCs observed morphologically in Figure 2.
Hepatocytes Go through an Oval Cell-like Stage en route to Becoming LDPCs
To gain further insight into the process of dedifferentiation of hepatocytes to LDPCs and to establish a stem/progenitor cell hierarchy, we examined the expression of several oval cell markers during the culture period. We considered the possibility that hepatocytes could be transitioning through an oval cell-like stage en route to becoming LDPCs. This was based on the phenotypic similarities between oval cells and LDPCs suggesting a potential lineage relationship. Therefore, we studied the expression of OV-6, CK7 and gamma glutamyl transaminase (GGT) during the dedifferentiation of hepatocytes into LDPCs. For this purpose, LDPC cultures initiated with pure hepatocytes were subjected to RT-PCR and IF staining for the aforementioned markers at predetermined time points during the culture period. RT-PCR analysis showed that CK7 expression, which was absent in the beginning first appeared around day 4, peaked on day 6 and then gradually declined and was undetectable in LDPCs by day 14. GGT first became detectable around day 6 and progressively increased in intensity only to become undetectable in LDPCs on day 14 (Fig. 4A). IF staining for these markers showed a very similar pattern to that seen with RT-PCR data with the exception that some GGT protein expression was detectable in LDPCs on day 14. Oval cell specific protein OV-6, on the other hand, was first detected by IF staining on Day 6, reached a peak on day 8 after which it rapidly decreased becoming virtually undetectable in LDPCs (Fig. 4B). The expression pattern of these markers correlated well with the morphological changes we observed in culture. Oval cell markers were upregulated as hepatocytes were in the process of transforming into progressively smaller cells and down-regulated as the LDPCs became the dominant cell type. In order to demonstrate that these changes took place in the same cell population, we performed co-staining for oval cell marker OV-6 and LDPC markers CD45 and LMO2 and found that on day 8, most of the cells co-expressed oval cell and LDPC markers (Fig. 4C). Taken together, these data strongly suggested that hepatocytes passed through an oval cell-like stage en route to becoming LDPCs.
Figure 4.
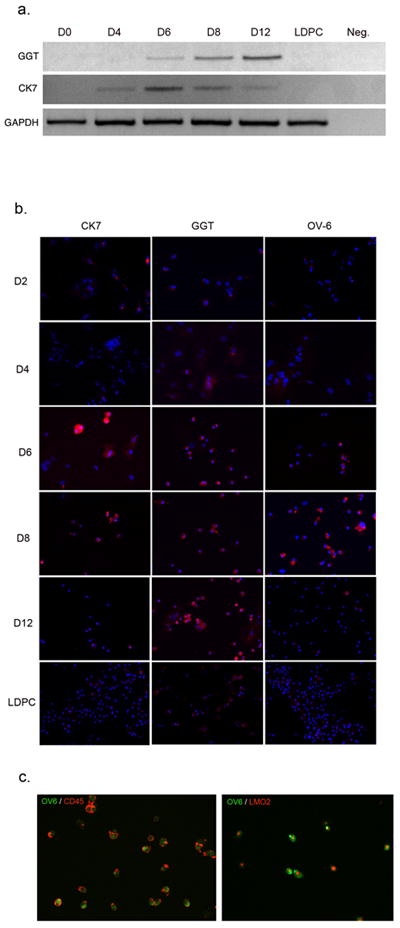
Transition of dedifferentiating hepatocytes through an oval cell-like stage en route to LDPCs. (A) RT-PCR analysis of two oval cell markers (also expressed by biliary ductal cells) CK7 and GGT at various time points during LDPC culture period. Both markers, which were not expressed initially, became detectable between days 4 and 6 at about the same time as the loss of hepatocyte-specific markers shown in Figure 3. They then peaked and subsequently became undetectable in “pure” LDPCs collected on day 14. The lane marked Neg. shows the negative control reaction. (B) IF analysis of the LDPC cultures during the same period for CK7, GGT and oval cell-specific protein OV-6. The expression pattern for CK7 and GGT confirms the pattern seen with RT-PCR analysis with the exception that GGT expression persisted in LDPCs at low level (most likely previously transcribed protein). OV-6 expression, on the other hand became detectable on day 6, reached its peak on day 8 and then gradually declined to undetectable levels in LDPCs on day 12 (original magnification, 100x). (C) Co-staining of the cells for LDPC markers CD45 and LMO2 and oval cell marker OV-6 on day 8 of the culture. Virtually all cells co-expressed LDPC markers and oval cell marker indicating a transient overlap between oval cell-like stage and LDPC stage. The combined data from RT-PCR and IF staining strongly suggests that hepatocytes transition through an oval cell-like stage before they turn into LDPCs (original magnification: 100x).
AlbCreXRosa26 Transgenic Mice Express β-galactosidase Exclusively in Hepatocytes
To provide additional evidence for the origin of LDPCs from hepatocytes in culture, we generated a double transgenic mouse strain by crossing AlbCre and Rosa26 mouse strains. As predicted, the resulting AlbCreXRosa26 mice expressed the enzyme, β-galactosidase only in the liver by western blot analysis (Fig. 5A). The hepatocyte-specific expression of this marker, which labeled albumin-expressing cells permanently, was confirmed by X-gal staining and IF staining for β-galactosidase. The results showed that expression of the reporter construct was restricted to hepatocytes (Fig. 5B).
Figure 5.
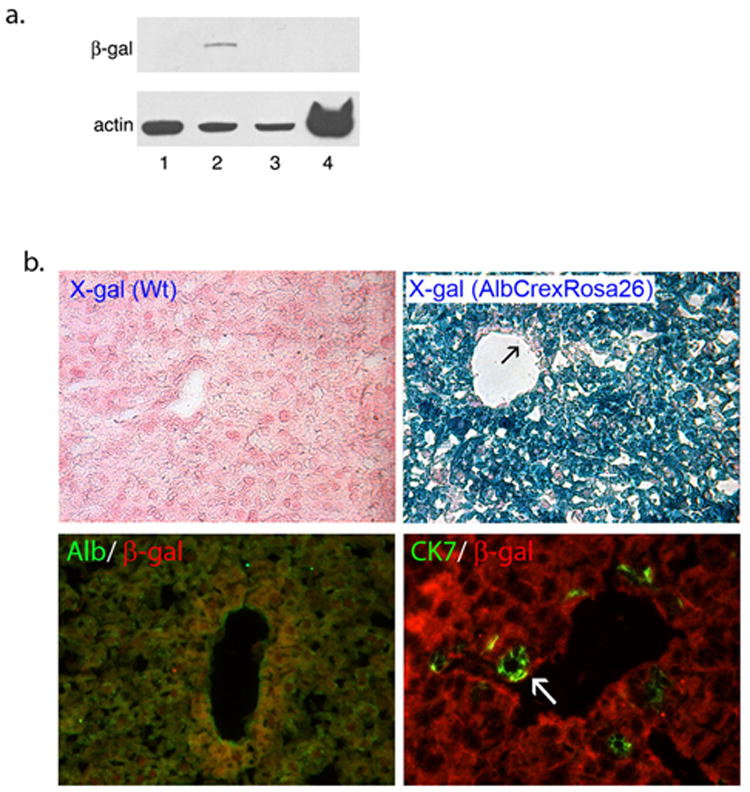
β-galactosidase expression in double transgenic AlbCreXRosa26 mice. (A) Western blot analysis of β-galactosidase expression. The blot showed that double transgenic kidney (lane 1), double transgenic spleen (lane 4) and wild-type liver (lane 3) did not express β-galactosidase where as double transgenic liver (lane 2) was positive. (B) X-gal and β-galactosidase IF staining of the double transgenic liver. X-gal staining of the wild-type liver did not show any reaction (left upper panel, original magnification: 100x) where as double transgenic liver strongly reacted. X-gal positivity appeared to be restricted to hepatocytes of the double transgenic mice as some cells including vascular endothelium (arrow) were x-gal negative (right upper panel, original magnification: 100x). Immunofluorescent co-staining for β-galactosidase and albumin (left lower panel, original magnification: 100x) showed complete overlap of the markers. On the other hand CK7 positive ductal cells (arrow) did not express β-galactosidase (right lower panel, original magnification: 200x). Overall data were consistent with tissue-specific (liver) and cell-specific (hepatocyte) expression of β-galactosidase in AlbCreXRosa26 mice.
Hepatocytes from AlbCreXRosa26 Mice Dedifferentiate into β-galactosidase Positive LDPCs
The next step was to examine LDPCs generated from AlbCreXRosa26 mice for β-galactosidase expression. The LDPC cultures of hepatocytes from double transgenic mice were subjected to X-gal staining at various time points which strongly suggested hepatocytes as the source of LDPCs (Fig.6A). In order to ensure that the small, round cells that appeared in the cultures were LDPCs, we performed co-staining for β-galactosidase and LDPCs markers, CD45 and LMO2. Virtually all cells co-expressed β-galactosidase and LDPC markers thus confirming the identity of the mouse hepatocyte-derived LDPCs (Fig. 6B).
Figure 6.
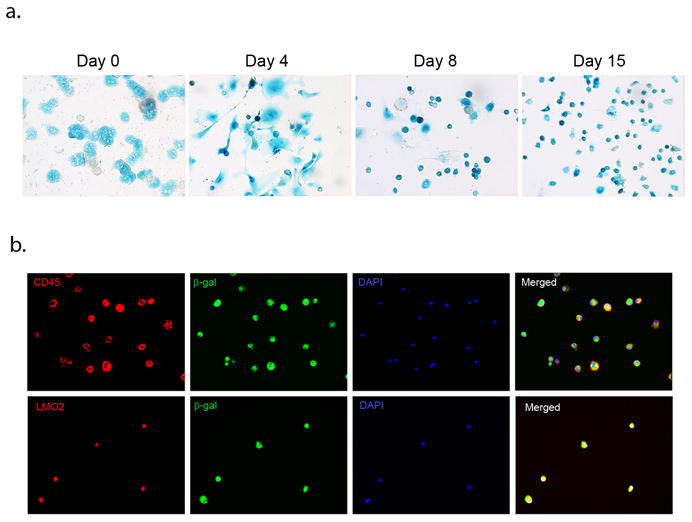
Analysis of β-galactosidase expression in LDPCs derived from double transgenic AlbCreXRosa26 mice. (A) X-gal staining of LDPC cultures on day 0 showed that nearly all hepatocytes were x-gal positive. On Day 4, dedifferentiating hepatocytes began to give rise to small x-gal positive cells which become more prominent by Day 8. On Day 15, virtually all cells in the culture were x-gal positive LDPCs (original magnification in all panels: 100x). (B) Confirmation of β-galactosidase positive cells as LDPCs. The round small cells that emerged in the cultures set up with hepatocytes from AlbCreXRosa26 mice were subjected to co-staining for β-galactosidase and LDPC markers CD45 and LMO2 (original magnification in all panels: 100x). The results showed complete overlap of the β-galactosidase and LDPC markers confirming that the cells were indeed LDPCs expressing β-galactosidase. These findings strongly support the hypothesis that LDPCs directly originate from hepatocytes.
LDPCs engraft and differentiate into hepatocytes in vivo
To underscrore the biological relevance of LDPCs, we performed transplantation experiment using rat LDPCs generated from male Fischer344 rats. We did a flow cytometric analysis of the harvested LDPCs using CD45 as a marker of LDPC purity which was > 97% (Sup.Fig.4A). Therefore, we proceeded with transplantation of these cells into female Fischer344 rats after LDPCs had been labeled with fluorescent dye PHK26 for tracking purposes. After two months, we examined the liver samples of the surviving three rats under fluorescence microscope before and after performing IF staining for albumin and FISH for Y-chromosome. Unstained sections revealed the presence of PKH+ cell clusters (approximately 1% of all cells) morphologically consistent with biliary ductal cells and hepatocytes (Fig. 7A, B, C). To confirm this finding, we then proceeded with IF staining for albumin and FISH for Y-chromosome which showed the presence of male hepatocytes (Fig. 7D, E, F and Sup.Fig.4C) in approximately 0.2 % of the cells examined. This is not an insignificant number in view of the fact that even in our positive control, male rat liver, (Sup.Fig.4B) only 2 % of the cells were positive for Y-chromosome. Taken together, our findings strongly suggested that LDPCs had engrafted and differentiated into hepatocytes in the recipient animals.
Figure 7.
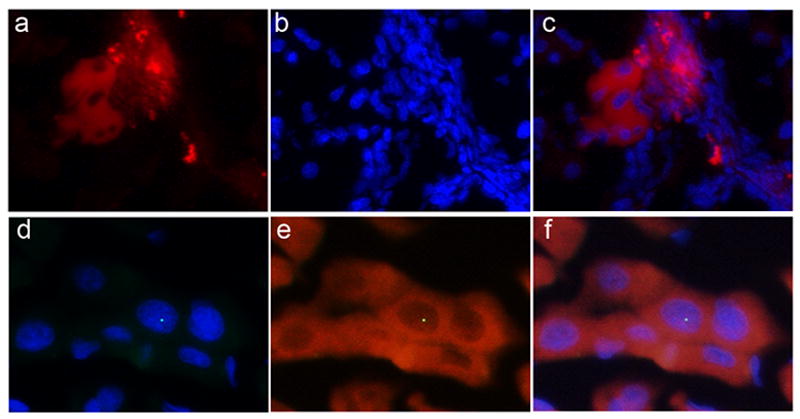
Hepatic differentiation of LDPCs in vivo. Fluorescent-labeled (PKH26) LDPCs from male Fischer rats were transplanted into female Fischer rats that had undergone retrorsine pretreatment and partial hepatectomy. Frozen liver section obtained from the recipient rats two months after the transplantation were examined under fluorescence microscope. (a) Unstained section showing PKH + cells, (b) DAPI nuclear staining, (c) merged images of a and b (original magnification: 100x). They showed foci of PKH+ cells morphologically consistent with biliary ductal cells and hepatocytes. To confirm that male LDPCs had engrafted and differentiated into hepatocytes in the recipients, we performed IF staining for albumin and FISH for Y-chromosome. (d) Merged images of DAPI nuclear staining and FISH (green dot) for Y-chromosome, (e) merged images of albumin staining (red) and FISH for Y-chromosome, (f) merged images of d and e (original magnification: 400x). These findings showed that transplanted LDPCs had engrafted in the liver and differentiated into hepatocytes
Discussion
The main objective of this study was to identify the origin of Liver Derived Progenitor Cells, which are unique bipotential adult hepatic progenitors first isolated and characterized by us (18). LDPCs, which are capable of forming mature hepatocytes both in vitro and in vivo, are generated in culture from normal liver tissues that have not been exposed to chemicals or any type of injury. This is in contrast to the many published protocols used to generate the quintessential hepatic progenitor oval cells. Therefore, LDPCs have a unique clinical application potential in humans. However, the source or the origin of these cells and therefore their lineage relationship to other cells in the liver is essentially unknown.
It is now well established that many adult tissues harbor stem cells or progenitors, which are capable of generating some or all of the cell types found in that particular tissue. Commonly referred to as “tissue-specific stem cells”, these cells have been identified in tissues including but not limited to heart, skin, brain, small intestine, mammary gland and teeth (26-31). In the adult liver, however, the situation is a bit more complex. This results from an extensive proliferative capacity of mature hepatocytes, which can regenerate the original liver mass even after 90% hepatectomy. However, when the degree of liver injury is very severe or when the liver has been exposed to certain toxins or chemicals, hepatocytes are unable to proliferate. It is under these conditions that the hepatic stem/progenitor compartment is activated. The most-widely known and characterized liver progenitors are oval cells. They are believed to have primary hepatic origin and thought to reside in small numbers in the terminal bile ducts. It is widely accepted, and perhaps even assumed, that hepatocytes do not contribute directly to this progenitor or stem cell compartment.
Our earlier observations on the production of LDPCs, and the rapid replacement of primary hepatocytes by hepatic progenitors in cultures suggested that hepatocytes might in fact, be the original source of the hepatic progenitor cells. To test this hypothesis, we designed a series of experiments generating LDPCs from two different species. First, we obtained an extremely pure population of hepatocytes from rat liver. Then, by fluorescently labeling and documenting the evolution of LDPC cultures set up with these hepatocytes, we were able to show that the LDPCs were also fluorescently labeled, strongly suggesting that they were directly derived from hepatocytes. The quantitative flow cytometric analysis of the initial and the resultant cell populations indicated that hepatocytes were the only logical source for LDPCs.
Next, we wanted to show the origin of LDPCs in a transgenic mouse model, which would allow us to track the fate of hepatocytes. To accomplish this, we generated a double transgenic AlbCreXRosa26 mouse strain, which was previously generated and published by others (32). Again, using several different techniques, we showed that these animals activated and expressed β-galactosidase only in hepatocytes. Using purified hepatocytes from these animals, we found that the LDPCs were also β-galactosidase positive supporting the conclusion that they were derived directly from hepatocytes. These studies also showed two potentially unique mechanisms by which hepatocytes dedifferentiate into hepatic progenitors. One is by cell condensation or shrinkage, which is morphologically similar to cells undergoing apoptosis; the other is budding off from multinucleated cell clusters reminiscent of cell division in yeast (33, 34). It will be important to confirm these observations by future studies. Another interesting finding was the up-regulation of mesenchymal markers CD44 and vimentin during this dedifferentiation process suggesting an epithelial-mesenchymal-transition. This is particularly intriguing given the known role of EMT in liver development and regeneration (35-37). However, the confirmation of EMT in generation of LDPCs requires further studies. The other somewhat unexpected finding was the transient expression of some oval cell markers as hepatocytes dedifferentiated into LDPC. Based on co-expression of OV-6 and LDPC markers by the majority of cells in culture, we believe that there is a single population of hepatic progenitors that are generated through a transient oval cell-like stage from hepatocytes.
Having shown the ability of mature hepatocytes to dedifferentiate into LDPCs in vitro, we tested the potential biological relevance by demonstrating their ability to regenerate hepatocytes in an animal model. Therefore, we proceeded with a well-established transplant protocol consisting of retrorsine pretreatment and partial hepatectomy in rats. The presence of both PKH-labeled and Y-chromosome positive hepatocytes in the recipient animals convincingly showed that transplanted LDPCs were able to engraft and re-differentiate into hepatocytes in vivo. This finding raises the question whether, under certain circumstances where the microenvironment is not conducive to proliferation of hepatocytes, the process of dedifferentiation of hepatocytes into progenitors followed by proliferation and re-differentiation is a potential mechanism for organ regeneration in the liver. It is also conceivable that the process of dedifferentiation of hepatocytes via EMT may also be playing a role in tumor formation under pathological conditions (38). Even though, we do not have any direct evidence, some published studies provide indirect clues for the presence of LDPC-like cells in vivo. Sell et al. reported proliferation of small, OV-6 negative intraportal progenitors termed “null cells” as a restitutive response to allyl alcohol-induced periportal necrosis in the rat liver (39, 40). In these studies, null cells later began to express OV-6 and eventually differentiated into mature hepatocytes. Based on morphological similarities between null cells and LDPCs, and the sequence of phenotypic changes null cells undergo to become hepatocytes, it is tempting to speculate that null cells may be the in vivo counterpart of LDPCs.
In summary, the combination of studies using primary rat and transgenic mouse hepatocytes has allowed us to trace the putative origin of LDPCs, a population of liver progenitors. Our results strongly suggest that they arise from direct dedifferention of mature hepatocytes in culture. This finding has a number of major implications. First, it shows that hepatocytes are far more plastic than previously thought and potentially capable of contributing directly to the stem/progenitor cell pool of the liver. Second, it confirms that a fully mature, terminally differentiated somatic cell can acquire a stem/progenitor cell phenotype without genetic or epigenetic manipulation. While our studies have demonstrated this in liver, similar differentiation and dedifferentiation properties may exist in other organs and tissues. Finally, it has a significant potential impact on the treatment of liver diseases requiring liver or hepatocyte transplantation.
Supplementary Material
S. Figure 1. (A) Cell kinetics of LDPC cultures. In addition to significant morphological changes, the cells in the LDPC cultures underwent drastic changes in number. Greater than 80% of the cells died by Day 6 followed by rapid repopulation of the culture by LDPCs with the cell number being nearly restored back to the original by Day 14. (B) Flow cytometric analysis of total fluorescence (PKH2) of the cells on day 1 and day 14. Total fluorescence of the cells on day 1 was assigned an arbitrary unit of 1.0 of which < 1% was due to non-hepatocytes. On the same scale, the total fluorescence of LDPCs on day 14 was 0.5 (average of three experiments) which was 50-fold higher than the fluorescence of the non-hepatocyte fraction on day 1. This implies that most, if not all, of the LDPCs originated from hepatocytes in the initial culture.
S. Figure 2. Expression of several mature and immature hepatic markers at certain time points during LDPC culture period. (A) In order to better understand the changes that took place during the dedifferentiation process and better characterize the cell populations present, we examined the expression of some markers found in mature hepatocytes (HNF4α and HepPar1), ductal cells and some liver progenitors (CK19) and those found in immature hepatocytes (Liv2) and embryonic liver cells (Sall4) by IF staining. (B) Quantitative analysis of the IF studies for the same markers. Quantification for Liv2 and CK19 showed they were not detectable in the cultures at any point. Overall expression pattern suggested that initial cell population consisted of mature hepatocytes that rapidly dedifferentiated into a cell population which had a liver progenitor phenotype supporting our findings described in Figures 2, 3 and 4.
S. Figure 3. Negative control images for various markers examined in IF studies. These are merged IF images of the marker of interest with the DAPI staining of the same field except for the two nuclear markers (HNF4α and LMO2) which were not stained with DAPI.
S. Figure 4. Transplantation of LDPCs from male Fischer rats into female recipients. (A) Flow cytometric analysis of transplanted cells for CD45 expression revealed that > 97% of the cells were positive indicating it was an extremely pure LDPC population (light and dark grey histograms show unstained and isotype control samples respectively). (B) Control samples for Y-chromosome FISH labeling showed that female rat liver did not show any labeling whereas male liver was positive (green dots) as expected. The images on the left are FISH for Y-chromosome, the images in the middle shows DAPI staining of the same field and the images on the right are merged images of the two (original magnification: 200x). (C) Additional Y-chromosome FISH images of the female recipient livers showing the presence of Y-chromosome containing, albumin positive hepatocytes as in Figure 7 (original magnification: 400x). This clearly shows that transplanted male LDPCs engrafted and differentiated into hepatocytes.
Acknowledgments
Financial Support Y.C and M.B.S. were supported by intramural research funds from the Department of Medicine, University of Minnesota. P.P.W, L.S. and C.J.S. were supported, in part by National Institutes of Health ARRA Grant R01 DK081865-01.
List of Abbreviations
- ES
Embryonic Stem Cells
- iPS
Induced Pluripotent Stem Cells
- c-kit
Stem cell receptor, CD117
- Thy-1
Cell surface antigen, CD90
- PBS
Phosphate Buffered Saline
- SSC
Saline Sodium Citrate buffer
- LMO2
Lim domain only-2 transcription factor
- PE
Phycoerythrin
- OV-6
Oval cell-specific surface antigen
- PKH2
Green fluorescent cell linker
- FITC
Fluoroisothiocyanate, green fluorescent marker
- Sall4
Sal-like zinc-finger transcription factor 4
- HepPar1
Hepatocyte Parafin-1 antigen
References
- 1.Stocker E, Heine WD. Regeneration of liver parenchyma under normal and pathological conditions. Beitr Pathol. 1971;144:400–408. [PubMed] [Google Scholar]
- 2.Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Poli V, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 3.Van Eyken P, Sciot R, Desmet V. Intrahepatic bile duct development in the rat: a cytokeratin immunohistochemical study. Lab Invest. 1988;59:52–59. [PubMed] [Google Scholar]
- 4.Xiao JC, Ruck P, Adam A, Wang TX, Kaiserling E. Small epithelial cells in human liver cirrhosis exhibit features of hepatic stem-like cells: immunohistochemical, electron microscopic and immunoelectron microscopic findings. Histopathology. 2003;42:141–149. doi: 10.1046/j.1365-2559.2003.01544.x. [DOI] [PubMed] [Google Scholar]
- 5.Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, et al. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. The origin and liver repopulating capacity of murine oval cells. Proc Natl Acad Sci USA. 2003;100(Suppl. 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menthena A, Deb N, Oertel M, Grozdanov PN, Sandhu J, Shah S, Guha C, Shafritz DA, Dabeva MD. Bone marrow progenitors are not the source of expanding oval cells in injured liver. Stem Cells. 2004;22:1049–1061. doi: 10.1634/stemcells.22-6-1049. [DOI] [PubMed] [Google Scholar]
- 8.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 9.Petersen BE, Goff JP, Greenberger JS. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology. 1998;27:433–445. doi: 10.1002/hep.510270218. [DOI] [PubMed] [Google Scholar]
- 10.Matsusaka S, Toyosaka A, Nakasho K. The role of oval cells in rat hepatocyte transplantation. Transplantation. 2000;70:441–446. doi: 10.1097/00007890-200008150-00009. [DOI] [PubMed] [Google Scholar]
- 11.Hamazaki T, Iiboshi Y, Oka M, Papst PJ, Meacham AM, Zon LI, et al. Hepatic maturation in differentiating embryonic stem cells in vitro. FEBS Lett. 2001;497:15–19. doi: 10.1016/s0014-5793(01)02423-1. [DOI] [PubMed] [Google Scholar]
- 12.Chinzei R, Tanaka Y, Shimizu-Saito K, Hara Y, Kakinuma S, Watanabe M, et al. Embryoid-body cells derived from a mouse embryonic stem cell line show differentiation into functional hepatocytes. Hepatology. 2002;36:22–29. doi: 10.1053/jhep.2002.34136. [DOI] [PubMed] [Google Scholar]
- 13.Gai H, Nguyen DM, Moon YJ, Aguila JR, Fink LM, Ward DC, et al. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 2010;79:171–181. doi: 10.1016/j.diff.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, et al. Liver from bone marrow in humans. Hepatology. 2000;32:11–16. doi: 10.1053/jhep.2000.9124. [DOI] [PubMed] [Google Scholar]
- 15.Theise ND, Badve S, Saxena R, Henegariu O, Sell S, Crawford JM, et al. Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology. 2000;31:235–240. doi: 10.1002/hep.510310135. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, et al. Cell fusion is the principal source of bone marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- 17.Vassilopoulos G, Wang PR, Russell DW. Transplanted bone marrow regenerates liver by cell fusion. Nature. 2003;422:901–904. doi: 10.1038/nature01539. [DOI] [PubMed] [Google Scholar]
- 18.Sahin MB, Schwartz RE, Buckley SM, Heremans Y, Chase L, Hu WS, et al. Isolation and characterization of a novel population of progenitor cells from unmanipulated rat liver. Liver Transpl. 2008;14:333–345. doi: 10.1002/lt.21380. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Zhou H, Sarver AL, Zeng Y, Roy-Chowdhury J, Steer CJ, Sahin MB. Hepatic differentiation of liver-derived progenitor cells and their characterization by microRNA analysis. Liver Transpl. 2010;16:1086–1097. doi: 10.1002/lt.22111. [DOI] [PubMed] [Google Scholar]
- 20.Geerts A, Lazou JM, De Bleser P, Wisse E. Tissue distribution, quantitation and proliferation kinetics of fat-storing cells in carbon tetrachloride-injured rat liver. Hepatology. 1991;13:1193–1202. [PubMed] [Google Scholar]
- 21.Yokoi Y, Namihisa T, Kuroda H, Komatsu I, Miyazaki A, Watanabe S, et al. Immunocytochemical detection of desmin in fat-storing cells (Ito cells) Hepatology. 1984;4:709–714. doi: 10.1002/hep.1840040425. [DOI] [PubMed] [Google Scholar]
- 22.Hoyle GW, Hill RL. Molecular cloning and sequencing of a cDNA for a carbohydrate binding receptor unique to rat Kupffer cells. J Biol Chem. 1988;263:7487–7492. [PubMed] [Google Scholar]
- 23.Kinugasa Y, Nakashima Y, Matsuo S, Shono K, Suita S, Sueishi K. Bile ductular proliferation as a prognostic factor in biliary atresia: an immunohistochemical assessment. J Pediatr Surg. 1999;34:1715–1720. doi: 10.1016/s0022-3468(99)90652-8. [DOI] [PubMed] [Google Scholar]
- 24.Takashimizu I, Tanaka Y, Yoshie S, Kano Y, Ichikawa H, Cui L, Ogiwara N, Johkura K, Sasaki K. Localization of Liv2 as an immature hepatocyte marker in EB outgrowth. Scientific World Journal. 2009;9:190–9. doi: 10.1100/tsw.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136(3):1000–11. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 27.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Regeneration of a germinal layer in the adult mammalian brain. Proc Natl Acad Sci USA. 1999;96:11619–11624. doi: 10.1073/pnas.96.20.11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 30.Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 31.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A. 2003;100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisberg M, Yang C, Martino M, Duncan MB, Rieder F, Tanjore H, et al. Fibroblasts derived from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–23347. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 33.Hooser SB. Fulminant hepatocyte apoptosis in vivo following microcystin-LR administration to rats. Toxicol Pathol. 2000;28:726–733. doi: 10.1177/019262330002800513. [DOI] [PubMed] [Google Scholar]
- 34.Gladfelter AS, Hungerbuehler AK, Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J Cell Biol. 2006;172(3):347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sicklick J, Choi SS, Bustamante M, McCall SJ, Perez EH, Huang J, Li YX, et al. Evidence for epithelial-mesenchymal transitions in adult liver cells. Am J Physiol Gastrointest Liver Physiol. 2006;291:G575–G583. doi: 10.1152/ajpgi.00102.2006. [DOI] [PubMed] [Google Scholar]
- 36.Kaimori A, Potter J, Kaimori JY, Wang C, Mezey E, Koteish A. Transforming growth factor-beta1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro. J Biol Chem. 2007;282:220. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 37.Omenetti A, Yang L, Li YX, McCall SJ, Jung Y, Sicklick JK, Huang J, et al. Hedgehog-mediated mesenchymal-epithelial interactions modulate hepatic response to bile duct ligation. Lab Invest. 2007;87:499–514. doi: 10.1038/labinvest.3700537. [DOI] [PubMed] [Google Scholar]
- 38.Choi SS, Diehl AM. Epithelial to mesenchymal transitions in the liver. Hepatology. 2009;50(6):2007–2013. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yavorkovsky L, Lai E, Ilic Z, Sell S. Participation of small intraportal stem cells in the restitutive response of the liver to periportal necrosis induced by allyl alcohol. Hepatology. 1995;21:1702–1712. [PubMed] [Google Scholar]
- 40.Yin L, Lynch D, Sell S. Participation of different cell types in the restitutive response of the rat liver to periportal injury induced by allyl alcohol. J Hepatol. 1999;31:497–507. doi: 10.1016/s0168-8278(99)80043-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Figure 1. (A) Cell kinetics of LDPC cultures. In addition to significant morphological changes, the cells in the LDPC cultures underwent drastic changes in number. Greater than 80% of the cells died by Day 6 followed by rapid repopulation of the culture by LDPCs with the cell number being nearly restored back to the original by Day 14. (B) Flow cytometric analysis of total fluorescence (PKH2) of the cells on day 1 and day 14. Total fluorescence of the cells on day 1 was assigned an arbitrary unit of 1.0 of which < 1% was due to non-hepatocytes. On the same scale, the total fluorescence of LDPCs on day 14 was 0.5 (average of three experiments) which was 50-fold higher than the fluorescence of the non-hepatocyte fraction on day 1. This implies that most, if not all, of the LDPCs originated from hepatocytes in the initial culture.
S. Figure 2. Expression of several mature and immature hepatic markers at certain time points during LDPC culture period. (A) In order to better understand the changes that took place during the dedifferentiation process and better characterize the cell populations present, we examined the expression of some markers found in mature hepatocytes (HNF4α and HepPar1), ductal cells and some liver progenitors (CK19) and those found in immature hepatocytes (Liv2) and embryonic liver cells (Sall4) by IF staining. (B) Quantitative analysis of the IF studies for the same markers. Quantification for Liv2 and CK19 showed they were not detectable in the cultures at any point. Overall expression pattern suggested that initial cell population consisted of mature hepatocytes that rapidly dedifferentiated into a cell population which had a liver progenitor phenotype supporting our findings described in Figures 2, 3 and 4.
S. Figure 3. Negative control images for various markers examined in IF studies. These are merged IF images of the marker of interest with the DAPI staining of the same field except for the two nuclear markers (HNF4α and LMO2) which were not stained with DAPI.
S. Figure 4. Transplantation of LDPCs from male Fischer rats into female recipients. (A) Flow cytometric analysis of transplanted cells for CD45 expression revealed that > 97% of the cells were positive indicating it was an extremely pure LDPC population (light and dark grey histograms show unstained and isotype control samples respectively). (B) Control samples for Y-chromosome FISH labeling showed that female rat liver did not show any labeling whereas male liver was positive (green dots) as expected. The images on the left are FISH for Y-chromosome, the images in the middle shows DAPI staining of the same field and the images on the right are merged images of the two (original magnification: 200x). (C) Additional Y-chromosome FISH images of the female recipient livers showing the presence of Y-chromosome containing, albumin positive hepatocytes as in Figure 7 (original magnification: 400x). This clearly shows that transplanted male LDPCs engrafted and differentiated into hepatocytes.


