Abstract
Transplantation of human neural stem cells (hNSCs) is emerging as a viable treatment for stroke related brain injury. However, intraparenchymal grafts do not regenerate lost tissue, but rather integrate into the host parenchyma without significantly affecting the lesion cavity. Providing a structural support for the delivered cells appears important for cell based therapeutic approaches. The non-invasive monitoring of therapeutic methods would provide valuable information regarding therapeutic strategies but remains a challenge. Labeling transplanted cells with metal-based 1H-magnetic resonance imaging (MRI) contrast agents affects the visualization of the lesion cavity. Herein, we demonstrate that a 19F-MRI contrast agent can adequately monitor the distribution of transplanted cells, whilst allowing an evaluation of the lesion cavity and the formation of new tissue on 1H-MRI scans. Twenty percent of cells labeled with the 19F-agent were of host origin, potentially reflecting the re-uptake of label from dead transplanted cells. Both T2- and diffusion-weighted MRI scans indicated that transplantation of hNSCs suspended in a gel form of a xenogeneic extracellular matrix (ECM) bioscaffold resulted in uniformly distributed cells throughout the lesion cavity. However, diffusion MRI indicated that the injected materials did not yet establish diffusion barriers (i.e. cellular network, fiber tracts) normally found within striatal tissue. The ECM bioscaffold therefore provides an important support to hNSCs for the creation of de novo tissue and multi-nuclei MRI represents an adept method for the visualization of some aspects of this process. However, significant developments of both the transplantation paradigm, as well as regenerative imaging, are required to successfully create new tissue in the lesion cavity and to monitor this process non-invasively.
1. INTRODUCTION
Cell therapy for neurological conditions is currently emerging from the bench to the bedside [1]. However, merely injecting cells does not replace lost tissue, but these cells predominantly promote beneficial effects through paracrine effects and/or by replacing a small number of lost cells within a damaged tissue. Large areas of tissue loss, as can be seen in chronic stroke or traumatic brain injury, remain unpopulated by these cells [2].
Integration of cells with appropriate biomaterials can potentially further shift the cell therapy paradigm to a regenerative medicine therapeutic option for the brain [3, 4]. In situ remodeling of a cell-seeded bioscaffold is an attractive and potentially viable approach for functional tissue engineering of the central nervous system. This approach will not only support cells during the transplantation process, but will also provide a structural support system for the cells within the cavity upon implantation [5–8]. In the healthy brain, cells are embedded within extracellular matrix (ECM) and extracellular fluid (ECF) [9]. Within the infarct cavity, extracellular fluid is abundantly available, but there is a complete loss of ECM in addition to the loss of cells. Replacing this lost ECM can therefore potentially supply the transplanted cells with their “natural” microenvironment and facilitate the regeneration of lost tissue [10]. The ECM derived from decellularized allogeneic and xenogeneic tissues (from hereon referred to as ECM bioscaffold) has been successfully used to facilitate constructive remodeling of numerous tissues and organs, including skeletal muscle [11, 12], lower urinary tract [13, 14], esophagus [15, 16], myocardium [17, 18], and dura mater [19, 20]. In fact, such ECM bioscaffolds have been shown to promote neurogenesis in several tissues [21, 22].
In situ tissue engineering in the brain, however, would benefit greatly from the availability of non-invasive imaging that allows the targeted injection of cells and biomaterials, whilst simultaneously monitoring the processes involved in remodeling of the tissue [23]. Magnetic resonance imaging (MRI) is ideally suited for this application in pre-clinical animal models, as well as human clinical applications. MRI can provide a high resolution anatomical image of the stroke lesion that can provide the 3-dimensional stereotactic coordinates for transplantation. Pre- and post-transplantation T2-weighted MR images can provide proof that indeed the cavity was filled with biomaterial. However, these methods might only indicate that the density and ratio of cells/ECM is similar to “normal” brain, but are insufficient to demonstrate the formation of de novo tissue or remodeling of an ECM bioscaffold material. In contrast, diffusion MRI is very sensitive to the 3-dimensional movement of water molecules within tissues and the lesion cavity. If tissue is forming within the cavity, this movement will be increasingly restricted in distance and direction. Diffusion MRI therefore is an important additional imaging parameter that can inform on the regenerative process.
To visualize the presence and distribution of transplanted cells, it is important to independently image these cells. Imaging approaches that rely on 1H-MRI, such as iron oxide- or gadolinium-labeling of cells [24], will interfere with the imaging of the stroke pathology. Therefore using multi-nuclear MRI can provide an attractive alternative. Transplanted cells can be labeled using a 19F-MRI contrast agent prior to transplantation without interfering with T2- or diffusion-weighted imaging [25], hence affording the visualization of the stroke pathology, tissue regeneration and transplanted cells using serial MRI. The effects of a bimodal 19F-BODIPy contrast agent on human neural stem cells were investigated prior to implantation with decellularized matrix into animals with pathological cavities formed by a stroke. In vivo serial MRI demonstrates the proof-of-principle to monitor tissue formation non-invasively over one week.
2. MATERIAL & METHODS
2.1. Human neural stem cells
The cmyc-ERTAM conditionally immortalized human neural stem cell lines CTX0E03 and STROC05 (ReNeuron Ltd.) [26–28] were cultured in chemically defined media consisting of Dulbecco’s Modified Eagle’s Medium/Ham’s with F12 medium (DMEM:F12, Gibco) supplemented with a range of components[26]. To maintain proliferation through the conditional immortalization gene, 4-hydroxy-tamoxifen (4-OHT, Sigma-Aldrich) and growth factors (basic fibroblast growth factor-2, bFGF, 10ng/ml, and epidermal growth factor, EGF, 20 ng/ml, Peprotech) were added to media in laminin-coated tissue culture flasks (mouse, 1:100, 10 mg/ml, Trevigen). For cell differentiation, growth factors and 4-OHT were withdrawn.
2.2. Bimodal 19F-MRI contrast agent
A bimodal (i.e., MRI and fluorescence) tracer agent was used to label cells (CS-1000-DM-Red, Celsense, Inc., Pittsburgh, PA) in culture. This reagent is a perfluorocarbon emulsion that has a BODIPy dye conjugated directly to the perfluorocarbon. The excitation/emission maximum occur at 590/620 nm, respectively. The manufacturer data reports a mean droplet size of approximately 160 nm, and the perfluorocarbon oil component comprises approximately 12% (by weight) of the emulsion. The T1 and T2 values for the agent were determined to be 425 ms and 82 ms at 7T, respectively (Supplementary Figure 1A).
2.3. In vitro assays
Quantifying cell labeling using fluorescence
To ensure optimal labeling, a concentration (0, 1, 3 and 5 mg/ml) versus time (3, 6 and 24 h) experiment was conducted. Media containing the bimodal 19F agent, as well as 10% human serum albumin (HSA) were added to the tissue culture flasks and cells were incubated for 24 h at 37°C. HSA was added for labeling of cells as it improved cellular uptake and prevented the agent from sticking to the coated flasks (Supplementary Figure 2).
Cells were seeded onto laminin-coated coverslips at 5×104 immediately after labeling and grown overnight at 37°C. For each condition, 3 biological replicates with each having 3 technical replicates (i.e. coverslips) were analyzed. Cells were fixed for 15 min in 4% paraformaldehyde (PFA), before being rinsed 3×5 min in phosphate buffered saline (PBS). To visualize, the cell membrane, cells were counterstained for 2 h at room temperature (RT) with nestin (1:1000 rabbit anti-nestin, AB5968, Abcam) in 0.1% Triton/PBS (Sigma-Aldrich), followed by incubation for 2 h at RT with a secondary Alexa 488 goat anti-rabbit (1:100, A-11034, Invitrogen). After incubation, cells were rinsed 3×5 min in PBS and mounted with DAPI in Vectashield.
Five images were captured from each coverslip to give a total of 15 random images (x20 objective) for each agent concentration and incubation time. The color histogram plugin in Image J (version 1.37, National Institutes of Health, http://rsb.info.nih.gov/ij/) was used to quantify immunofluorescence in labeled cells. Images were captured at a constant exposure time across all conditions using a Zeiss Axioplan (Zeiss). Fluorescence emitted in the red channel was measured in arbitrary reflective fluorescence units (RFU) and divided by the total number of DAPI-positive cells (RFU/cell).
NMR analysis of cellular uptake
For further corroboration of the fluorescent measurements, 19F content per cell was also quantified by NMR. For this, a large quantity of cells was required and cells were therefore grown to 90% confluency in T25 flasks. Total cell number was determined by hematocytometer cell counts (average of 2.12×106 cells per sample, 3 biological replicates). Labeled cells were assayed for 19F content by NMR [29, 30]. Cells of a known number were placed in 0.2 mL lysis buffer and mixed with 2% trifluoroacetic acid (TFA) 19F standard. NMR spectroscopy of the cell pellets was performed at 470 MHz using a Bruker spectrometer (Bruker Biospin, Billerica, MA) with a delay time of 8 s and 32 averages. Two distinct peaks were observed at −76 ppm for TFA and −91.6 ppm for the perflurocarbon (Supplementary Figure 1B). The mean number of 19F per cell (Fc) was calculated using the following formula Fc=[(Ic/Ir)Nr]/Nc, where (Ic/Ir) is the ratio of the integrated values of the perfluorocarbon peak in the cell pellet at −91.58 ppm divided by the TFA reference peak at −76.0 ppm, Nr is the total number of 19F in the TFA reference sample, and Nc is the total cell number in the pellet.
Viability
To measure the effect of agent on cell viability, a Trypan Blue exclusion test (Invitrogen) was conducted immediately after labeling using the same conditions as those used for fluorescence intensity measurements.
Standard Protocol
Based on cellular uptake and viability, a concentration of 5mg/ml (with 10% HSA) for a labeling duration of 24 hours was determined to be an optimal condition for the clinical CTX0E03 cell line (1.96×1012 19F spins/cell with an overall viability of 82% compared to 86% for non-labeled controls). For this, cells were seeded at 5×105 into T25 laminin-coated culture flasks and grown to 90% confluence. Cells were harvested by incubation with TrypZean-EDTA (Lonza) for 5 min at 37°C, followed by addition of soybean trypsin inhibitor (Sigma-Aldrich) to block further enzymatic activity. Cell suspensions were centrifuged for 5 min at 1500 rpm and the cell pellet was re-suspended in PBS to remove excess 19F agent. After this washing step, cells were re-suspended for all further experiments, as described below.
Proliferation
Cells were seeded on coverslips and grown over 4 days in proliferation media (containing 4-OHT, bFGF and EGF). Each day samples were fixed using PFA and cells were stained with a rabbit anti-Ki67 (1:1000, ab5580, Abcam) antibody to quantify the number of dividing cells. Immunocytochemical staining followed the procedure as described above for nestin.
MTT assay
To quantify a potential impact on the cells’ mitochondrial function, the colorimetric 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay compared 19F-labeled versus control cells, as previously described[31]. The MTT solution (40 μl) was added to cells in a 24-well plate for four hours. The solubilization solution was then added overnight and absorbance was read on a Beckman Coulter microplate reader at 550 to 600 nm.
Differentiation
As differentiation is a key function of neural stem cells, we determined if 19F-labeling affects the cells’ multipotentiality. To evaluate multipotentiality, cells were seeded on coverslips and grown in differentiation media (i.e. media without growth factors or 4-OHT) for 7 days. Cells were fixed with PFA and stained for the neural stem cell marker nestin (rabbit anti-nestin, 1:1000, ab5968, Abcam), the astrocytic marker glial fibrillary acidic protein (mouse anti-GFAP, 1:1000, G3893, Sigma), and the neuronal marker β-III-tubulin (mouse anti-Tuj, 1:1000, ab7751, Abcam). Appropriate secondaries (1:500, goat anti-mouse 555, A21422; goat anti-rabbit 488, A11008, Invitrogen) were used to visualize the appropriate antigen.
Leakage and Re-uptake Experiment
To establish if there is a transfer of the contrast agent from labeled cells, 2.5×105 cells labeled with the bimodal 19F agent were co-culture with 2.5×105 cells labeled with the green fluorscent Cell Tracker (10 μM for 2 minutes, C2925, Invitrogen). Images were taken at 1 and 7 days. The number of red, green and yellow cells were counted to determine transfer of the red 19F agent into “non-labeled” (green) cells.
Evolution of 19F concentration
Establishing the presence of agent over time is an important aspect of its long-term detection. Comparing the evolution of 19F concentration by fluorescence can therefore provide an indication under which condition cells can be potentially detected using MRI. However, it is important to acknowledge that photobleaching, as well as degradation of the BODIPY within low pH cellular vesicles, can lead to an underestimation of 19F concentration. Nonetheless, the fluorescent intensity of 19F-labeled cells from the differentiation and proliferation experiments was quantified as described above.
2.4. Cell detection by 19F-MRI
Cell phantom preparation
19F phantoms consisted of a gradient of 0, 1.5×106, 2.5×106, or 4.5×106 cells in a 50 μl volume of 6% gelatin in PCR tubes. Samples were placed in a row to determine the signal threshold for cell detection. This procedure yields an estimate of the concentration of cells that can be detected upon transplantation given the particular hardware configuration, imaging methods employed, and the time constraints of in vivo imaging.
19F/T2 MR imaging
MR Images were acquired on a 7T Varian system. To localize the 19F signal, a custom-made dual-tune quadrature coil (David Herlihy, Imperial College) was used for receive and transmit. Co-localization and positioning of slices for the 19F scan were based on a 1H-based T2-weighted anatomical scan that was acquired using a Spin Echo Multi-Slice (SEMS) sequence (TR = 2 s, TE = 14.73 ms, averages = 8, FOV = 40 × 40 mm, matrix 128 × 128, single slice at 2 mm thickness). Samples were stored at 4°C and allowed to achieve room temperature (21°C) prior to imaging.
The 19F images were acquired at 282 MHz using a Fast Spin Echo (FSE) sequence (TR = 1500ms, echo train = 16 echoes, echo spacing = 8.53ms, averages = 600, FOV = 40 × 40 mm, matrix size = 32 × 32, single slice at 12 mm thickness, 60 minutes scan time). The measured phantom signal-to-noise was plotted as a function of its cell density per voxel. The minimum cell per voxel detection threshold was determined by extrapolating the SNR to a value of 1.25.
2.5. Extracellular matrix (ECM) bioscaffold production
Biologic scaffolds composed of ECM derived from porcine brain and urinary bladder were produced by enzymatic, chemical, and mechanical methods of decellularization [32]. Decellularization of these tissues was verified by multiple methods [33, 34]. Urinary bladder ECM (UBM) and brain ECM were lyophilized, ground into powder, solubilized and processed into an injectable gel form with a final protein concentration of 8.0 mg/ml (7.2 mg/ml UBM and 0.8 mg/ml brain ECM) by solubilizing with pepsin in 0.01 N HCl, neutralizing with 0.1 N NaOH, and diluting with PBS to the desired concentration [32]. The solubilized form was stored at −20°C and maintained at 4°C immediately prior to mixing with cells. The solubilized material formed a gel at 37°C, or stated differently, when in contact with body tissue.
2.6. Cell/ECM bioscaffold preparation for MRI and transplantation
The solubilized ECM bioscaffold was neutralized to pH 7.4 forty min. prior to preparation. After addition and suspension of 3.75×106 cells in 50 μl of neutralized ECM (i.e. 7.5×104 cells/μl), the soluble cell/ECM bioscaffold mixture was maintained on wet ice prior to intracerebral injection. Upon injection, this mixture formed a gel at body temperature (37°C) to keep cells and ECM bioscaffold in the transplant location [32].
2.7. Ex vivo Experiment
To compare the anatomical effects and MR-detection of the ECM bioscaffold, ECM+cells, as well as ECM+19F-labeled cells within a stroke cavity, animals with middle cerebral artery occlusion were transplanted two weeks following infarction and perfusion-fixed for ex vivo MRI 24 hours after injection.
This method afforded a direct comparison between the different components being injected. All procedures complied with the UK Animals (Scientific) Procedures Act (1986) and the Ethical Review Process of the Institute of Psychiatry.
Middle cerebral artery occlusion (MCAo)
Fifteen Sprague-Dawley rats (weight 270–320g, Harlan, UK) were maintained on a 12 h light/dark schedule with food available ad libitum. The right MCA was occluded for 70 min by insertion of a propylene filament (Doccol Corporation, USA), tip diameter with coating 0.33 +/− 0.02 mm, via the common carotid artery to the ostium of the MCA in the circle of Willis. Isoflurane (4% for induction, 2% for maintenance in 30% oxygen) was used for anesthesia during insertion and removal of the filament, with temperature held at 37±1°C by rectal probe and heating pad. During the recovery period of two weeks, animals were monitored for neurological deficits [35].
Pre-implantation MRI planning
Animals (n=15) were scanned for the presence or absence of a lesion 10 days post-MCAo surgery by using a spin echo T2-weighted MRI scan (TR = 4000 ms, TE = 60 ms, averages = 10, FOV = 40 × 40 mm, matrix = 128×128, 45 slices at 0.6 mm slice thickness). Two animals were excluded due to a lack of lesion. In those animals with a stroke lesion, MR images were compared to the rat brain atlas [36] and 3-dimensional stereotactic injection coordinates were derived to correspond to the center of the lesion cavity [6]. Injection volume and cell number was adapted according to lesion volumes that were calculated in the image analysis package JIM (version 4, Xinapse). 2.1–2.5×106 cells were injected per animal. A MCAo only (n=4) and a normal control group (n=3) were also included for comparison.
Stereotactic surgery
Two weeks after MCAo under isofluorane anesthesia (4% induction, 2% maintenance), animals either received ECM only (n=3), ECM+cells (n=3) or ECM+19F-labeled cells (n=3). Animals were placed in a stereotactic frame and injections (25–40 μL at a 2 μl/min) were made at appropriate coordinates for each animal using a 50 μl gastight Hamilton syringe with a 30G blunt tip needle.
Perfusion fixation
Animals were terminally anaesthetized 24 hours after transplantation by overdosing with sodium pentobarbital (60 mg/kg i.p.). A transcardial perfusion of 0.9% saline was followed by 4% ice-cold paraformaldehyde in 0.2 M phosphate buffered saline (PBS). Animals’ heads were maintained in paraformaldehyde until MRI scanning.
19F/T2 MR imaging
Co-localization and positioning of slices for the 19F scan were based on a short 1H-based T2-weighted anatomical scan (16 min) acquired using a fast spin echo (FSE) sequence (TR = 3000 ms, echo spacing = 15 ms, echo train = 4, effective echo = 60 ms, k0 = 4, averages = 8, FOV = 30 × 30 mm, matrix 128 × 128, 45 slices at 0.6 mm thickness).
The 19F images were acquired at 7T (282Mhz) using a Fast Spin Echo (FSE) sequence (TR = 500ms, echo spacing = 8.53, echo train = 16 echoes, effective echo = 17.06 ms, k0=2, averages = 600, FOV = 30 × 30 mm, matrix size = 32 × 32, 3 slices at 2 mm thickness, 45 minutes acquisition time). A vial of 1mM 19F agent was placed on top of the animals’ head to allow tuning to the 19F frequency and to provide an internal standard.
High resolution T2-weighted MRI
Due to the lower signal-to-noise of the dual-tune coil used for the 19F imaging, a high resolution T2-weighted MRI scan was acquired on the following day with a custom-build fixed tuned 1H quadrature coil (David Herlihy, Imperial College), using a Multi-Slice Multi-Echo (MSME) sequence (TR=4200 ms, echo spacing = 10 ms, echo train = 8, effective echo = 80 ms, averages = 4, FOV = 30x 30 mm, matrix = 192×192, 45 slices at 0.6 mm thickness). Multi-echo images were summed to an average echo image for co-registration and presentation purposes.
Diffusion MRI
In the same imaging session as the high resolution T2-weighted MR images, a diffusion-weighted data were acquired using a Stimulated Multi-Echo Trace (STEM-TRACE) sequence (TR = 3000 ms, TE = 24.63 ms, TM = 33.75 ms, averages = 4, diffusion gradient duration (δ) and separation (Δ) = 5 and 50 ms, diffusion gradient amplitudes = 350, 300, 250, 200, 150, 100, 50, 25 and 0 mT/m, maximum b-value 10,600 s/mm2, FOV = 30 × 30 mm, matrix 128 × 64, 45 slices at 0.6 mm thickness) was acquired to determine the impact of the ECM bioscaffold on the fluid in the lesion cavity. Images were reconstructed to a final 128 × 128 matrix size to allow co-registration with the high-resolution T2-weighted images. Apparent Diffusion Coefficient (ADC) maps were calculated by applying a non-linear least squares fit to a mono-exponential diffusion model using in-house developed software.
Image co-registration
High-resolution T2-weighted images were manually co-registered to the low-resolution 1H T2 images used to localize 19F data. Co-registration was performed using MANGO software package (Jack L. Lancaster & Michael J. Martinez, available at http://ric.uthscsa.edu/mango) by applying rigid transformations. The same transformations were then applied to ADC maps.
Immunohistochemistry
After completion of MR imaging, intact brains were removed following craniotomy and immersed in cryoprotective solution (30% sucrose in PBS). Once these brains were cryoprotected, they were rapidly frozen on dry ice. Coronal (40 μm) sections were cut on a freezing cryostat (Leica, Germany) directly onto glass slides to prevent dislodgement of the ECM/cell mixture from the lesion cavity.
For immunohistochemistry, sections were incubated in 15% of the appropriate blocking sera and 0.3% PBS-Triton for 40 min prior to overnight incubation with primary antibodies (diluted in 10% normal serum and 0.3% PBS-Triton) at 4°C. Primary antibodies specifically detecting human cells consisted of mouse anti-human nuclei marker (SC101, 1:200, AB-101-U-050, Stem Cell Inc.), rabbit anti-heat shock protein 27 (HSP27, 1:200, SPA-803F, Cambridge Bioscience), as well as rabbit anti-GFAP (1:1000, Z0334, DAKO) detecting astrocytes and rat anti-CD11b (1:200, 6332-100, Abcam) detecting microglia/macrophages. After incubation with primary antibodies, sections were washed 3×5 min in PBS and then incubated with a secondary antibody for 2 h at RT (21°C). Secondary antibodies consisted of appropriate fluorescent Alexa488 or Alexa647 antibodies (1:500, Invitrogen, UK). Sections were rinsed 3×5 min in PBS before application of Vectashield with DAPI (Vector Labs, UK).
To visualize the injected ECM within the lesion cavity histologically, sections were incubated in Bouin’s solution at 60°C (60 min), cooled and stained using Weigert’s hematoxylin (10 min), Biebrich scarlet solution (2 min), phosphomolybdic-phosphotungstic acid (15 min) and aniline blue (5 min). In between steps, sections were rinsed with dH2O, as well as acid/alcohol and xylene substitution prior to coverslips being applied. Images were captured using Axiovision on a Zeiss Axioplan (Zeiss, Germany).
2.8. In vivo experiment
To establish the evolution of the ECM bioscaffold and 19F-labeled cells within the lesion cavity, 3 animals underwent serial MR imaging pre-transplantation, as well as 1 and 7 days post-transplantation. Animals, cell/ECM transplantation, MRI, perfusion fixation and immunohistochemistry followed the same methods as described for the ex vivo experiment.
2.9. Statistical Analyses
Statistical analysis of cell counts and measurements were performed on mean values by using two-way analysis of variance (ANOVA) followed by Bonferroni post-hoc analysis and expressed as means ± standard error of means (SEM) with Prism 4 (GraphPad). A P value of <.05 was considered significant.
3. RESULTS
3.1. Cell labeling and viability
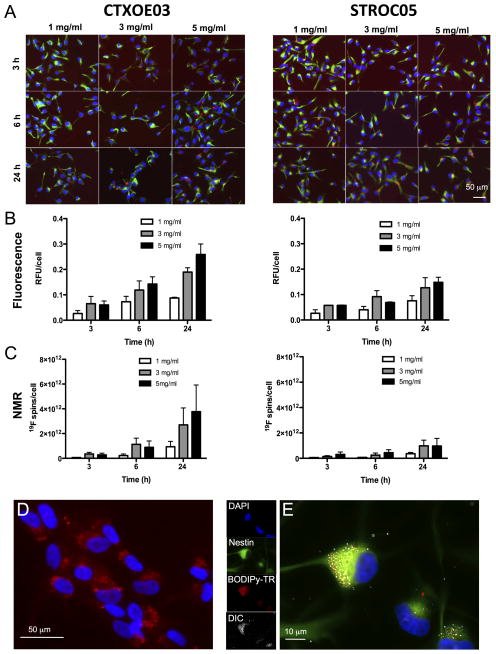
The MRI detection of transplanted cells independently from the monitoring of pathology requires multi-nuclei MRI. Thus, a 19F-MRI cell labeling agent was employed. To ensure that cells retain their normal properties that are important to promote behavioral recovery, it is essential to establish an efficient labeling of cells without affecting cellular function. A time versus concentration curve of the bimodal 19F agent in two clinical-grade human neural stem cell lines, CTX0E03 and STROC05 (Figure 1A) indicates that higher concentrations of agent over longer times in culture medium results in more uptake, as determined by fluorescence (Figure 1B) and NMR spectroscopy (Figure 1C). After 6 and 24 hours of incubation, the CTX0E03 cells incorporate significantly more agent compared to the STROC05 cells. This label uptake highlights that not all neural stem cells will incorporate agent at the same rate. However, an efficient labeling of >97% of cells can be achieved (Figure 1D) with agent being contained within the cell’s cytoplasm (Figure 1E). Importantly no significant amount of extracellular or membrane-bound agent can be observed using this approach.
Figure 1.
Labeling of human neural stem cells with bimodal 19F agent. A. The human neural stem cell lines CTX0E03 and STROC05 were labeled with the 19F agent at different concentrations (1, 3, 5 mg/ml) for different amounts of time (3, 6, and 24h). Based on these arrays, uptake of the 19F agent was determined by both fluorescence microscopy (B) and 19F -nuclear magnetic resonance (NMR) (C). Both methods of detection indicated that CTX0E03 incorporated significantly more agent than STROC05 cells. Higher concentrations of agent provide the greatest agent uptake in a dose-dependent fashion. The 5mg/ml for 24h condition therefore provided the most signal of 19F in this population of cells with 1.96×1012 19F spins/cell (D). A higher magnification of the agent’s cellular incorporation indicates a clear intracellular labeling of the CTX0E03 cells (green = nestin, red is BODIPy-TR) (E). The agent inside the cell is also visible through differential interference contrast (DIC).
3.2. Minor effects of 19F labeling on cell fate
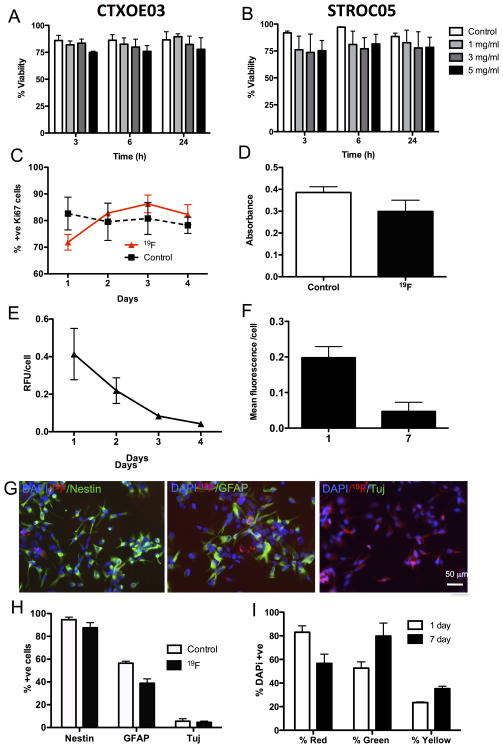
Labeling of cells with the 19F agent resulted in a minor decrease in viability (4–12%). CTX0E03 cells exhibited a better tolerance to the labeling procedure compared to STROC05 cells. Labeling over 24 hours with 5 mg/ml of 19F agent did not result in a significant decrease of viability for the CTX0E03 (Figure 2A) or STROC05 cells (Figure 2B). Based on the labeling efficiency and viability results, labeling of CTX0E03 with 5mg/ml for 24 hours was adapted as the standard protocol.
Figure 2.
Effects of 19F on cell fate. Incorporation of the 19F agent had minimal effect on cell viability in CTX0E03 (A) and STROC05 cells (B) compared to control conditions (no contrast agent added to media). However, viability in STROC05 cells was more variable after agent incorporation compared to CTX0E03 cells. A decrease in viability with the 5 mg/ml concentration was evident after 3 hours of incubation, but no significant decrease in viability was observed for longer times of incubation. There was also an effect of labeling on the number of proliferating (Ki67-positive) CTXOE03 cells at 1 day, but at subsequent time points both labeled and control cells showed similar levels of proliferation (C). A MTT assay further indicated that there was a slight reduction in mitochondrial activity after labeling, but this failed to reach statistical significance, further indicating a minimal effect on CTXOE03 cell behavior (D). Label retention in cells was high 24 hours after labeling in differentiated (E) and proliferating (F) cells. However, over time the amount of label present within the cells decreased significantly. The 19F agent did not affect CTXOE03 cells’ expression of nestin, but significantly reduced the cells’ differentiation into astrocytes (GFAP) (G&H). No effect on neuronal differentiation (Tuj, β-III-tubulin) was observed. Co-culturing of differentiated cells labeled either with the bimodal 19F (red fluorescence) with cells labeled with PKH67 (green fluorescence) indicate that approximately 20% of cells contain both labels at 1 day versus 30% at 7 days (I).
A cardinal feature of stem cells is their ability to proliferate and simultaneously produce more stem cells. This function was transiently affected in CTX0E03 cells within 24 hours of labeling as manifest by a 10% reduction in proliferating cells (Figure 2C). This was also reflected in mitochondrial activity (Figure 2D). Proliferation of CTXOE03 gradually decreased the amount of 19F agent present within a single cell, resulting in almost complete loss of detection after 4 days of proliferation (Figure 2E). Differentiating CTXOE03 labeled with the 19F agent also showed a decrease in label, but retained a higher amount after 7 days of differentiation (Figure 2F). It is important to note that differentiating will continue to dividefor a period of time before becoming post-mitotic. Cells retained their normal phenotypic characteristics (Figure 2G) with nestin expression >90% in undifferentiated cells (Figure 2H). Although no significant effect of cell labeling was observed on neuronal differentiation, a significant decrease in astrocytic differentiation was evident.
The decrease of label in a cell population may be the result of cell divisions. Therefore, despite label retention within cells, a greater quantity of cells are needed for detection using the same amount of label. It is, however, also conceivable that some label is exocytosed from the cells (i.e. leakage) and is taken-up again by other cells. To evaluate this possibility, red 19F-labeled cells were co-cultured with green Cell Tracker-labeled cells (Figure 2I). At day 1 of co-culture, approximately 20% of cells were yellow indicating that some label transfer occurred. This could either be from live cells, from dead cells that contained the 19F agent or from extracellular agent that was not sufficiently cleared by washes. Over 7 days, there was a significant 10% increase in yellow cells. Therefore at least some transfer of the agent occurs over 7 days following in vitro labeling. The presence of transplanted cells needs to be carefully evaluated to determine to what degree this also occurs in vivo.
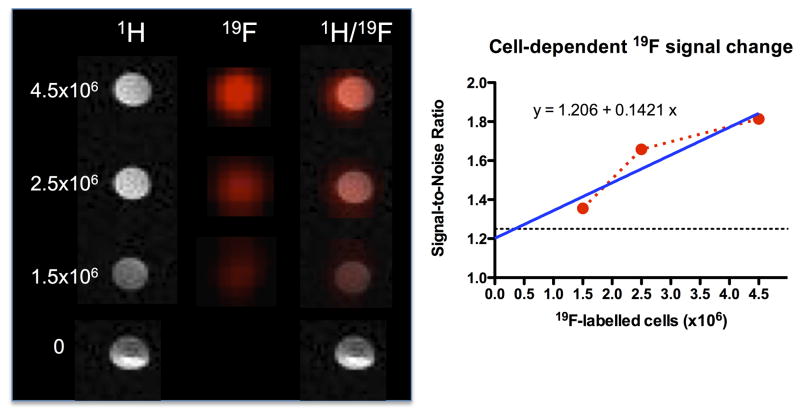
3.3. Establishing a cell detection threshold
An in vitro titration of 19F-labeled CTXOE03 cells was used to measure the 19F signal and contrast this with the signal for noise (i.e. 0 cells present). As expected, the 19F signal (and hence signal-to-noise) increased linearly with the number of cells (Figure 3). A minimum of 1.7×104 cells/voxel is required for detection at the 1.25 SNR level. Therefore, a significant amount of cells is required within each voxel to allow detection. In the case of tissue engineering inside an infarct cavity caused by a stroke, a significant amount of cells is required to completely occupy the approximately 40 μl volume of lost tissue. Hence 19F MRI is appropriate for this application.
Figure 3.
Detection threshold of 19F-labeled cells. Different concentrations (0, 1.5×106, 2.5×106, 4.5×106) of labeled cells were suspended in 6% gelatin to measure detection by 1H- and 19F-MRI. A weak 19F signal can be detected with 1.5×106 cells (35% of signal above no label/noise), whereas a good (60% signal increase) detection is evident with 2.5×106 cells. The linear increase in signal was plotted and analyzed using a linear regression. Considering a detection threshold of 1.25 SNR, a minimum of 3.1×105 cells is required for detection here. Based on the cell pellet and voxel size, this results in a detection of 1.7×104 cells/voxel.
3.4. Non-invasive imaging of 19F-labeled cells and ECM bioscaffold
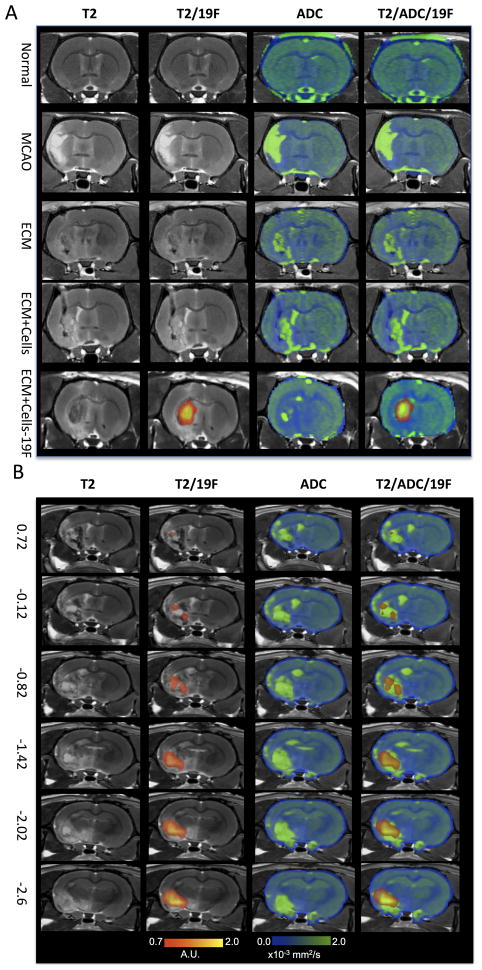
To enable tissue formation within the stroke cavity, the transplanted cells, however, require structural support and a compatible microenvironmental niche. As in normal tissue, such an environment can be provided by the ECM bioscaffold. Upon implantation into the stroke cavity the ECM+cells construct reduced the T2-hyperintensity that reflects the stroke pathology and also reduced the diffusion of water within the cavity (Figure 4A). Addition of CTXOE03 cells to the ECM bioscaffold did not further change the characteristics indicating that the ECM bioscaffold itself has the most dramatic effect upon the lesion cavity. Importantly, lack of tissue integrity of the ECM bioscaffold in the lesion cavity is still clearly visible on the apparent diffusion coefficient (ADC) maps, whereas boundaries between pathology and intact tissue on the T2-weighted image are more difficult to distinguish on these ex vivo MR images. Only transplanted cells labeled with the 19F agent produce a signal on the 19F MRI scan. Transplanted cells are distributed throughout the lesion cavity. However, especially anteriorly, small areas of infarction appear not to contain high levels of transplanted cells based on the 19F images (Figure 4B).
Figure 4.
Ex vivo 19F- and diffusion-MR imaging. A. Middle cerebral artery occlusion (MCAo) induces a hyperintense signal on a T2-weighted MR image compared to a normal control. In the absence of 19F-labeled cells, no signal is detected in the 19F scan. The apparent diffusion coefficient (ADC) image shows, as expected, an increase in diffusion in the area of stroke infarction. Injection of extracellular matrix (ECM) derived from decellularized tissue affects both the hyperintense T2-weighted signal, but also the diffusion scan. Importantly, on the T2-weighted image, the lesion cavity almost completely disappeared, although the ADC image still exhibits areas of high diffusivity. T2-weighted scans only represent the composition of the “tissue”, whereas the diffusion scan detects if the cellular and extracellular barriers to water diffusion are present/reestablished. Only 19F-labeled cells can be detected using the 19F channel. It is evident here that injection of the ECM and 19F F-labeled cells extensively cover the lesion cavity. Transplantation of unlabeled cells with the ECM did not significantly affect the ADC compared to injection of just ECM, indicating that the ECM was primarily responsible for significant changes in the lesion cavity’s characteristics. B. The coverage of the lesion cavity by the ECM and 19F-labeled cells is further illustrated along the anterior-posterior slices using stereotactic coordinates from the Rat Brain Atlas. Cells and ECM extensively cover the lesion, but it is also evident here that there are defined areas on the T2-weighted scans that are not covered by the ECM (or insufficient material is found here to change the signal to a more isointense signal). There are also areas where 19F-labelled cells do not cover the whole of the cavity (especially anterior regions).
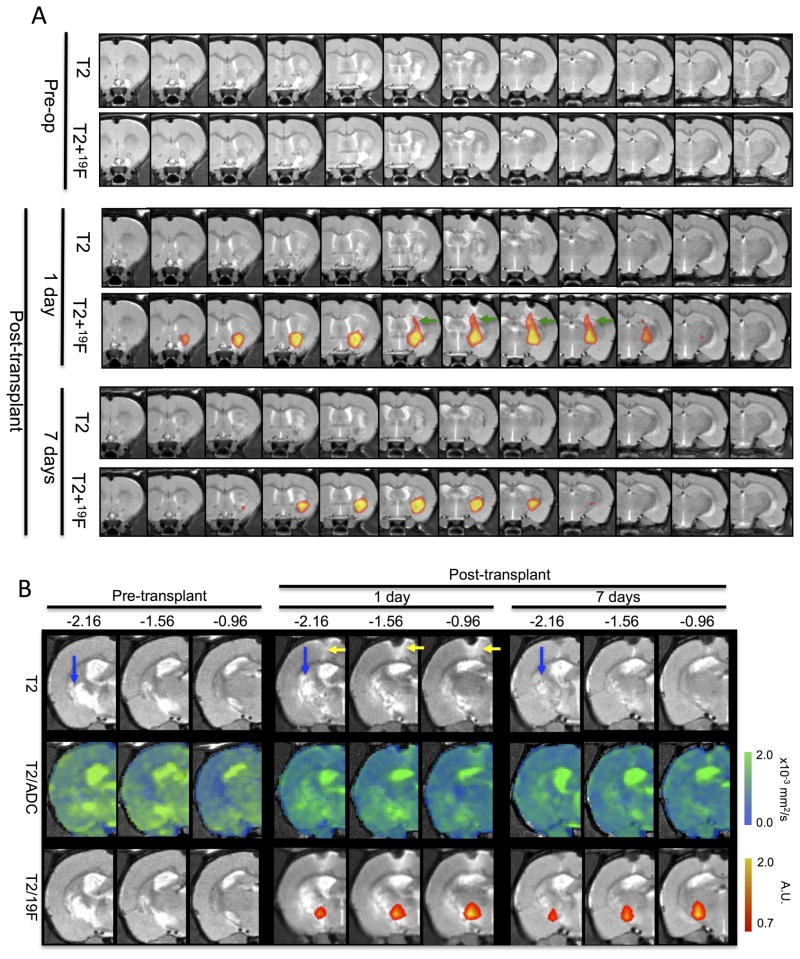
A serial in vivo study further highlighted the potential of the ECM+cells construct to promote in situ tissue formation. 19F- and diffusion MRI are important tools to monitor this process. 19F-labeled cells target the lesion cavity based on stereotactic coordinates that are derived from pre-transplant MRI scans (Figure 5A). The injection tract is also visible 1 day following transplantation using 19F-MRI, but this is no longer visible 1 week post-transplantation. However, the presence of cells in the core of the lesion cavity is very clear, although the area covered by the 19F signal significantly decreased between 1 and 7 days. In one animal, dramatic changes in the posterior areas of the lesion cavity were apparent (Figure 5B). Pre-transplantation, a robust lesion cavity was visible on both the T2- and diffusion-MRI. After transplantation, some attenuation of this signal was apparent and based on the 19F MRI scan, transplanted cells were detected in the ventral part of the lesion. By 7 days following transplantation, the T2- and diffusion-MRI scans indicated a dramatically reduced infarction with transplanted cells still present within the ventral part of the lesion. These scans indicate that dramatic changes in the lesion cavity can be achieved by the transplantation of ECM bioscaffold seeded with human neural stem cells.
Figure 5.
In vivo serial 19F- and diffusion-MR imaging. A. The distribution of 19F-labelled human neural stem cells within de-cellularised extra-cellular matrix encompass the lesion cavity. 19F-labelled cells can also be seen along the injection tract (green arrow) that crosses through “intact” overlying cortex on day 1. However, by 7 days post-transplantation, the signal from the injection tract is no longer visible, although the cells distributed throughout the lesion cavity can still be reliably detected, although the extent of distribution is reduced. B. In the posterior regions of the cavity in 1 animal, dramatic changes were observed on both the T2- and diffusion-weighted MR images. The pre-transplant images indicate a clear cavity with high diffusion (blue arrows), but 1 day post-injection of ECM+19F-labeled cells, this area gradually reduces. By 7 days post-transplantation, this area mostly exhibits a normal signal, although diffusion has not completed restored to a normal signature. It is further evident that 19F-labeled cells remain within this region without a decrease or spreading out of the signal over time.
3.5. Histologic validation of 19F-MRI cell detection
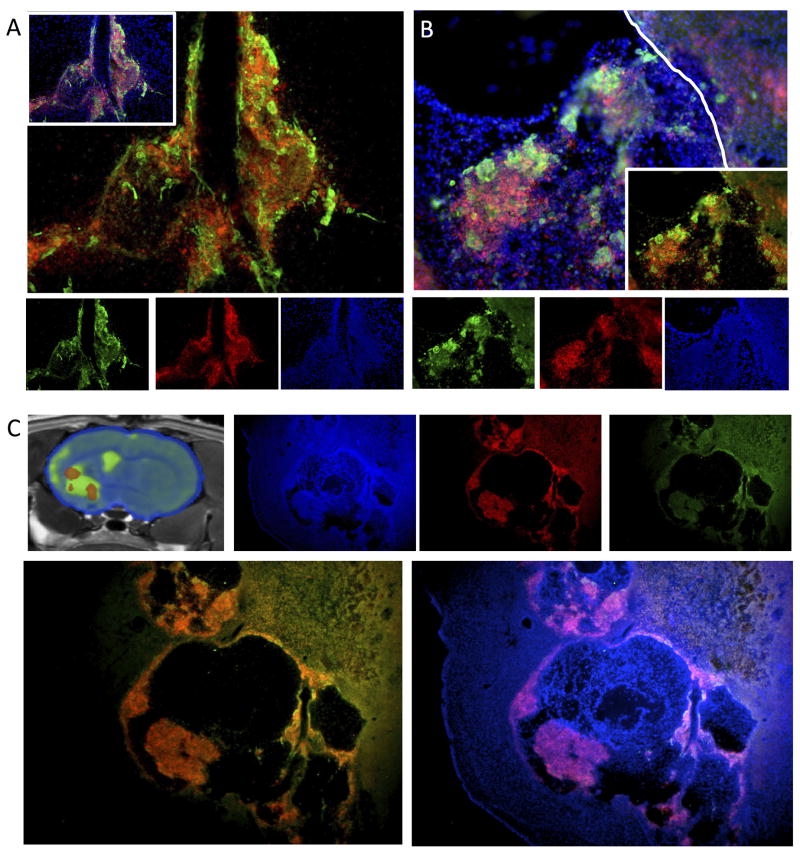
As indicated by the 19F-MR images, at day 1 transplanted 19F-labeled CTXOE03 cells were contained within the injection tract. Immunohistochemistry validated this observation with labeled cells restricted to the injection tract as it penetrated through the tissue (Figure 6A). There was a good correspondence between the distribution of the 19F label and transplanted cells macroscopically, but at the cellular level it was evident that 19.2(±4)% of cells were host cells labeled with the 19F agent. It is unclear here if this was due to a transfer from live cells or a consequence of re-uptake from labeled dead cells also contained within the transplant. A small number of transplanted cells also did not contain detectable levels of the 19F agent. Similar observations were also found within the lesion cavity (Figure 6B), where the overall number and mass of transplanted cells was dramatically higher compared to the injection tract. As indicated by the 19F MR images, within the lesion cavity there were areas that are void of transplanted cells (Figure 6C). However, these areas contained the ECM bioscaffold material. Host cells can be seen to infiltrate the implanted ECM bioscaffold and there was some transfer of the 19F label to these host cells. Nevertheless, the agent does not diffuse out of the area of transplanted cells and therefore is a good marker of the topological distribution of these cells within the host brain.
Figure 6.
Histological validation of the 19F-labeled cells. A. In the injection tract, 19F-labeled cells corresponded well to transplanted cells, but there was also a clear discrepancy between the 19F-label, transplanted and host cells. A fluorescent immunohistochemical investigation validated these observations demonstrating areas with transplanted human neural stem cells (detected using the anti-SC101 antibody with Alexa488 in green) and the BODIPy-TR red fluorescence of the 19F agent. Approximately 20% of 19F-labelled cells were host cells. However, a few transplanted cells also did not appear to contain the 19F agent. A similar distribution of 19F-label was also observed within the lesion cavity (B). The topology of distribution between transplanted cells and 19F-labelled cells was very consistent and no 19F-labelled cells were found outside of the immediate area of injection/cavity. C. 19F-MRI indicate that there is a distinct topology to the distribution of transplanted cells within the lesion cavity with areas containing 19F-labeled cells, but also areas that did not contain 19F-labelled cells (or these were below the detection threshold). Transplanted cells in some instances were clumped together and host cells infiltrated the injected ECM from decellularized tissue. This pattern of 19F-labelled cells therefore corresponded to the 19F-MR images.
3.6. ECM aids in situ tissue formation
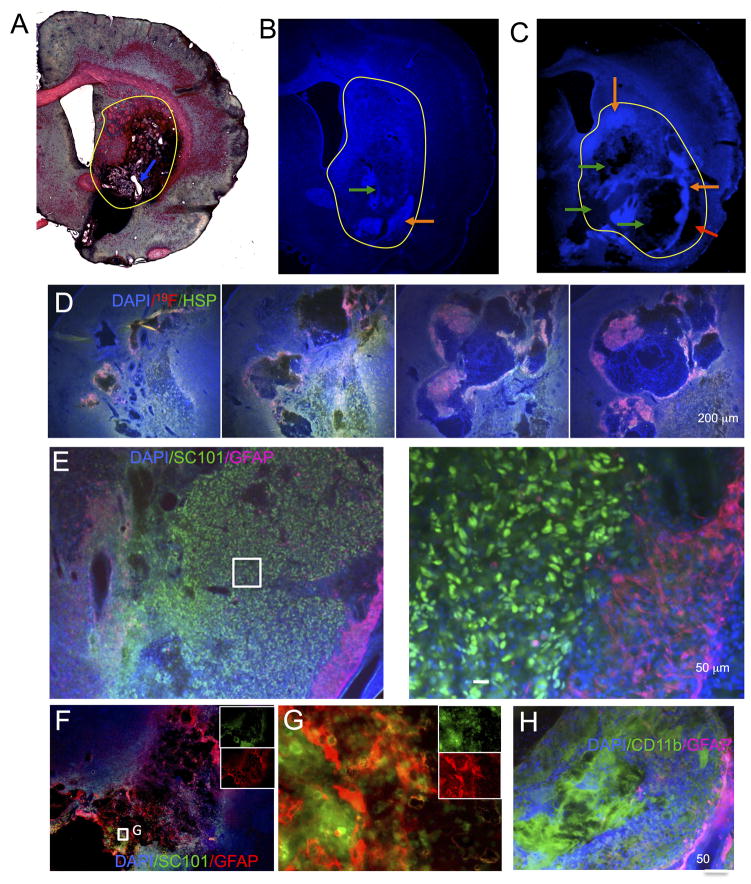
Injection of the ECM bioscaffold filled the lesion cavity (Figure 7A), but small pockets of unfilled cavity remained, indicating that gelation and viscosity of ECM bioscaffold may require some optimization to achieve a complete and uniform filling of the cavity. Human neural stem cells (CTXOE03) mixed with the ECM bioscaffold prior to transplantation also exhibited a reasonably homogenous distribution, but cellular density was variable within the ECM bioscaffold(Figure 7B). Additionally, there were areas that were devoid of cells within the ECM bioscaffold (Figure 7C). Although host cells invaded the ECM bioscaffold, a better distribution of transplanted cells is desirable. In some locations, the ECM bioscaffold did not interface with the host tissue (Figure 7C). This is likely a consequence of ECM gelation of the ECM bioscaffold, which may have prevented homogeneous distribution or been affected by mixing with the extracellular fluid. It is important to note that the lesion cavity is a large volume to fill completely and homogenously. It is conceivable that surrounding host environment exerted some influence over the presence of cells within the ECM bioscaffold, as there was a clear posterior to anterior difference in cellular distribution and lesion coverage by the ECM bioscaffold and transplanted cells (Figure 7D). The ECM bioscaffold provided a structural support to the transplanted cells, and these cells mostly remained within the grafted area with excellent survival (Figure 7E). However, the cytoarchitecture of the de novo tissue is amorphous and distinct from normal striatal tissue. Very few transplanted cells were observed to migrate into areas of the ECM bioscaffold that attracted host cells (Figure 7D), and host cells appeared to preferentially migrate into areas of the ECM bioscaffold void of transplanted cells (Figure 7D). Host astrocytes infiltrate the ECM bioscaffold (Figure 7F) and nested between grafted cells (Figure 7G), but very few transplanted cells differentiated into astrocytes. No neuronal differentiation of transplanted cells was observed. Cells infiltrating the ECM bioscaffold were predominantly of the microglia/macrophage phenotype (Figure 7H). This was evident in both ECM bioscaffold and ECM+cell transplants. Interestingly, microglia/macrophages were infrequently found in areas of transplanted cells but appeared to selectively infiltrate the unpopulated ECM bioscaffold.
Figure 7.
Histological validation of de-cellularised extracellular matrix in stroke cavity. A. A trichrome staining reveals the area with the injected ECM as a dark pinkish area. The ECM injection mostly filled up the lesion cavity (yellow circle), but small areas (white spots, blue arrow) indicate that in some areas no ECM was present. A similar assessment can be seen in fluorescence, where the DAPI stain indicates in some animals a complete coverage of the lesion (B), whereas in others a more complex pattern of cells and matrix within the lesion is evident (C). Areas of a high cellular density (orange arrow) and areas devoid of cells (green arrows) can be found throughout the injected area. It is also evident here that the gelation of the ECM left an area of the lesion cavity uncovered by ECM and cells (red arrow) (C). It is evident that the cells and ECM mixture throughout the lesion is not always homogenous (D). Posterior regions (left) often were more homogenous than anterior regions within the same animal (right). However, overall graft survival was excellent (E) with a large number of cells surviving (as marked in green here with SC101), although only very few penetrated the host glial scar (GFAP in pink). Only a very small percentage (<5%) of transplanted cells differentiated in astrocytes. In contrast, host astrocytes were found to invade some aspects of injected ECM providing “bridges” between edges of host tissue (F). Transplanted cells were interspersed with host astrocytes in these circumstances (G). The vast majority of cells infiltrating the ECM consisted of CD11b-positive cells representing cells from the microglia/macrophage phenotype (H).
4. DISCUSSION
Improving behavioral/functional deficits after stroke remains a major challenge. Although cell transplantation can promote improvements in behavioral deficits, recovery often remains incomplete and a large tissue cavity remains within the brain. In situ tissue engineering is an exciting new opportunity to potentially regenerate some of the lost tissue after a stroke. However, a structural support is required to allow neural stem cells to remain within the cavity and interconnect to form a de novo tissue. We herein demonstrate that an extracellular matrix (ECM) bioscaffold provides an appropriate microenvironment for these cells to remain within the lesion cavity and to form a primitive tissue. However this tissue lacks an appropriate cytoarchitecture and phenotypic differentiation. We further demonstrate here that non-invasive imaging can be used to monitor the process of construct remodeling and tissue regeneration. The distribution of transplanted cells within the lesion cavity can be visualized using a 19F-MRI contrast agent that retains the ability to monitor the evolution of the lesion environment by T2- and diffusion-weighted MRI. Diffusion MRI is of particular interest because it is sensitive to tissue integrity and the re-establishment of diffusion barriers that are found within normal tissue. In contrast, restoration of a T2 signal comparable to normal striatum is not necessarily evidence of a restoration of striatal tissue, but a reflection of similar tissue density established through the grafting of cells and ECM bioscaffold within the stroke-induced cavity. This study therefore highlights the potential to promote tissue growth within a stroke cavity and that the ability to monitor lesion remodeling processes non-invasively using 19F- and diffusion-MRI.
4.1. In situ tissue engineering in the brain
Biomaterials have been extensively used to promote spinal cord repair [37–39]. However, little progress has been achieved to implement these approaches into the brain. The main reason for this lack of progress is the more restricted nature of access through the skull and inadequate delivery. The use of MRI and appropriately engineered biomaterials that can be injected directly into the brain through a thin needle overcome these limitations [6]. However, the lesion cavity offers certain advantages over implanting cells into the parenchyma. Notably, it affords implantation of larger cell volumes and combination with biomaterials which improves overall graft survival and diminishes the inflammatory response [8, 40]. A very extensive graft survival was observed here within the lesion cavity, although the distribution of cells was not homogenous throughout the ECM bioscaffold. Lack of construct homogeneity indicates that further development is needed to prepare a homogenous mixture of biomaterials and cells and to retain this consistency upon implantation. Thermoreversible gelation of biomaterials is attractive for transplantation [41] and provides an anchoring of cells within the cavity. Cellular distribution throughout the infarct area might be improved by decreasing gelation time and mitigating the combination of the biomaterial/cell mixture with the ECM bioscaffold in the lesion cavity. Invasion of the host parenchyma was also minimal and demands improvement to assure an integration of the de novo tissue with surrounding functional host tissue. It is also worth noting that there was no significant phenotypic differentiation of transplanted cells, potentially indicating that there is a lack of sufficient salient signaling that would guide the cells towards a particular regional and phenotypic identity.
This local isolation of the graft, however, is potentially an advantage of tissue-specific ECM. The ECM provides components that define the structural microenvironment. It is conceivable that tissue-specific ECM could be harvested from differentiated cells in vitro [42] and then prepared for used with undifferentiated hNSCs for transplantation. This tissue-specific ECM information could then cue transplanted cells by defining their regional identity given the absence of appropriate cues within the lesion cavity. It is known that regeneration of tissue through implantation of ECM requires digestion by inflammatory cells. Specifically, the M2 subtype of microglia/macrophages from the myeloid lineage are thought to be essential to this process [43]. Indeed, a dramatic infiltration of microglia/macrophages into the ECM was observed in the present study. Interestingly, areas with transplanted cells generally did not see this infiltration suggesting that transplanted cells may secrete factors preventing this infiltration or digest the ECM themselves and thereby attenuate or eliminate signals inducing the microglia/macrophage infiltration.
It is conceivable that in the absence of transplanted cells, M2 sub-type myeloid cells are required to digest some ECM and thereby release chemoattractant factors that provoke an infiltration of host stem cells that then repopulate the remaining ECM [44]. This phenomenon would allow an approach that does not require the transplantation of cells, but instead relies on the recruitment of the host’s endogenous stem cells. Strategies to improve this recruitment process into the lesion cavity have been shown to attract endogenous cells from the sub-ependymal zone [45, 46]. Reports of growth factors being secreted from biomaterials inside the cavity suggest that a dramatic improvement in recovery can be produced even without the transplantation of cells [47, 48]. However, it is unclear if this is indeed due to a repair of the lesion cavity or by promoting changes in the host parenchyma. Nevertheless, implantation of biomaterials into the stroke cavity offers the opportunity to deliver large quantities of neuroprotective factors locally in the area of damage, potentially avoiding side effects if these factors were delivered systemically [49]. The combination of functionalized biomaterials and transplanted cells introduces novel opportunities to potentially repair stroke-induced tissue damage.
4.2. Non-invasive imaging of a regenerating tissue
The implantation of cells with biomaterials introduces new challenges to visualize and monitor the repair process non-invasively. Although the visualization of transplanted cells by MRI has been widely used, this cellular MRI relies on contrast agents that affect the 1H MRI signal [24]. This interferes with the detection of the lesion cavity on T2-weighted images and prevents the monitoring of a potential regeneration of tissue within this area [5]. A method of regenerative imaging is therefore required that can distinguish the various components involved in regenerating a functional tissue.
Multi-nuclear MRI can detect contrast agents that do not affect the 1H signal of the lesion cavity. Specifically, 19F nuclei have a sensitivity of 83% compared to 1H. Labeling of cells with a 19F contrast agent therefore affords the non-invasive visualization of transplanted cells overlaid on a 1H anatomical image [25]. A dual-modal 19F contrast agent here only minimally affected cell behaviors in vitro and provided a faithful representation of the distribution of transplanted cells within the lesion cavity without interfering with the detection of stroke damage or de novo tissue formation. It is, however, important to note that 19.2% of cells containing the 19F contrast agent were host cells rather than transplanted cells. The use of a reliable antibody against human cells in the rat, as well as the use of a bimodal agent, here allowed reliable assessment of the label transfer. It is likely that dead transplanted cells (15%) released the agent which was then taken-up by host cells. It is also conceivable that some agent was transferred to host cells, but this is less likely to have resulted in a significant host labeling. However, the low detection threshold also indicates that this approach is unlikely to afford the detection of a small number of cells generally used to track the migration of transplanted cells [31]. In the case of in situ tissue regeneration, the tracking of a few migrating cells is not as important as visualizing the presence and distribution of transplanted cells involved in the formation of new tissue. In this case, the most relevant information is to define the main distribution of transplanted cells and this study demonstrates that 19F-labelling is adequate for this purpose.
Being able to define where most of the transplanted cells are located then requires multiple imaging methods to characterize the processes involved in regenerating the lost tissue. T2-weighted MR imaging indicates changes in the lesion based on the spin density that differs markedly between extracellular fluid filling the cavity versus the presence of cells and ECM. Some of the T2-weighted images collected in this study indicate a MR signal in the cavity similar to that of surrounding intact tissue, but diffusion MRI reveals that the tissue had not yet completely established the same integrity observed in a normal tissue. The ECM with cells produced remarkable changes in the diffusion MRI signal that indicated a disappearance of the extracellular fluid-filled cavity caused by the stroke. It will be important to expand on the current study with longer time points that could see further changes in tissue formation.
The general lack of blood vessel infiltration into the lesion cavity is a major concern for the long-term survival of new tissue observed in this study. Transplanted cells are likely to survive if there are insufficient barriers to the perfusion of the cavity with extracellular fluid. As the extracellular matrix and cellular connections mature, this wide-spread perfusion with extracellular fluid will decrease, resulting in a lack of nutrient support to sustain cell survival. Formation of blood vessels within the ECM and newly-formed tissue will be required to guarantee long-term cell survival. Supplementing biomaterials with, for instance, vascular endothelial growth factor (VEGF) could attract blood vessels and ensure long-term survival [48].
Ideally, regenerative imaging will be able to incorporate imaging paradigm that can visualize the distinct processes involved in regenerating de novo tissue within the stroke cavity. Not only will this need to encompass the detection and distribution of transplanted cells and general characteristics of tissue structure (T2, diffusion), but also allow a thorough assessment of blood supply to the area (perfusion MRI), as well as its region-specific differentiation (MR spectroscopy). One of the major technical challenges for this regenerative imaging is to develop efficient scanning protocols that would allow an incorporation of all of these elements within a single scanning session and provide an integrated visualization of all the processes, enabling control and coordination of post-stroke de novo brain tissue development.
5. Conclusion
In situ tissue engineering affords an exciting new opportunity to apply regenerative medicine approaches in the injured brain. Herein, we demonstrate that extracellular matrix has potential as a substrate for transplanted cells leading to the formation of de novo tissue. However, it is not yet clear whether de novo tissue formed from matrix and cells can exhibit normal cytoarchitecture of the tissue it is replacing, whether it integrates functionally with the rest of the brain, and whether this approach will ultimately yield behavioral recovery. Visualizing the processes involved in de novo tissue formation will be key to monitor and better understand these processes. As shown here, regenerative imaging must not only visualize transplanted cells, but must also detect the extent of angiogenesis, the establishment of a normal tissue structure and the differentiation of cells into region-specific phenotypes.
Supplementary Material
A. Relaxivity measurements of 19F agent in solution. A phantom consisting of 219.4 mg/ml in 1 ml distilled H2O was placed in the scanner and both T1 and T2 measurements were taken. For T1 measurements, a spin echo multi-slice (SEMS) sequence (TE=14.5 ms; TR array = 19.76, 25, 40, 60, 100, 200, 500, 100, 2000, 4000, 10000 ms; averages = 16; FOV = 30 × 10 mm; matrix = 128 × 32; single slice at 8 mm thickness) was used to produce a map in Image J from which the R1 of the 19F agent was calculated to be 425 ms. For T2 measurements, a SEMS sequence (TR = 5000 ms; TE array = 14.52, 16, 24, 50, 100, 200, 300; averages = 16; FOV = 30 × 10 mm; matrix = 128 × 32; single slice at 8 mm thickness) indicated a R2 value of 82 ms. B. NMR measurement of 19F spins in labeled CTX0E03 cells (5mg/ml for 24 h). One main peak can be detected at −76 ppm (C). A defined concentration of TFA in the sample is used as a means to quantify the fluorine signal. A second, much weaker, peak of the 19F agent is observed at −91.5 ppm (D).
A. The bimodal 19F agent is attaching to the laminin-coated flask, with most of the agent (red fluorescence) after PFA fixation being on the surface of the cells or attached the culture flask. B. To achieve a consistent intracellular labeling and minimize extracellular attachment of the agent, 10% human serum albumin was added to the culture media. C. The “stickiness” of the agent also constitutes a problem to detach cells from the culture vessel for transplantation. Although a cell scraper can be used to increase the yield of cells from the vessel, this dramatically reduces the viability of the cells. In contrast, trypsination of the cells produces an acceptable level of cell viability for transplantation (>85%).
Acknowledgments
Dr Saga Johansson for pilot studies using the 19F MRI agent, Dr Jelena Janjic for preliminary versions of the 19F agent, Mr Christopher Medberry for capturing images of stained tissue sections. This study was funded through a NIBIB Quantum Grant (1 P20 EB007076-01), a translation stem cell grant by the UK Medical Research Council (G0802552) and the 7th framework from the European Union (201842-ENCITE). The authors also would like to thank Dr Po-Wah So, the manager of the preclinical imaging unit at KCL, and the British Heart Foundation for funding the MRI scanner.
Footnotes
Conflict of Interest Statement
Mr Anthony Bladucci is an employee of Celsense, the producer of the 19F MRI contrast agent. Dr Eric T. Ahrens is a consultant to Celsense.
Authors’ contribution
Ellen Bible: Design of study, conducted all in vitro experiments, cell and ECM integration, gel phantom preparation, MCAo surgery, cell transplantation, acquired T2 MRI scans, perfusion-fixation, immunohistochemistry, data analysis. Flavio Dell’Acqua: Design of study, set-up of diffusion MRI, acquisition of ex vivo and in vivo diffusion MRI, data processing for diffusion MRI, co-registration of MRI data sets. Bhavana Solanky: Design of study, set-up of 19F imaging, 19F imaging of gel phantom, 19F imaging of ex vivo and in vivo experiments. Anthony Balducci: Synthesis and quality control of 19F agent, 19F NMR of cell uptake. Peter Crapo: Development and preparation of extracellular matrix, integration of ECM with cells. Stephen Badylak: Design of study, development of ECM, financial support. Eric Ahrens: Design of study, development of 19F agent and 19F NMR of cell uptake. Mike Modo: Conceived study, data collection, data processing, financial support, drafted manuscript, final approval of manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aboody K, Capela A, Niazi N, Stern JH, Temple S. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta Stone. Neuron. 2011;70(4):597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Modo M, Beech JS, Meade TJ, Williams SC, Price J. A chronic 1 year assessment of MRI contrast agent-labelled neural stem cell transplants in stroke. Neuroimage. 2009;47 (Suppl 2):T133–142. doi: 10.1016/j.neuroimage.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orive G, Anitua E, Pedraz JL, Emerich DF. Biomaterials for promoting brain protection, repair and regeneration. Nat Rev Neurosci. 2009;10(9):682–692. doi: 10.1038/nrn2685. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Cooke MJ, Shoichet MS. Creating permissive microenvironments for stem cell transplantation into the central nervous system. Trends Biotechnol. 2011 Aug 8; doi: 10.1016/j.tibtech.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Bible E, Chau DY, Alexander MR, Price J, Shakesheff KM, Modo M. The support of neural stem cells transplanted into stroke-induced brain cavities by PLGA particles. Biomaterials. 2009;30(16):2985–2994. [Google Scholar]
- 6.Bible E, Chau DY, Alexander MR, Price J, Shakesheff KM, Modo M. Attachment of stem cells to scaffold particles for intra-cerebral transplantation. Nat Protoc. 2009;4(10):1440–1453. doi: 10.1038/nprot.2009.156. [DOI] [PubMed] [Google Scholar]
- 7.Kubinova S, Sykova E. Nanotechnologies in regenerative medicine. Minim Invasive Ther Allied Technol. 2010;19(3):144–156. doi: 10.3109/13645706.2010.481398. [DOI] [PubMed] [Google Scholar]
- 8.Park KI, Teng YD, Snyder EY. The injured brain interacts reciprocally with neural stem cells supported by scaffolds to reconstitute lost tissue. Nat Biotechnol. 2002;20(11):1111–1117. doi: 10.1038/nbt751. [DOI] [PubMed] [Google Scholar]
- 9.Agnati LF, Guidolin D, Fuxe K. The brain as a system of nested but partially overlapping networks. Heuristic relevance of the model for brain physiology and pathology. J Neural Transm. 2007;114(1):3–19. doi: 10.1007/s00702-006-0563-x. [DOI] [PubMed] [Google Scholar]
- 10.Baeten KM, Akassoglou K. Extracellular matrix and matrix receptors in blood-brain barrier formation and stroke. Dev Neurobiol. 2011;71(11):1018–1039. doi: 10.1002/dneu.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentin JE, Turner NJ, Gilbert TW, Badylak SF. Functional skeletal muscle formation with a biologic scaffold. Biomaterials. 2010 Oct;31(29):7475–7484. doi: 10.1016/j.biomaterials.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner NJ, Yates AJ, Jr, Weber DJ, Qureshi IR, Stolz DB, Gilbert TW, et al. Xenogeneic extracellular matrix as an inductive scaffold for regeneration of a functioning musculotendinous junction. Tissue Eng Part A. 2010;16(11):3309–3317. doi: 10.1089/ten.TEA.2010.0169. [DOI] [PubMed] [Google Scholar]
- 13.Kropp BP, Ludlow JK, Spicer D, Rippy MK, Badylak SF, Adams MC, et al. Rabbit urethral regeneration using small intestinal submucosa onlay grafts. Urology. 1998;52(1):138–142. doi: 10.1016/s0090-4295(98)00114-9. [DOI] [PubMed] [Google Scholar]
- 14.Kropp BP, Rippy MK, Badylak SF, Adams MC, Keating MA, Rink RC, et al. Regenerative urinary bladder augmentation using small intestinal submucosa: urodynamic and histopathologic assessment in long-term canine bladder augmentations. J Urol. 1996;155(6):2098–2104. doi: 10.1016/s0022-5347(01)66117-2. [DOI] [PubMed] [Google Scholar]
- 15.Badylak S, Meurling S, Chen M, Spievack A, Simmons-Byrd A. Resorbable bioscaffold for esophageal repair in a dog model. J Pediatr Surg. 2000;35(7):1097–1103. doi: 10.1053/jpsu.2000.7834. [DOI] [PubMed] [Google Scholar]
- 16.Badylak SF, Vorp DA, Spievack AR, Simmons-Byrd A, Hanke J, Freytes DO, et al. Esophageal reconstruction with ECM and muscle tissue in a dog model. J Surg Res. 2005;128(1):87–97. doi: 10.1016/j.jss.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Badylak S, Obermiller J, Geddes L, Matheny R. Extracellular matrix for myocardial repair. Heart Surg Forum. 2003;6(2):E20–26. doi: 10.1532/hsf.917. [DOI] [PubMed] [Google Scholar]
- 18.Kochupura PV, Azeloglu EU, Kelly DJ, Doronin SV, Badylak SF, Krukenkamp IB, et al. Tissue-engineered myocardial patch derived from extracellular matrix provides regional mechanical function. Circulation. 2005;112(9 Suppl):I144–149. doi: 10.1161/CIRCULATIONAHA.104.524355. [DOI] [PubMed] [Google Scholar]
- 19.Cobb MA, Badylak SF, Janas W, Boop FA. Histology after dural grafting with small intestinal submucosa. Surg Neurol. 1996;46(4):389–393. doi: 10.1016/s0090-3019(96)00202-9. discussion 393-384. [DOI] [PubMed] [Google Scholar]
- 20.Cobb MA, Badylak SF, Janas W, Simmons-Byrd A, Boop FA. Porcine small intestinal submucosa as a dural substitute. Surg Neurol. 1999;51(1):99–104. doi: 10.1016/s0090-3019(97)00475-8. [DOI] [PubMed] [Google Scholar]
- 21.Boruch AV, Nieponice A, Qureshi IR, Gilbert TW, Badylak SF. Constructive remodeling of biologic scaffolds is dependent on early exposure to physiologic bladder filling in a canine partial cystectomy model. J Surg Res. 2010;161(2):217–225. doi: 10.1016/j.jss.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal V, Brown BN, Beattie AJ, Gilbert TW, Badylak SF. Evidence of innervation following extracellular matrix scaffold-mediated remodelling of muscular tissues. J Tissue Eng Regen Med. 2009;3(8):590–600. doi: 10.1002/term.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modo M. Understanding stem cell-mediated brain repair through neuroimaging. Curr Stem Cell Res Ther. 2006;1(1):55–63. doi: 10.2174/157488806775269106. [DOI] [PubMed] [Google Scholar]
- 24.Modo M, Hoehn M, Bulte JW. Cellular MR imaging. Mol Imaging. 2005;4(3):143–164. doi: 10.1162/15353500200505145. [DOI] [PubMed] [Google Scholar]
- 25.Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat Biotechnol. 2005;23(8):983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 26.Johansson S, Price J, Modo M. Effect of inflammatory cytokines on major histocompatibility complex expression and differentiation of human neural stem/progenitor cells. Stem Cells. 2008;26(9):2444–2454. doi: 10.1634/stemcells.2008-0116. [DOI] [PubMed] [Google Scholar]
- 27.Pollock K, Stroemer P, Patel S, Stevanato L, Hope A, Miljan E, et al. A conditionally immortal clonal stem cell line from human cortical neuroepithelium for the treatment of ischemic stroke. Exp Neurol. 2006;199(1):143–155. doi: 10.1016/j.expneurol.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 28.El-Akabawy G, Medina LM, Jeffries A, Price J, Modo M. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the hedgehog pathway. Stem Cells Dev. 2011;20(11):1873–1887. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 29.Janjic JM, Srinivas M, Kadayakkara DKK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. Journal of the American Chemical Society. 2008;130(9):2832–2841. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 30.Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn Reson Med. 2007;58:725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- 31.Brekke C, Morgan SC, Lowe AS, Meade TJ, Price J, Williams SC, et al. The in vitro effects of a bimodal contrast agent on cellular functions and relaxometry. NMR Biomed. 2007;20(2):77–89. doi: 10.1002/nbm.1077. [DOI] [PubMed] [Google Scholar]
- 32.Freytes DO, Martin J, Velankar SS, Lee AS, Badylak SF. Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix. Biomaterials. 2008;29(11):1630–1637. doi: 10.1016/j.biomaterials.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32(12):3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodde J. Naturally occurring scaffolds for soft tissue repair and regeneration. Tissue Eng. 2002;8(2):295–308. doi: 10.1089/107632702753725058. [DOI] [PubMed] [Google Scholar]
- 35.Modo M, Stroemer RP, Tang E, Veizovic T, Sowniski P, Hodges H. Neurological sequelae and long-term behavioural assessment of rats with transient middle cerebral artery occlusion. J Neurosci Methods. 2000;104(1):99–109. doi: 10.1016/s0165-0270(00)00329-0. [DOI] [PubMed] [Google Scholar]
- 36.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Amsterdam, NL: Academic Press; 2007. [Google Scholar]
- 37.Perale G, Giordano C, Bianco F, Rossi F, Tunesi M, Daniele F, et al. Hydrogel for cell housing in the brain and in the spinal cord. Int J Artif Organs. 2011;34(3):295–303. doi: 10.5301/ijao.2011.6488. [DOI] [PubMed] [Google Scholar]
- 38.Kubinova S, Sykova E. Nanotechnology for treatment of stroke and spinal cord injury. Nanomedicine (Lond) 2010;5(1):99–108. doi: 10.2217/nnm.09.93. [DOI] [PubMed] [Google Scholar]
- 39.Teng YD, Lavik EB, Qu X, Park KI, Ourednik J, Zurakowski D, et al. Functional recovery following traumatic spinal cord injury mediated by a unique polymer scaffold seeded with neural stem cells. Proc Natl Acad Sci U S A. 2002;99(5):3024–3029. doi: 10.1073/pnas.052678899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong J, Chan A, Morad L, Kornblum HI, Fan G, Carmichael ST. Hydrogel matrix to support stem cell survival after brain transplantation in stroke. Neurorehabil Neural Repair. 2010;24(7):636–644. doi: 10.1177/1545968310361958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osanai T, Kuroda S, Yasuda H, Chiba Y, Maruichi K, Hokari M, et al. Noninvasive transplantation of bone marrow stromal cells for ischemic stroke: preliminary study with a thermoreversible gelation polymer hydrogel. Neurosurgery. 2010;66(6):1140–1147. doi: 10.1227/01.NEU.0000369610.76181.CF. discussion 1147. [DOI] [PubMed] [Google Scholar]
- 42.Wolchok JC, Tresco PA. The isolation of cell derived extracellular matrix constructs using sacrificial open-cell foams. Biomaterials. 2010;31(36):9595–9603. doi: 10.1016/j.biomaterials.2010.08.072. [DOI] [PubMed] [Google Scholar]
- 43.Stefater JA, 3rd, Ren S, Lang RA, Duffield JS. Metchnikoff’s policemen: macrophages in development, homeostasis and regeneration. Trends Mol Med. 2011;17(12):743–752. doi: 10.1016/j.molmed.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal V, Tottey S, Johnson SA, Freund JM, Siu BF, Badylak SF. Recruitment of progenitor cells by an extracellular matrix cryptic peptide in a mouse model of digit amputation. Tissue Eng Part A. 2011;17(19–20):2435–2443. doi: 10.1089/ten.tea.2011.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooke MJ, Wang Y, Morshead CM, Shoichet MS. Controlled epi-cortical delivery of epidermal growth factor for the stimulation of endogenous neural stem cell proliferation in stroke-injured brain. Biomaterials. 2011;32(24):5688–5697. doi: 10.1016/j.biomaterials.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Cooke MJ, Lapitsky Y, Wylie RG, Sachewsky N, Corbett D, et al. Transport of epidermal growth factor in the stroke-injured brain. J Control Release. 2011;149(3):225–235. doi: 10.1016/j.jconrel.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Tian WM, Hou SP, Xu QY, Spector M, Cui FZ. An experimental test of stroke recovery by implanting a hyaluronic acid hydrogel carrying a Nogo receptor antibody in a rat model. Biomed Mater. 2007;2(4):233–240. doi: 10.1088/1748-6041/2/4/005. [DOI] [PubMed] [Google Scholar]
- 48.Emerich DF, Silva E, Ali O, Mooney D, Bell W, Yu SJ, et al. Injectable VEGF hydrogels produce near complete neurological and anatomical protection following cerebral ischemia in rats. Cell Transplant. 2010;19(9):1063–1071. doi: 10.3727/096368910X498278. [DOI] [PubMed] [Google Scholar]
- 49.Nicholls F, Gorenkova N, Bible E, Franco C, Chau DY, Tolias C, et al. Improving the treatment of stroke through nanotechnology. In: Heiss JD, Kateb B, editors. Nanoneurosurgery. Boca Raton, FL: CRC Press; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Relaxivity measurements of 19F agent in solution. A phantom consisting of 219.4 mg/ml in 1 ml distilled H2O was placed in the scanner and both T1 and T2 measurements were taken. For T1 measurements, a spin echo multi-slice (SEMS) sequence (TE=14.5 ms; TR array = 19.76, 25, 40, 60, 100, 200, 500, 100, 2000, 4000, 10000 ms; averages = 16; FOV = 30 × 10 mm; matrix = 128 × 32; single slice at 8 mm thickness) was used to produce a map in Image J from which the R1 of the 19F agent was calculated to be 425 ms. For T2 measurements, a SEMS sequence (TR = 5000 ms; TE array = 14.52, 16, 24, 50, 100, 200, 300; averages = 16; FOV = 30 × 10 mm; matrix = 128 × 32; single slice at 8 mm thickness) indicated a R2 value of 82 ms. B. NMR measurement of 19F spins in labeled CTX0E03 cells (5mg/ml for 24 h). One main peak can be detected at −76 ppm (C). A defined concentration of TFA in the sample is used as a means to quantify the fluorine signal. A second, much weaker, peak of the 19F agent is observed at −91.5 ppm (D).
A. The bimodal 19F agent is attaching to the laminin-coated flask, with most of the agent (red fluorescence) after PFA fixation being on the surface of the cells or attached the culture flask. B. To achieve a consistent intracellular labeling and minimize extracellular attachment of the agent, 10% human serum albumin was added to the culture media. C. The “stickiness” of the agent also constitutes a problem to detach cells from the culture vessel for transplantation. Although a cell scraper can be used to increase the yield of cells from the vessel, this dramatically reduces the viability of the cells. In contrast, trypsination of the cells produces an acceptable level of cell viability for transplantation (>85%).