Abstract
The production of virus by infected cells is an essential process for the spread and persistence of viral diseases, the effectiveness of live-viral vaccines, and the manufacture of viruses for diverse applications. Yet despite its importance, methods to precisely measure virus production from cells are lacking. Most methods test infected-cell populations, masking how individual cells behave. Here we measured the kinetics of virus production from single cells. We combined simple steps of liquid-phase infection, serial dilution, centrifugation, and harvesting, without specialized equipment, to track the production of virus particles from BHK cells infected with vesicular stomatitis virus. Remarkably, cell-to-cell differences in latent times to virus release were within a factor of two, while production rates and virus yields spanned over 300-fold, highlighting an extreme diversity in virus production for cells from the same population. These findings have fundamental and technological implications for health and disease.
Keywords: single-cell sampling, virus-cell interactions, cell heterogeneity, kinetics, stochastic, intracellular resources, vesicular stomatitis virus, virus yield
INTRODUCTION
When a virus infects a cell, it hijacks resources of the cell to manufacture and release a new generation of progeny virus particles. The resulting cycle of virus production at the cellular level is an essential part of diverse natural and engineered processes, including: the spread and persistence of infectious diseases such as AIDS, influenza, and the common cold (Flint et al., 2009; Levin et al., 1999); the activation of immune responses within virus-infected hosts (Murphy et al., 2008); the design of attenuated viruses for vaccines (Lim et al., 2006); the use of viruses to treat cancers (Melcher et al., 2011), and the large-scale production of viruses for health applications (Lohr et al., 2010). Yet despite its central importance, methods to precisely quantify virus production at the cellular level are lacking. Most approaches measure the production of virus by sampling from a population of infected cells, providing average values that mask the potentially wide-ranging and significant behaviors of individual cells.
Within a laboratory culture of virus-infected cells, or within a tissue of an infected host, individual cells can diverge in behavior from the average or majority of infected cells. However, these rare cells may nevertheless contribute importantly to the long-term behavior of the infection, well beyond their initial encounter with the virus. For example, in the presence of an anti-viral drug, some cells may produce higher than average levels of virus progeny, behavior that can arise from infection by drug-resistant virus (Larder et al., 1990). Over multiple cycles of growth, selection of such drug-resistant viruses can enable them to dominate the virus population (Condra et al., 1995), which may contribute toward the progression and spread of disease. Alternatively, in culture only a small fraction of virus-infected dendritic cells appear to trigger an initial innate defensive response, which is subsequently amplified to create an anti-viral state across a majority of cells (Hu et al., 2011). Moreover, the activation of such innate immune responses in vivo shapes the development of adaptive immune responses that impact recovery from infection and establishment of long-term immunity (Guidotti and Chisari, 2001). Further, some infected cells may adopt a state of non-progressive infection or latency, permitting them to evade clearance by immune defenses until re-activation by environmental or other cues (Kutsch et al., 2002). These examples suggest how a minority of initial infected cells, in vitro or in vivo, may deviate from the average infected-cell behavior yet contribute disproportionately, over the longer-term, to the extent or nature of infection spread. Because these rare-cell behaviors are generally obscured during average-cell measures of infection, they highlight the need for single-cell measures of behavior that can be readily performed on many individual cells to reveal the extent of cell heterogeneity.
To date, limited studies have focused on the production of virus from single infected cells. In a classic study Burnet incubated and plated diluted phage stocks with excess bacteria and quantified phage released from different samples at different time points, providing early evidence for the step-like production of phage from their host cells (Burnet, 1929). Further, the study of phage-infected bacterial cells allowed measures of total ‘burst sizes’ of phage progeny from multiple individual cells, revealing a broad distribution from below 50 to over 1000 phage per cell (Delbrück, 1945). We extended these approaches to a mammalian RNA virus, vesicular stomatitis virus (VSV), by infecting single cells with single virus particles, and we found infected cells produced yields from below 50 to over 8000 virus particles (Zhu et al., 2009b). Further, Cuevas et al studied the kinetics of VSV production from individual infected cells that were sequentially sampled and analyzed, providing initial release profiles of virus progeny (Cuevas et al., 2005). However, this study allowed for production of virus from secondary infections, limiting interpretation of single-cell behavior only to the earliest stages of virus release.
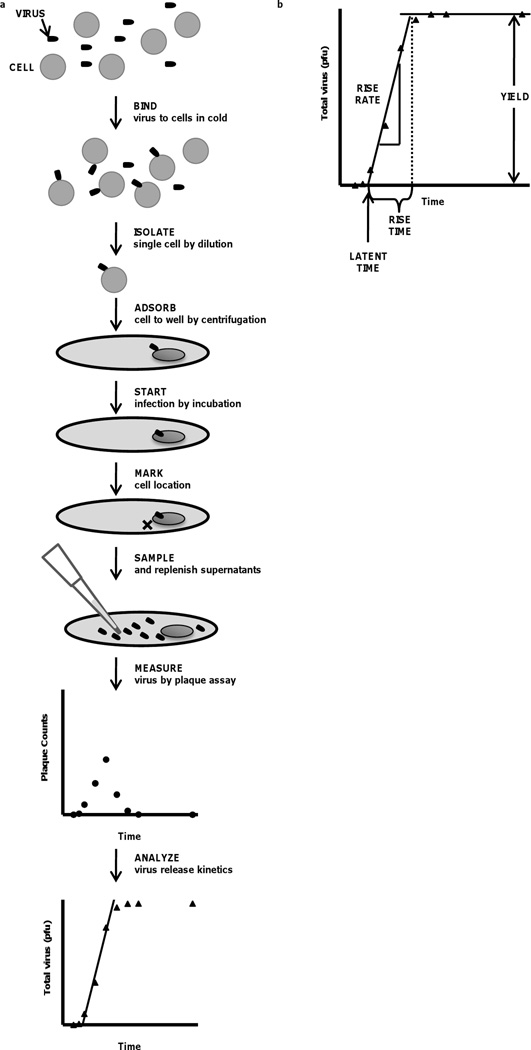
Here we demonstrate a method to measure the kinetics of virus production from individual cells, without confounding effects from secondary infections. Our method combines a series of simple steps that can be performed in any cell biology or virology facility, without reliance on specialized equipment (Fig. 1a). We isolate single infected cells, perform multiple harvest-and-replenish steps on the liquid medium in the cell’s environment, followed by measurement of infectious virus titer by plaque counting, enabling us to construct a kinetic profile of virus production for each cell. Integration of the virus counts gives a cumulative production or “one-step” curve, from which we calculate quantitative features of virus production from each cell (Fig. 1b): LATENT TIME – the time interval between entry of adsorbed virus into its host cell and initial release of virus progeny, RISE TIME – the time interval over which the cell produces virus, YIELD – the cumulative total of virus produced by the infected cell, and RISE RATE – the average rate of virus production from a cell (YIELD / RISE TIME). We demonstrate our method by infecting BHK cells with vesicular stomatitis virus (VSV), a model RNA virus. VSV is the focus of on-going studies that are advancing an integrated understanding of how virus-cell interactions impact the kinetics of virus growth and infection spread, as well as the evolutionary dynamics of virus populations (Cuevas et al., 2005; Haseltine et al., 2008; Lam et al., 2005; Lim et al., 2006; Novella et al., 1995; Turner and Elena, 2000; Zhu et al., 2009b).
Figure 1.
Kinetics of virus release from single infected cells. (a) Our method combines steps of virus infection, cell immobilization by adsorption, multi-time sampling, and analysis, and (b) one-step virus release profiles, from which one can estimate parameters of single-cell virus production: latent time, rise rate, yield and rise time.
RESULTS
Virus production rates from single cells
BHK cells were released from their culture surfaces by treatment with trypsin, counted, cooled on ice, and mixed with excess VSV (MOI 5) in the cold, to enable virus to bind to cell surfaces without fusion and entry. Infected cells were serially diluted so an aliquot containing on average less than one cell per well could be added to each well of a 96-well plate, plates were subjected to centrifugation to adhere cells to the bottom surface of each well, and wells containing single cells were identified by microscopy. Cell locations within each well were marked to indicate areas to avoid disruption during sample-replenish steps, and plates were incubated to initiate infections. A total harvest and replenishment of liquid media was performed at set intervals to capture the released virus progeny from each cell, and virus within each sample was measured by plaque assay.
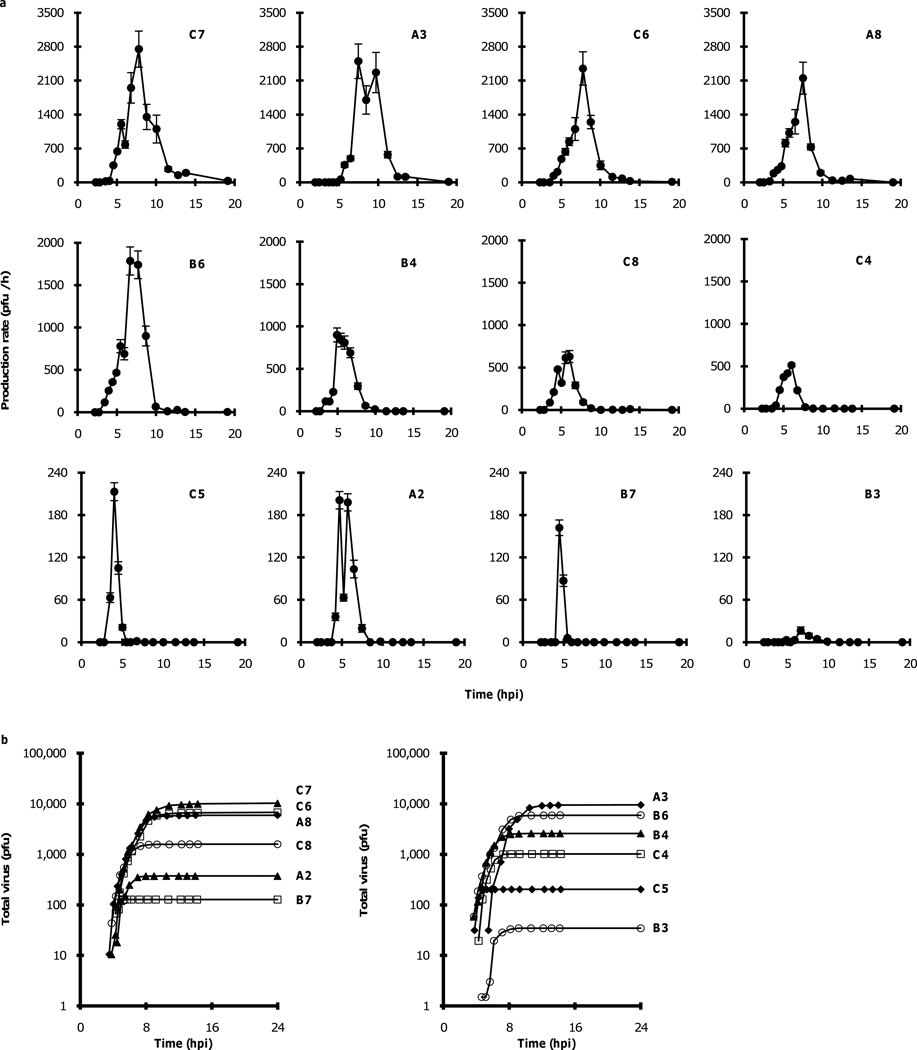
From a sampling of twenty-four wells, each containing a single cell, 12 wells produced plaque counts from which rates of virus release were calculated over the full time-course of the virus growth cycle (Fig. 2a). Cells initiated release of virus between 3.5 and 6.5 hours post infection (hpi), and once initiated, different cells released virus progeny at vastly different rates, spanning from 10 to 3000 plaque-forming units (pfu) per hour, with an average maximum production rate of 1200 ± 1000 pfu per hour (± 1 s.d.). Generally, cells produced virus between 4 and 12 hpi, and in most cases (8 of 12) virus production rates exhibited a single-peaked value. Possible mechanisms for two-peak production, apparent in Fig. 2a (C7, A3, C8, A2), would include infections initiated from a cell that subsequently divided, so supernatant sampling could reflect the sum of different virus production profiles from two cells. Alternatively, the two-peak behavior might reflect the complex coupling between intracellular processes that promote virus production and host cellular defenses that may inhibit virus production. By calculating the cumulative release of virus particles at each sample time we generated one-step virus production curves for single cells (Fig 2b). Here, 12 infected cells produced an average yield of 3700 ± 3800 pfu (± 1 s.d.), with virus production from individual cells spanning from 30 to 10,000 pfu, a range of more than 300-fold.
Figure 2.
Virus production profiles for virus-infected cells. (a) Production rate profiles for 12 cells drawn from the same initial population of infected cells, and (b) one-step production of virus from single cells.
Although each cell was exposed to an excess of five infectious virus particles (MOI 5), not all cells were productive. Several factors may be relevant. First, our conditions for solution-phase adsorption of virus particles to cells may be insufficient to ensure that every cell is infected. Second, virus that initiates infection may not carry through the process to production of progeny. We have previously found that cells infected by a recombinant GFP-virus could produce GFP without producing detectable virus (Zhu et al., 2009b), indicating infections may terminate after initial gene expression. Finally, cells may have been inadvertently removed during sampling. Our method, which aims to minimize loss or removal of cells by sampling distant from their initial adsorption location, may nevertheless be too disruptive to retain each cell over the entire sampling period.
Integrated kinetics of virus production
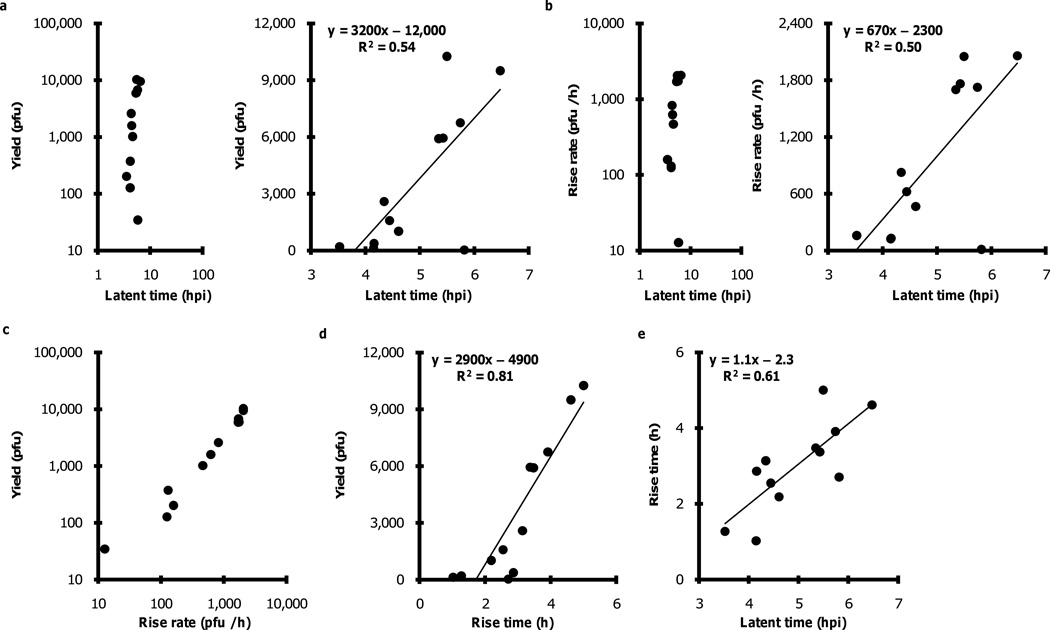
For each of 12 cells we estimated the latent time, rise rate, yield and rise time from its one-step production curve. Plots of yield or rise rate versus latent time show orders of magnitude variation in yield and rise rate, while latent times were narrowly distributed (Figs. 3a/b, left panels). On linear plots longer latent times corresponded to both higher yields and higher rise rates (Figs. 3a/b, right panels). Yields strongly correlated with rise rates (Fig. 3c), a result that follows from the definition of rise rate (yield per rise time), the very broad distribution of yields, and the relatively narrow distribution of rise times. Yields correlated with rise times, where higher yields corresponded with longer rise times (Fig. 3d). Here it was also apparent that more productive cells were not as variable in their rise times as the less productive cells; specifically, cells producing fewer than 1000 pfu spanned a broader range of rise times than cells producing higher yields. Finally, cells that initiated release of virus progeny later also spent more time actively producing virus (Fig. 3e).
Figure 3.
Correlations among virus-release traits from single cells. Cells that produce virus earlier (shorter latent times) tend to produce fewer virus particles at slower rates as indicated by (a) yield vs latent time and (b) rise rate vs latent time; (c) yields and rise rates are correlated owing to relatively narrow distributions among rise times, (d) larger yields require a longer period of virus production, and (e) earlier producers tend to be productive for shorter periods than later producers.
Error propagation in the plaque assay
Our estimated propagation of error from pipetting and plaque counting was comparable to the observed error over a wide range of virus concentrations that included more than 90 percent of the measurements in this study. Observed errors only exceeded estimated errors at lowest concentrations, below 4 pfu per 100 µl, representing less than 10 percent of the one-step infection measurements in this study. The single-cell one-step infections were minimally affected by this result. An exception was the single-cell that produced only 34 pfu, where the actual yield could be up to 40 percent higher.
Dependence of single-cell yield on multiplicity of infection
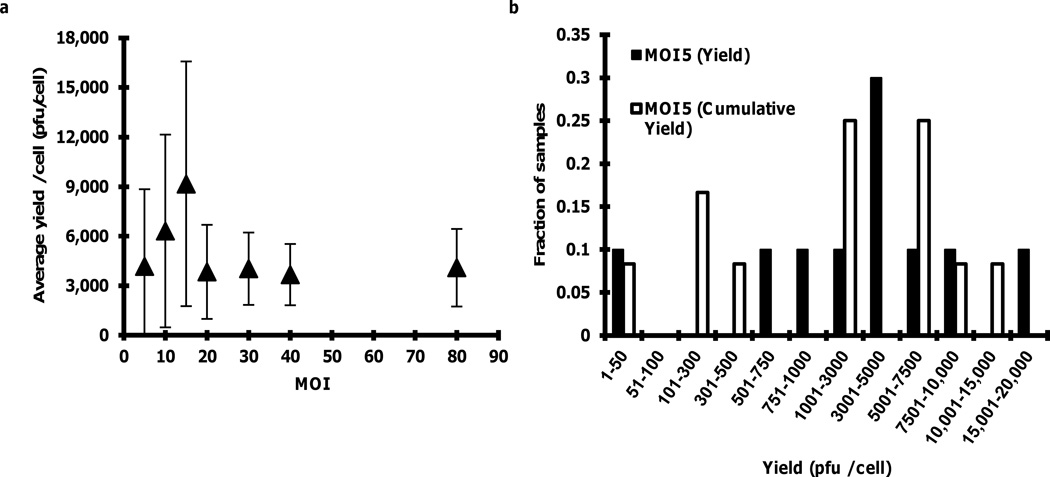
BHK cells were infected in solution at MOI 5, 10, 15, 20, 30, 40, and 80 pfu per cell. At each MOI, 10-to-12 wells containing single-cells were identified and sampled. In a few cases the wash step caused the infected cell to dislodge from the surface of the well; for example, three out of ten cells infected at MOI 80 were dislodged from their wells during the washing step, so these cells were not included in the analysis. For each MOI, the mean of the yields from at least seven cells was determined, as shown in Fig. 4a. The error bars represent one standard deviation about the mean. Yields at MOI 5 and MOI 15 were not significantly different (p-value 0.076).
Figure 4.
Virus yields from single infected cells. (a) Dependence of single-cell yields on MOI. Average single-cell yields at MOI 5 and MOI 15 did not differ (p-value 0.076, Student’s t-test, non-equal variance, 95 percent confidence interval). Error bars at each MOI reflect one standard deviation of the average yield. (b) Yield distribution from single-cells from the MOI-series experiment (MOI 5 Yield) and the kinetic experiment (MOI 5 Cumulative Yield) where the average yields did not differ (p-value 0.60, Student’s t-test, non-equal variance, 95 percent confidence interval).
The average yield measured at MOI 5 (Fig. 4a) was found to be consistent with the average of the cumulative yields from the single-cell kinetic measures (Fig. 2b). The average yield measured at MOI 5 was 4170 ± 4680 pfu per cell, which not significantly higher than the average cumulative yield from the kinetic measures (p-value 0.60). The yield distributions from both experiments spanned a similar range (Fig 4b).
DISCUSSION
Single infected cells drawn from the same population produced virus particles spanning a broad range --- from 30 to 10,000 plaque forming units (pfu). This broad distribution in virus production could be attributed to variation across the virus population, since RNA viruses such as VSV are known for their mechanisms for generating extreme genetic heterogeneity (Domingo et al., 1985), and we previously found low-yield behaviors that were transferable from one generation to the next (Zhu et al., 2009b). We thought it possible that the distribution of yields could reflect different numbers of virus particles that adsorb to individual cells when they are treated with excess virus (MOI 5), but the average yield from single-cells infected at higher MOIs did not change significantly (Fig. 4a). The broad distribution may well reflect different stages of the cell cycle, which could account for six-fold differences in yield (Zhu et al., 2009b). Further, computational modeling of VSV early kinetics suggests that the intrinsic noise or stochasticity associated with early intracellular replication can give rise to further six-fold differences in genomic template levels (Hensel et al., 2009), but how such differences impact virus production remain to be determined. In a study of phage T7, a DNA virus that infects bacteria, we found that richer nutrient conditions promoted faster cell growth, as well as earlier and faster production of phage particles within cells, and computational modeling highlighted links between phage production and capacity of the cellular resources for protein synthesis (You et al., 2002). Taken together, these studies suggest that the broad range of virus yields from different cells reflects potentially diverse factors: viral genetic variation, cell-to-cell variation in resources, and the noisy kinetics associated with small numbers of virus molecules present during the early stages of growth.
While our method for the study of virus release from single cells offers a new perspective on diversity of kinetic profiles and virus yields, generalization of the method for the study of other viruses will need to address several points. First, we have synchronized infections by using excess virus and adsorbing them to cells in the cold, a process that holds adsorbed virus in a pre-fusion (pre-infection) state that may be used to simultaneously trigger infection upon incubation. Viruses that enter their host cells by other mechanisms may require other approaches to synchronize their infections. Second, we used centrifugal forces to drive cell contact and adhesion to culture plates, but other approaches that use specific or non-specific chemical interactions between cells and the culture surfaces may be needed for other systems. Third, we employed the standard plaque assay to quantify the release of viable virus particles. Other approaches might well use the polymerase chain reaction (PCR) to quantify viral genomes or biochemical analyses of viral constituents to quantify other correlates of virus production. Finally, in its current implementation our method is likely to be too labor intensive for high-throughput analysis of hundreds or thousands of individual cells. Less time- and labor-intensive approaches may emerge from advances in microscale technologies that, for example, employ flow-enhanced quantification of infections initiated by single infected cells (Zhu et al., 2009a; Zhu and Yin, 2007).
CONCLUSIONS
We have shown that it is possible to study viral release kinetics from single infected cells. Such single-cell measures reveal details about the virus-host biology that are not possible to extract from population-level measures. Specifically, by performing such measures on multiple cells one can gain an appreciation for how traits of virus growth may be distributed and correlated. These observations motivate new questions on how genetic, environmental and other factors impact virus-host interactions.
MATERIALS & METHODS
1. Cell type and virus strain
Baby hamster kidney cells (BHK-21) adapted for infection, originally from Isabel Novella, were used in all experiments. Cells were passaged approximately every two days and maintained in media composed of 1X Minimum Essential Medium with Earle salts (MEM, Cellgro) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals) and 2 mM GlutaMAX (Gibco). The media used for infections was MEM supplemented with 2% FBS and 2 mM GlutaMAX. The VSV strain used is a recombinant strain (N1) encoding VSV genes in the same order as the wild-type virus (Wertz et al., 1998), generously provided by Gail Wertz.
2. Single cell infections
2.1 General protocol
Monolayers of BHK-21 cells grown in 75 cm2 cell culture flasks were released with 2 ml 0.05%trypsin/0.53 mM EDTA (Cellgro) after washing with Dulbecco’s phosphate buffered saline (DPBS, Gibco). The trypsin was neutralized with 8 ml infection media and the cell suspension was placed in a 15 ml conical tube. Cell counts were determined by hemacytometer. The cell suspension was put on ice, cooled virus solution was added, and the suspension was mixed thoroughly every 5 minutes over 30 minutes to minimize cell aggregation and allow for virus attachment to cells without entry. The cell suspension was then serially diluted in cold media to less than 1 cell per 100 ul and then added to a 96-well plate (100 µl/well). The plate was centrifuged at 1000 rpm and 4°C for 3 minutes to promote cell deposition and attachment to the plate, and the plate was incubated at 37°C allowing initiation of infection by endocytosis (Miller and Lenard, 1980).
2.2 Virus stock preparation
Virus stock was created using virus produced from a single cell. Following the general protocol for single-cell infections, BHK-21 cells were infected with VSV at an MOI of 5 and an extra 100 µl of media was added to each well. At 13.5 hours post infection (hpi), samples were taken from 5 wells containing single cells. 50 µl was used to determine the titer of the sample and the rest was reserved for a future infection. A high titer sample was used to infect a monolayer of cells at low MOI (~ 0.0003), supernatant was collected at 16 hpi, centrifuged to remove cell debris, filtered, aliquoted, and stored at −80°C for future use.
2.3 Determination of effective virus titer
An effective titer for the single-cell stock virus was determined by following a procedure similar to the single-cell infection protocol. A 100-fold dilution of stock virus was added to a concentrated cell suspension (1.85 × 106 cells/ml), giving about two virus particles per cell, and this solution was maintained on ice for 30 min. This concentration was added to mimic the high multiplicity conditions of the actual infections, but reduce the probability that single cells would be infected by multiple virus particles. The solution was mixed thoroughly every five minutes, to minimize cell aggregation. The virus-attached cell suspension was serially diluted in cell suspensions of equal density, so that only the virus-attached cell density was reduced. These cell suspensions were warmed for 3 minutes in a water bath set to 37°C to allow attached virus to endocytose. The suspensions were centrifuged at 4°C and 1000 rpm for 5 minutes to pellet the cells. The supernatant containing any unattached virus was decanted and the pellets were washed gently with DPBS. The cell pellets were re-suspended and added to wells of a 6-well plate. The cells were allowed 1 hour to settle to attach to the bottom of the well. At this time, most of the cells were adhered. The monolayers were washed with DPBS and overlaid with an agar solution (0.6% Difco Agar Noble dissolved in sterile water which makes up 10% of the total volume and 90% infection media by volume). After incubating for 22 hours, the agar gels were removed and the cells were fixed with a paraformaldehyde (PFA) solution (4% paraformaldehyde and 5% sucrose in 10 mM phosphate buffered saline (PBS, Sigma)), rinsed twice with PBS, and stained with 0.1% crystal violet in 20% ethanol. The plaques were counted and the effective titer of the original stock solution was determined, taking into account all the serial dilutions and the initial amount of virus added to the cell solution.
Typically, for these experiments, the density of the cell solution was about 106 cells/ml. A small volume of concentrated virus solution was added so that the cell solution would not significantly diluted when adding a high multiplicity of virus. In this case 86 µl of virus was added to 1 ml of cell solution. Sufficient virus was added to infect approximately 60 percent of the cells in the original suspension.
2.4 Single-cell one-step infection
The general protocol for single-cell infections was followed. The cell solution was infected at MOI 5 based on the effective titer of the single-cell virus stock. This MOI was chosen to ensure a large percentage of cells would become infected. The virus-attached cell solution filled three 96-well plates. One row of one plate was used for control samples. The plates were removed one at a time from the 37°C incubator and placed in an environmentally controlled chamber (LiveCell from Pathology Devices) maintaining the cells at 37°C, 5% CO2, and approximately 75% humidity, on a Nikon Eclipse TE300 microscope. The plates were scanned for wells containing individual cells, and the location of the cell in the well was recorded. To limit time outside the incubator for sampling, only eight wells were chosen per plate. The first sample was taken at two hours post infection (2 hours post warming to 37°C) well before the release of any virus progeny. To minimize perturbation of the cell, all 100 µl of infection media was slowly removed by pipette, distant from the cell, and then slowly replaced with fresh warm infection media at the same location. This procedure was repeated for each of the 24 selected wells. Each sampling removed plates from the incubator for 6–7 minutes. Samples were taken from 2 to 24 hours post infection. After the final sample was taken an additional wash (100 µl of media) was added and mixed thoroughly to dislodge any virus that had adhered to the well. All samples were stored at −80°C immediately after sampling until virus detection with plaque assay. Control samples and washes from the control wells were collected from 2 to 14 h after addition of virus.
3. Quantification of virus production
Virus in each sample was measured by plaque assay. The low levels of virus and small volumes of sample in the single-cell infections excluded the possibility for replicate measures. In all cases 200 µl of various sample dilutions were used to infect BHK cell monolayers in 6-well plates. The virus was permitted 1 hour to adsorb at 37°C, the monolayers were washed with 2 ml DPBS, and then overlaid with an agar solution. The plates were incubated 24 hours before fixation and staining. Samples were diluted 1:3 and then serially diluted 1:10 to ensure that very small amounts of virus could be detected.
Error propagation was estimated based on contributions from pipetting and plaque counting. The error in plaque counting, approximated as the square root of the total plaques counted, and measurement error from our pipettes, as supplied by the manufacturer, were propagated and compounded as described elsewhere (Hughes and Hase, 2010). The results of the error propagation calculations should represent the maximum possible error. To confirm that this error exceeds the actual error, a stock of virus (9700 pfu/100 µl) was made and serially diluted, by factors of three down to approximately 1 pfu/100 µl. These stocks were separated into three aliquots and frozen at −80°C until quantification with plaque assay. The procedure for diluting the aliquots and quantifying by plaque assay was done in the same manner as the one-step samples, but duplicate measures were taken and the stock aliquots were quantified on three separate occasions.
To test for correlations between early and middle stages of virus release and the total virus yield, we assembled cumulative one-step curves for virus production and estimated the yield, rise rate, latent time, and rise time. The yield is simply the cumulative number of progeny virus produced by a single cell. The rise rate is estimated from the slope of the cumulative virus production curve, based on three data points through the half-yield level. Extrapolation of the line through these data to the time axis provides the latent time, indicating the time scale for the earliest release of progeny virus. Finally, the rise time is the length of the period over which virus particles are released (yield per rise rate).
4. Dependence of single-cell yield on multiplicity of infection
Cells were infected in solution across a range of MOI 5-to-80 PFU per cell following the procedure described in section 2.1. Excess virus not adsorbed was removed 2.5 hpi (before release of progeny virus) and samples were collected 24 hpi and quantified as described in section 3. Student’s t-test was used to determine if the differences in average yields at MOIs 5 and 15 were statistically significant. At MOI 5 and MOI 15 there were 11 data points and the variances were not assumed to be equal.
Research Highlights.
> A new method enables measurement of virus production kinetics from single-cells. > The method was used to study vesicular stomatitis virus production from BHK cells. > Virus yields varied 300-fold, while latent times were within two-fold. > Such measures reveal how traits of virus production may be distributed and correlated.
Supplementary Material
Error Propagation in the Plaque Assay. (a) A virus stock of approximately 9700 PFU/100 µl was serially diluted by factors of 3 down to approximately 1 PFU/100 µl. Measured plaque counts are the average of 3 duplicate measures of each serial dilution. Error bars represent the standard deviation of those averages. The measured counts are accurate down to approximately 4 PFU/100 µl. (b) Parity plot of all one-step infection measures with the measured plaque counts from a. Green error bars represent the standard deviation calculated using error propagation for each sample and the black error bars are the standard deviation describe in a. This plot shows that the actual plaque assay error is comparable to the error propagation calculations at all concentrations.
ACKNOWLEDGEMENTS
We thank Paul Ahlquist and Jim Rawlings for thoughtful feedback on a draft of the manuscript. Ankur Gupta, Rishi Srivastava, Adam Swick and Collin Timm provided helpful comments and discussions. We are grateful for support from the National Institutes of Health (AI071197).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Burnet F. A Method for the Study of Bacteriophage Multiplication in Broth. Brit. J. Expt. Path. 1929;10(2):109–114. [Google Scholar]
- Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374(6522):569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- Cuevas JM, Moya A, Sanjuan R. Following the very initial growth of biological RNA viral clones. J Gen Virol. 2005;86(Pt 2):435–443. doi: 10.1099/vir.0.80359-0. [DOI] [PubMed] [Google Scholar]
- Delbrück M. The burst size distribution in the growth of bacterial viruses (bacteriophages) J. Bact. 1945;50:131–135. doi: 10.1128/JB.50.2.131-135.1945. [DOI] [PubMed] [Google Scholar]
- Domingo E, Martinez-Salas E, Sobrino F, Torre JCdl, Portela A, Ortin J, Lopez-Galindez C, Perez-Brena P, Villanueva N, Najera R, VandePol S, Steinhauer D, DePolo N, Holland J. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance- a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Flint SJ, Enquist LW, Racaniello VR. Principles of Virology. 3rd ed. Washington DC: ASM Press; 2009. [Google Scholar]
- Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu Rev Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- Haseltine EL, Lam V, Yin J, Rawlings JB. Image-Guided Modeling of Virus Growth and Spread. Bulletin of Mathematical Biology. 2008 April; doi: 10.1007/s11538-008-9316-3. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel SC, Rawlings JB, Yin J. Stochastic kinetic modeling of vesicular stomatitis virus intracellular growth. Bull Math Biol. 2009;71(7):1671–1692. doi: 10.1007/s11538-009-9419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Nudelman G, Shimoni Y, Kumar M, Ding Y, Lopez C, Hayot F, Wetmur JG, Sealfon SC. Role of cell-to-cell variability in activating a positive feedback antiviral response in human dendritic cells. PLoS One. 2011;6(2):e16614. doi: 10.1371/journal.pone.0016614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes I, Hase TPA. Measurements and their uncertainties : a practical guide to modern error analysis. New York: Oxford University Press, Oxford; 2010. [Google Scholar]
- Kutsch O, Benveniste EN, Shaw GM, Levy DN. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol. 2002;76(17):8776–8786. doi: 10.1128/JVI.76.17.8776-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam V, Duca KA, Yin J. Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnol Bioeng. 2005;90(7):793–804. doi: 10.1002/bit.20467. [DOI] [PubMed] [Google Scholar]
- Larder B, Chesebro B, Richman D. Susceptibilities of zidovudine-susceptible and -resistant human immunodeficiency virus isolates to antiviral agents determined by using a quantitative plaque reduction assay. Antimicrobial Agents and Chemotherapy. 1990;34(3):436–441. doi: 10.1128/aac.34.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Lipsitch M, Bonhoeffer S. Population biology, evolution, and infectious disease: convergence and synthesis. Science. 1999;283(5403):806–809. doi: 10.1126/science.283.5403.806. [DOI] [PubMed] [Google Scholar]
- Lim KI, Lang T, Lam V, Yin J. Model-based design of growth-attenuated viruses. PLoS Comput Biol. 2006;2(9):e116. doi: 10.1371/journal.pcbi.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr V, Genzel Y, Behrendt I, Scharfenberg K, Reichl U. A new MDCK suspension line cultivated in a fully defined medium in stirred-tank and wave bioreactor. Vaccine. 2010;28(38):6256–6264. doi: 10.1016/j.vaccine.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Melcher A, Parato K, Rooney CM, Bell JC. Thunder and lightning: immunotherapy and oncolytic viruses collide. Mol Ther. 2011;19(6):1008–1016. doi: 10.1038/mt.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DK, Lenard J. Inhibition of vesicular stomatitis virus infection by spike glycoprotein. Evidence for an intracellular, G protein-requiring step. J Cell Biol. 1980;84(2):430–437. doi: 10.1083/jcb.84.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Travers P, Walport M, Janeway C. Janeway's immunobiology. 7th ed. New York: Garland Science; 2008. [Google Scholar]
- Novella I, Duarte EA, Elena SF, Moya A, Domingo E, Holland JJ. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA. 1995;92:5841–5844. doi: 10.1073/pnas.92.13.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Elena SF. Cost of host radiation in an RNA virus. Genetics. 2000;156(4):1465–1470. doi: 10.1093/genetics/156.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz G, Perepelitsa V, Ball L. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci U S A. 1998;95(7):3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Suthers P, Yin J. Effects of Escherichia coli physiology on the growth of phage T7 in vivo and in silico. Journal of Bacteriology. 2002;184(7):1888–1894. doi: 10.1128/JB.184.7.1888-1894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Warrick JW, Haubert K, Beebe DJ, Yin J. Infection on a chip: a microscale platform for simple and sensitive cell-based virus assays. Biomed Microdevices. 2009a doi: 10.1007/s10544-008-9263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yin J. A quantitative comet assay: Imaging and analysis of virus plaques formed with a liquid overlay. J Virol Methods. 2007;139:100–102. doi: 10.1016/j.jviromet.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yongky A, Yin J. Growth of an RNA virus in single cells reveals a broad fitness distribution. Virology. 2009b;385(1):39–46. doi: 10.1016/j.virol.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Error Propagation in the Plaque Assay. (a) A virus stock of approximately 9700 PFU/100 µl was serially diluted by factors of 3 down to approximately 1 PFU/100 µl. Measured plaque counts are the average of 3 duplicate measures of each serial dilution. Error bars represent the standard deviation of those averages. The measured counts are accurate down to approximately 4 PFU/100 µl. (b) Parity plot of all one-step infection measures with the measured plaque counts from a. Green error bars represent the standard deviation calculated using error propagation for each sample and the black error bars are the standard deviation describe in a. This plot shows that the actual plaque assay error is comparable to the error propagation calculations at all concentrations.