SUMMARY
Pro-inflammatory cytokine TNFα plays critical roles in promoting malignant cell proliferation, angiogenesis, and tumor metastasis in many cancers. However, the mechanism of TNFα-mediated tumor development remains unclear. Here, we show that IKKα, an important downstream kinase of TNFα, interacts with and phosphorylates FOXA2 at S107/ S111, thereby suppressing FOXA2 transactivation activity, leading to decreased NUMB expression and further activates the downstream NOTCH pathway and promotes cell proliferation and tumorigenesis. Moreover, we found that levels of IKKα, pFOXA2 (S107/111), and activated NOTCH1 were significantly higher in hepatocellular carcinoma tumors than in normal liver tissues and that pFOXA2 (S107/111) expression was positively correlated with IKKα and activated NOTCH1 expression in tumor tissues. Therefore, dysregulation of NUMB-mediated suppression of NOTCH1 by TNFα/IKKα-associated FOXA2 inhibition likely contributes to inflammation-mediated cancer pathogenesis. Here, we report TNFα/IKKα/FOXA2/NUMB/NOTCH1 pathway that is critical for inflammation-mediated tumorigenesis and may provide a target for clinical intervention in human cancer.
INTRODUCTION
The link between inflammation and cancer has long been observed (Balkwill, 2001), and increasing epidemiological evidence has shown that chronic inflammation, such as liver infection or bowel inflammation, increases the risk of cancer development in certain organs (Castello et al., 2010; Danese, 2010). In addition, numerous studies have shown that inflammatory mediators play critical roles in cancer-related inflammation and promote malignant cell proliferation, angiogenesis, and tumor metastasis in many cancers (Allavena et al., 2008).
Recently, many non-IκB targets of IKKα and IKKβ have been identified. Previous studies showed that IKKα can regulate several target genes involved in cell transformation, tumor progression, and angiogenesis (Carayol, Wang, 2006, Luo et al., 2007, Huang et al., 2007). Moreover, upon activation by TNFα stimulation, IKKα accumulates in the nucleus and phosphorylates histone H3 to regulate transcription of target genes (Anest et al., 2003; Yamamoto et al., 2003). However, how IKKα regulates distinct pathways independent to traditional IKKα/IKKβ complex have not yet been clearly identified.
FOXA proteins have been shown to play important roles in regulating a wide spectrum of biological processes (Wolfrum et al., 2003; Lee et al., 2005) and many FOXA2 target genes have been identified from global location analysis (Wederell et al., 2008) and conditional gene ablation. Previous reports showed critical roles of FOXA2 in liver-associated diseases. (Bochkis et al., 2008, Lehner et al., 2007) and that inflammatory stimuli-induced loss of FOXA2 is involved in inflammation-associated obstructive pulmonary diseases (Jeffrey et al., 2011). Although dysregulation of FOXA2 has been directly linked to the progression of certain cancers, the role of FOXA2 in tumor progression is not clear.
NOTCH proteins play fundamental roles in cell fate decisions (Hsieh et al., 1996; Kimble and Crittenden, 2007; Tanigaki and Honjo, 2007). Once released from the extracellular part of the molecule, the NOTCH intracellular domain (NICD) translocates into the nucleus to activate transcription of target genes (Iso et al., 2003). Aberrant expression of the dominant active cytoplasmic domain of NOTCH receptors through chromosomal translocations or mutations in hematopoietic cells leads to cell-autonomous oncogenic activation of NOTCH (Medyouf et al., 2010).
NUMB is an important determinant of asymmetric cell division in mammalian development. Recent studies have demonstrated that NUMB acts as a tumor suppressor by inhibiting NOTCH signaling and that a loss of NUMB leads to increased NOTCH activity and confers a NOTCH-dependent proliferative advantage in several cancers (Westhoff et al., 2009; Karaczyn et al., 2010;).
In this study, we demonstrated a relationship between two well-defined cancer-associated pathways, NOTCH and TNFα signaling, and showed that expression of IKKα, but not of IKKβ, is associated with NOTCH1 activation. We showed that in response to TNFα stimuli, IKKα interacts with and phosphorylates FOXA2 at S107/111. We found that both in vivo and in vitro, phosphorylated FOXA2 loses its transactivation activity toward its downstream target gene, NUMB, and therefore promotes cell growth and tumorigenesis by activating NOTCH signaling. Importantly, our analysis showed that expression levels of IKKα, pFOXA2 (S107/111), and activated NOTCH1 are significantly higher in clinical hepatocellular carcinoma tumor specimens than in paired normal liver tissues and that phosphorylated FOXA2 expression is positively correlated with IKKα and activated NOTCH1 expression in tumor tissues. Therefore, dysregulation of NUMB-mediated suppression of NOTCH1 by TNFα/IKKα-associated FOXA2 inhibition likely contributes to inflammation-mediated cancer pathogenesis.
RESULTS
TNFα-Induced NOTCH1 Activation Requires IKKα
The pro-Inflammatory factor TNFα and NOTCH1 signaling are known to be essential in cell proliferation and tumorigenesis. However, the critical regulatory mechanism linking TNFα to NOTCH1 activation in inflammation-induced HCC progression remains unclear. To address a potential relationship between these two components in inflammation-induced HCC tumorigenesis, we examined the level of p-IKK, which is an important downstream kinase of TNFα, and activated NOTCH1 in 100 human primary HCC tumor specimens by immunohistochemical (IHC) staining. We detected NICD in 27 of the 47 specimens with high p-IKK expression, indicating a significant positive correlation between p-IKK and NICD (P = 0.003, Figures 1A and 1B). In addition, both p-IKK and NICD had higher expression levels in higher-grade tumors (Figures 1C and 1D). To determine whether this relationship holds in other cancers, we examined human primary tumor specimens of colon, lung, and prostate cancer by IHC staining. Similar to our finding in liver tumors, we also found a positive correlation between p-IKK and NICD (Figure S1 available online).
Figure 1. TNFα-Induced NOTCH1 Expression Requires IKKα.
(A) Correlation between levels of p-IKKα/β and activated NOTCH1 (NICD) in 100 human primary HCC tumor specimens stained with antibodies specific to NICD and p-IKK (P = 0.003, Pearson’s chi-square test).
(B) Two representative specimens from (A); arrows point to p-IKK and NICD.
(C) Correlation between tumor grade and p-IKK expression. High p-IKK expression correlated significantly with higher tumor grade (P = 0.035, Pearson’s chi-square test).
(D) Correlation between expression of p-IKK, NICD and tumor grade. High NICD expression correlated significantly with higher tumor grade (P = 0.01, Pearson’s chi-square test).
(E) Top: WT MEFs were treated with or without TNFα (20 ng/ml) for 24 h, and cell lysates were subjected to western blot analysis for NICD expression. Bottom: IKKα and IKKβ were transfected into IKKα−/− MEFs and IKKβ−/− MEFs, respectively. After treatment with TNFα (20 ng/ml) for 24 h, cell lysates were subjected to western blotting. Lysates from MEFs without TNFα treatment served as control. α-Tubulin was used as loading control.
(F) RBP-Jk Luc and IKKα siRNA were transfected into Hep3B cells for 48 h. After transfection, WT and IKKα-knockdown Hep3B cells were treated with TNFα for 24 h. Cell lysates were subjected to western blotting and luciferase reporter assay. Cells were transfected with nontargeting siRNA as control. Error bars represent SD (n = 3). * indicates, P < 0.05.
The clinical correlations from Figures 1A to 1D led us to ask whether TNFα/IKK might upregulate NICD expression. To validate the relationship between TNFα and NICD expression, we treated mouse embryonic fibroblasts (MEFs) with TNFα (Figure 1E). Indeed, using a specific antibody recognizing only NICD and not its NOTCH1 precursor, we found that TNFα increased NICD expression in MEFs (Figure 1E, top). To further investigate whether IKKα and/or IKKβ regulate NICD expression, we treated wild-type (WT), IKKα−/−, and IKKβ−/− MEFs with TNFα and compared their NICD expression. Interestingly, the NICD level was substantially lower in IKKα−/− MEFs than in WT MEFs (Figure 1E, bottom; compare lanes 1 and 2). Re-expression of IKKα in the IKKα−/− MEFs rescued NICD expression (Figure 1E, bottom; compare lanes 2 and 3). However, the NICD level was not affected by re-expression of IKKβ in IKKβ−/− MEFs (Figure 1E, bottom; compare lane 1 with lanes 4 and 5). Together, these results indicate that TNFα-induced NICD expression requires IKKα but not IKKβ, suggesting that TNFα may stimulate NOTCH1 activation through an IKKα-dependent pathway.
To further confirm that IKKα is required for TNFα-mediated increase of NICD expression, we performed a RBP-Jk reporter luciferase assay, which is designed to monitor the activity of NOTCH1 signal transduction pathways in cultured cells (McGill and McGlade, 2003; Carroll et al., 2006). To ensure the efficiency of IKKα siRNA knockdown, we first analyzed IKKα expression by western blot analysis (Figure 1F, top). RBP-Jk reporter luciferase assay showed that NOTCH1 is activated by TNFα stimulation and that knocking down IKKα blocked TNFα-induced RBP-Jk reporter activation (Figure 1F).
TNFα/IKKα-Induced NOTCH1 Activation Requires Inactivation of FOXA2 and Downregulation of NUMB
Next, we asked how TNFα/IKKα regulates NOTCH1 activation. As NUMB is known to inhibit NOTCH1 activation, we first tested whether NUMB is involved in TNFα/IKKα-mediated NOTCH1 activation. We found that TNFα treatment upregulated NICD expression and downregulated NUMB expression in both Huh-7 and Hep3B in HCC cell lines (Figure 2A). Because we observed similar results from both cell lines in most of the experiments, we only present results from Hep3B cells. Consistent with our results from IKKα- or IKKβ-knockout MEFs (Figure 1E, bottom), ectopic expression of IKKα but not IKKβ increased NICD expression and diminished NUMB expression (Figure S2). Knockdown of IKKα by siRNA impaired TNFα-mediated NUMB downregulation and subsequent NOTCH1 activation (Figure 2B, compare lanes 2 and 4). Knockdown of both IKKα and NUMB restored NICD expression (Figure 2B, compare lanes 4 and 6), suggesting that TNFα/IKKα-mediated NOTCH1 activation involves NUMB downregulation. We observed similar results when we used two different siRNAs individually targeting IKKα and NUMB to exclude potential off-target effects of siRNA (data not shown). To further elucidate the relationships among IKKα, NUMB, and NICD, we analyzed the MEFs by western blotting and found higher NUMB expression and lower NICD expression in IKKα−/− MEFs than in WT MEFs (Figure 2C). Re-expression of IKKα in IKKα−/− MEFs downregulated NUMB expression and rescued NICD expression accordingly (Figure 2C).
Figure 2. IKKα-Mediated NOTCH1 Activation Requires NUMB.
(A) Hep3B and Huh-7 cells were treated with TNFα (20 ng/ml) for 24 h. Endogenous NICD and NUMB expression levels were examined by western blotting. α-Tubulin was used as loading control.
(B) Knockdown of IKKα or double knockdown of IKKα and NUMB by siRNA in Hep3B cells. Nontargeting siRNA was used as control. After 24 h of TNFα treatment, cell lysates (30 µg) were subjected to western blotting to detect endogenous NICD and NUMB expression. α-Tubulin was used as loading control.
(C) Western blot analysis of endogenous NICD and NUMB expression in WT MEFs and IKKα−/− MEFs with or without reconstituted IKKα. α-Tubulin was used as loading control.
(D) Hep3B cells were transfected with WT IKKα or KD IKKα. Endogenous NICD and NUMB expression was analyzed by western blotting. α-Tubulin was used as loading control.
(E) RBP-Jk Luc and NUMB were transfected into Hep3B cells for 48 h. After transfection, Hep3B cells were treated with TNFα (20ng/ml) for 24 h. Cell lysates were subjected to the luciferase reporter assay. NUMB expression was analyzed by western blotting with 10 µg of total lysates. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(F) Hep3B cells were infected with retrovirus expressing IKKα or IKKα and NUMB. RNA extracts were purified from the cell lysates and quantitated with real-time PCR. The NOTCH1 target gene mRNA levels were normalized to the mRNA levels of target genes in vector-infected cells. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(G) Hep3B and Huh-7 cells were treated with TNFα (20 ng/ml) for 24 h. NUMB mRNA level was detected with qPCR. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(H) The TRANSFAC 7.0 and PROMO 8.3 programs were used to predict transcriptional factors that bind to the NUMB promoter. Five HNFs were identified from the prediction.
(I) Each of the five indicated HNFs was transfected alone or together with IKKα into Hep3B cells. NUMB mRNA level was detected with real-time PCR. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
As IKKα is a well-known kinase, we next asked whether the kinase activity of IKKα is required for IKKα-mediated NUMB repression. We found that WT IKKα, but not kinase-dead (KD) IKKα mutant, downregulated NUMB expression (Figure 2D), suggesting that IKKα kinase activity is required for IKKα-mediated NUMB downregulation and NOTCH1 activation. In addition, the results of the RBP-Jk luciferase assay showed that TNFα-induced NOTCH1 activation was blocked by overexpression of NUMB (Figure 2E). To determine whether IKKα-induced NOTCH1 activation affects transcriptionally regulated NOTCH1 downstream target genes (e.g. c-Myc, p21, p27, and the HEY family, we measured the change in RNA levels of c-Myc, p21, p27, Hes6, Hey1, and Hey2 by real-time PCR. The mRNA levels of NOTCH1 downstream targets increased with IKKα but were attenuated in the presence of NUMB (Figure 2F). Together, these results indicate that TNFα/IKKα activates NOTCH1 through downregulation of NUMB.
To investigate how TNFα/IKKα downregulates NUMB expression, we measured NUMB mRNA level with quantitative PCR (qPCR) in response to TNFα treatment in both Huh-7 and Hep3B cells. We found that TNFα treatment also downregulated NUMB at the mRNA level (Figure 2G). Next, we asked whether TNFα/IKKα signaling might suppress NUMB promoter activity. Analysis of the NUMB promoter with the PROMO 8.3 program (Messeguer et al., 2002; Farre et al., 2003) revealed five types of hepatocyte nuclear factor (HNF) binding sites located within the NUMB promoter (Figure 2H). As HNFs have been implicated in regulating cell growth in multiple human cancers (Hayashi et al., 1999), we further investigated whether HNFs regulate NUMB transcription. Co-transfecting each of the five types of HNFs indeed enhanced the NUMB mRNA level (Figure 2I). Interestingly, when IKKα was co-expressed in the transfection experiments, only FOXA2 (HNF3β)-upregulated NUMB mRNA level was reduced, indicating that FOXA2-activated NUMB transcription can be inhibited by IKKα (Figure 2I). Collectively, these results indicate that FOXA2 increases NUMB expression and that inhibition of FOXA2 activity by TNFα results in downregulation of NUMB and activation of NOTCH1.
IKKα Interacts with and Phosphorylates FOXA2 at S107 and S111
To determine how IKKα inhibits FOXA2 activity, we first tested whether IKKα interacts with FOXA2. Reciprocal co-immunoprecipitation showed that IKKα physically associated with FOXA2 (Figure 3A). We also observed this interaction between endogenous IKKα and FOXA2 using specific antibodies against IKKα and FOXA2 (Figure 3B). In addition, GST or His pull-down assays further supported the association of IKKα, but not of IKKβ, with FOXA2 (Figure 3C). Given the physical interaction between IKKα and FOXA2, we examined whether FOXA2 is a physiological substrate of IKKα. An in vitro kinase assay demonstrated that GST-FOXA2, but not GST protein, was phosphorylated by purified recombinant IKKα (Figure 3D, compare lanes 1 and 2). To determine which amino acid residue is phosphorylated by IKKα, we analyzed the GST-pFOXA2 fusion protein by mass spectrometry (MS) and found that FOXA2 was phosphorylated by IKKα at S107 and S111 (Figure 3E). Mutation of these two sites from serine to alanine abolished IKKα-mediated FOXA2 phosphorylation (Figure 3D, lane 5). Taken together, these data demonstrate that IKKα interacts with and phosphorylates FOXA2 at S107/111 in vitro.
Figure 3. IKKα Interacts with and Phosphorylates FOXA2 In Vivo and In Vitro.
(A) IKKα and FOXA2 were transfected into 293T cells and analyzed by reciprocal co-immunoprecipitation and immunoblotting using indicated antibodies.
(B) Lysates of Hep3B cells were analyzed by reciprocal co-immunoprecipitation and immunoblotting using indicated antibodies.
(C) In vitro transcribed FOXA2 proteins (labeled with S35) were incubated with commercially available recombinant His-IKKβ and GST-IKKα and then pulled down with His/GST beads. IVT: in vitro transcription and translation. S35 concentration in cell lysates was determined by film exposure.
(D) FOXA2 phosphorylation was identified by in vitro kinase assay. GST-FOXA2 was pulled down by GST beads from Hep3B cells and incubated with commercially available IKKα for 30 min.
(E) Mass spectrum of FOXA2 phosphorylation sites by IKKα using samples (without isotope) from (D).
(F) Hep3B cells were co-transfected with FOXA2 and WT IKKα or KD IKKα. pFOXA2 expression was detected using an antibody specific to pFOXA2 (S107/111). Cell lysates (10 µg) wereloaded for western blotting.
(G) IKKα was co-transfected with WT FOXA2 or FOXA2-SSAA. pFOXA2 (S107/111) was detected using an antibody specific to pFOXA2 (S107/111). Total FOXA2 expression and IKKα expression are also shown.
(H) Western blot analysis of pFOXA2 with an antibody specific to pFOXA2 (S107/111) using samples (without isotope) from (D). Total FOXA2 expression and IKKα expression are also shown.
(I) Hep3B cells were treated with TNFα and harvested at 10, 30, 60, and 120 min after treatment. Cell lysates (40 µg) were used for western blot analysis of pFOXA2 with an antibody specific to pFOXA2 (S107/111). Total FOXA2 expression and IKKα expression are also shown.
(J) IKKβ−/− MEFs were treated with TNFα for 45 min. Cell lysates were subjected to cell fractionation. After immunoprecipitation by the IKKα antibody, the nuclear and cytoplasmic fractions were subjected to western blot analysis with indicated antibodies. Lamin and tubulin were used as cell fractionation controls.
To determine whether these phosphorylations also occur in vivo, we treated Hep3B cells with TNFα and isolated endogenous FOXA2 for MS analysis. We identified the same two phosphorylation sites (Figure S3). In addition, we raised mouse polyclonal antibodies against pFOXA2 at S107/111 and used the antibody to study FOXA2 phosphorylation status. The antibody recognized specifically pFOXA2 (S107/111) phosphorylated by WT IKKα but not by KD IKKα (Figure 3F). The pFOXA2 antibody also recognized phosphorylated WT FOXA2 but not the double mutant with both serine residues mutated to alanine (FOXA2-SSAA), further supporting its specificity (Figure 3G). In addition, the pFOXA2 (S107/111) antibody detected pFOXA2 only in the presence of IKKα in an in vitro kinase assay (Figure 3H). Time course assays to study the effect of TNFα showed that levels of pFOXA2 (S107/111) were high at 10–30 min and declined thereafter (Figure 3I), consistent with the kinetics of TNFα-induced IKKα activation as measured by degradation of IκBα.
TNFα stimulates IKKα translocation into the nucleus (Anest et al., 2003; Yamamoto et al., 2003).Thus, to determine whether IKKα phosphorylates FOXA2 in the nucleus, we treated IKKβ−/− MEFs with TNFα and isolated the nuclear and cytoplasmic fractions from the cells for immunoprecipitation by the IKKα antibody. We found that stimulation of IKKβ−/− MEFs with TNFα increased nuclear accumulation of IKKα, the amount of FOXA2 bound to IKKα, and phosphorylation of FOXA2 (S107/111). However, we did not detect either pFOXA2 or FOXA2 expression in the cytoplasm after TNFα treatment, suggesting that TNFα increased the nuclear accumulation of IKKα for phosphorylation of FOXA2 (Figure 3J).
FOXA2 Phosphorylation by IKKα Suppresses FOXA2 Transactivation Activity
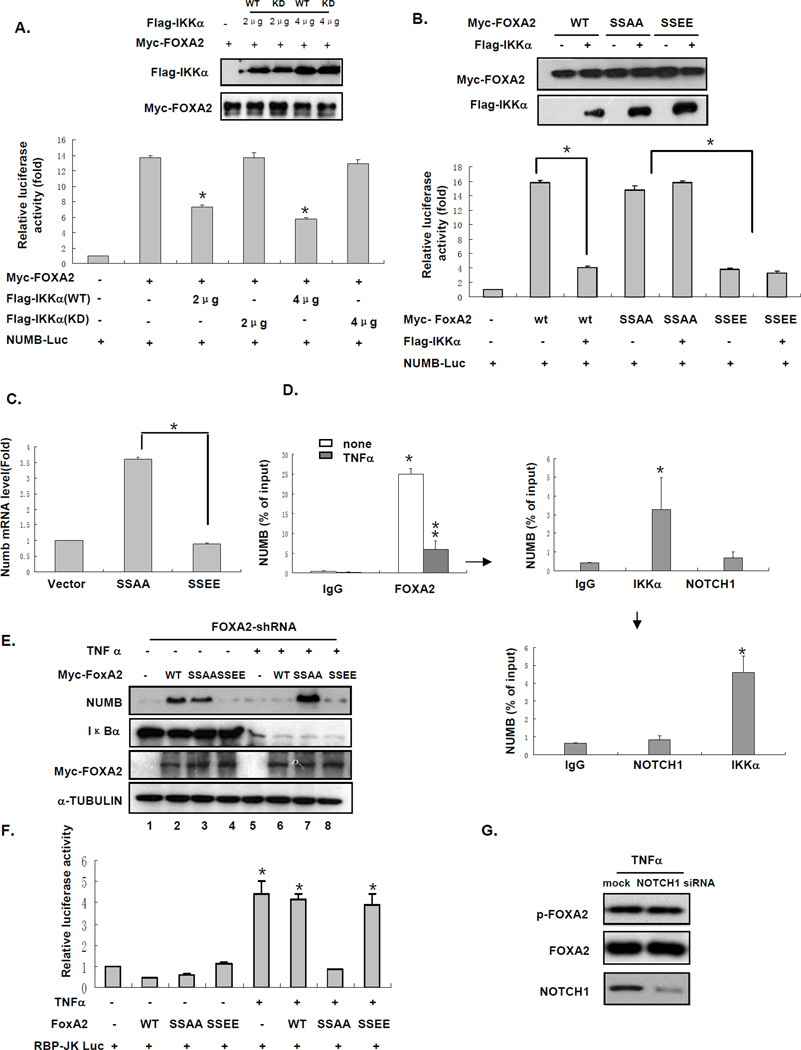
On the basis of our observations that IKKα phosphorylates FOXA2 (Figure 3) and inhibits NUMB expression (Figure 2), we hypothesized that FOXA2 phosphorylation by IKKα suppresses FOXA2 transactivation activity and therefore inhibits NUMB expression and activates the NOTCH pathway. To test this hypothesis, we examined the effect of IKKα on the FOXA2-dependent transcriptional activation of a NUMB promoter containing FOXA2 binding sites that drives the luciferase reporter gene. Only WT IKKα, not KD IKKα, inhibited FOXA2-induced NUMB promoter activity (Figure 4A). We obtained similar results when a construct containing FOXA2-responsive elements (FRE) to drive the luciferase reporter was introduced into 293T cells (Figure S4A).
Figure 4. FOXA2 Phosphorylation by IKKα Suppresses FOXA2 Transactivation Activity.
(A) NUMB promoter luciferase reporter and FOXA2 in the presence of different doses of WT IKKα or KD IKKα were transfected into 293T cells for 48 h. Cell lysates were subjected to luciferase reporter assays after transfection. Expression levels of WT IKKα, KD IKKα, and FOXA2 were analyzed by western blotting. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(B) Co-transfection of NUMB-promoter Luc and WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE plus the indicated Flag-tagged IKKα in 293T cells for 48 h. Lysates of 293T cells were subjected to luciferase assays. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05). Expression levels of IKKα, WT FOXA2, and FOXA2 mutants were detected by western blot analysis.
(C) Hep3B cells were infected with retrovirus expressing FOXA2-SSAA or FOXA2-SSEE. RNA extracts from the cells were isolated and subjected to real-time RT-PCR for NUMB mRNA expression. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(D) Hep3B cells were treated with TNFα for 24 h, and cell lysates were harvested and subjected to primary ChIP assay using the FOXA2 antibody. The purified DNA from the ChIP assay was subjected to real-time PCR using NUMB primers (to left of arrow). The immunoprecipitated products of Hep3B cells (with TNFα treatment) were desalted and re-ChIP-ed by IKKα and NOTCH1 antibody. Real-time PCR using NUMB primers was repeated using the purified DNA from the ChIP assay (to right of arrow). The products from the second immunoprecipitation were desalted again and re-subjected to re-ChIP assay by NOTCH1 and IKKα (as a positive control). The purified DNA from ChIP assay was subjected to real-time PCR using NUMB primers (below downward arrow). Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(E) Endogenous FOXA2 in Hep3B cells was knocked down by shRNA against the 5′-UTR. After shRNA knockdown of FOXA2, we transfected WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE into FOXA2−/− Hep3B cells, followed by TNFα treatment for 24 h. Cell lysates were analyzed by western blotting using indicated antibodies.
(F) RBP-Jk Luc and WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE were co-transfected into Hep3B cells for 24 h, followed by TNFα treatment for 24 h. Cell lysates were analyzed by luciferase reporter assay. Error bars represent SD (n = 3). * indicates statistical significance (P < 0.05).
(G) NOTCH1 expression was knocked down by NOTCH1 siRNA in Hep3B cells, which were treated with TNFα for 24 h, and lysates were analyzed by western blotting.
To test whether IKKα inhibits FOXA2 activity through the identified IKKα phosphorylation sites (S107/111), we generated two double mutants, FOXA2-SSAA and FOXA2-SSEE, which mimic the nonphosphorylated and phosphorylated state of FOXA2, respectively, by IKKα. FOXA2-SSAA, still activated the promoter activity, but FOXA2-SSEE lost its transactivation activity (Figure 4B). In addition, Unlike WT FOXA2, the transactivation activities of the two mutants were not affected by the presence of IKKα, suggesting that phosphorylation of FOXA2 at S107/111 impaired FOXA2 transactivation activity (Figures 4B and S4B). These results support that IKKα decreased FOXA2 transactivation activity by phosphorylating FOXA2 at S107/111. We then asked whether the decreased transactivation activity of FOXA2 by S107/111 phosphorylation also affects NUMB expression. We examined the NUMB mRNA level by transfecting various FOXA2 constructs expressing WT FOXA2, FOXA2-SSAA, FOXA2-SSEE, or vector control into Hep3B cells. We found that FOXA2-SSAA, but not FOXA2-SSEE, enhanced NUMB mRNA level (Figure 4C). These results further support the notion that S107/111 phosphorylation inhibits FOXA2 transactivation activity.
To investigate the molecular mechanism of IKKα-mediated inhibition of FOXA2 activity, we first examined the difference in the binding ability of FOXA2 to the NUMB promoter before and after TNFα treatment. We performed a chromatin immunoprecipitation (ChIP) assay using primers spanning the putative FOXA2 binding sites (see primer sequences in the Experimental Procedures section) in the NUMB promoter from Hep3B cell lysates with or without TNFα treatment. We found that binding of FOXA2 to the NUMB promoter was decreased by TNFα (Figure 4D, to left of arrow). In addition, we compared the binding ability of WT FOXA2, FOXA2-SSAA, and FOXA2-SSEE to the NUMB promoter by ChIP and found that WT FOXA2 and FOXA2-SSAA, but not FOXA2-SSEE, bound to the NUMB promoter (Figure S4C), suggesting that FOXA2 phosphorylation by IKKα suppressed FOXA2 transactivation activity on NUMB by decreasing FOXA2 accessibility to its promoter.
To further support the role of S107/111 phosphorylation of FOXA2 in the regulation of NUMB expression, we tested the function of FOXA2-SSAA and FOXA2-SSEE in FOXA2-knockdown Hep3B cells. In the absence of TNFα, both WT FOXA2 and FOXA2-SSAA, but not FOXA2-SSEE, stimulated NUMB protein level (Figure 4E). TNFα suppressed FOXA2-induced NUMB expression in the presence of WT FOXA2 (Figure 4E, compare lanes 2 and 6) but not of FOXA2-SSAA (compare lanes 3 and 7). Moreover, FOXA2-SSEE was insensitive to TNFα treatment (lane 8), suggesting that S107/111 phosphorylation of FOXA2 by TNFα-activated IKKα contributes to inactivation of FOXA2-induced NUMB expression. Consistently, NOTCH1 activity was also inhibited by FOXA2-SSAA as a result of NUMB expression, as measured by luciferase reporter assay (Figure 4F). Collectively, our results indicate that S107/111 phosphorylation of FOXA2 by IKKα suppresses FOXA2 transactivation activity by reducing FOXA2 ability to bind to the NUMB promoter, and therefore, decreases NUMB expression at the mRNA and protein level to activate the NOTCH1 pathway.
It is worth mentioning that NOTCH1 has been shown to associate with IKKα and is required at IKKα-stimulated promoters (Song et al., 2008). To investigate whether FOXA2 phosphorylation by IKKα could be affected by NOTCH1 binding, we treated NOTCH1-knockdown cells with TNFα. We detected no change in FOXA2 phosphorylation level (Figure 4G). In addition, we immunoprecipitated the complexes by FOXA2 with TNFα (from Figure 4D) and eluted them with re-ChIP buffer (please see the Experimental Procedures section for detailed ChIP-re-ChIP protocol). We found that IKKα and FOXA2, but not NOTCH1, bound to the same NUMB promoter (Figure 4D, to right of arrow). To ensure that the IKKα detected in the second ChIP was free of NOTCH1, we re-immunoprecipitated the eluate from the second ChIP by NOTCH1 antibody and IKKα antibody as a positive control (Figure 4D, below downward arrow). We found that IKKα, but not NOTCH1, bound to the FOXA2/NUMB promoter complex. Together, these results suggest that the FOXA2/IKKα complex, without NOTCH1, binds to the NUMB promoter.
FOXA2 Phosphorylation by IKKα De-Represses FOXA2-Mediated Repression of Cell Growth In Vitro
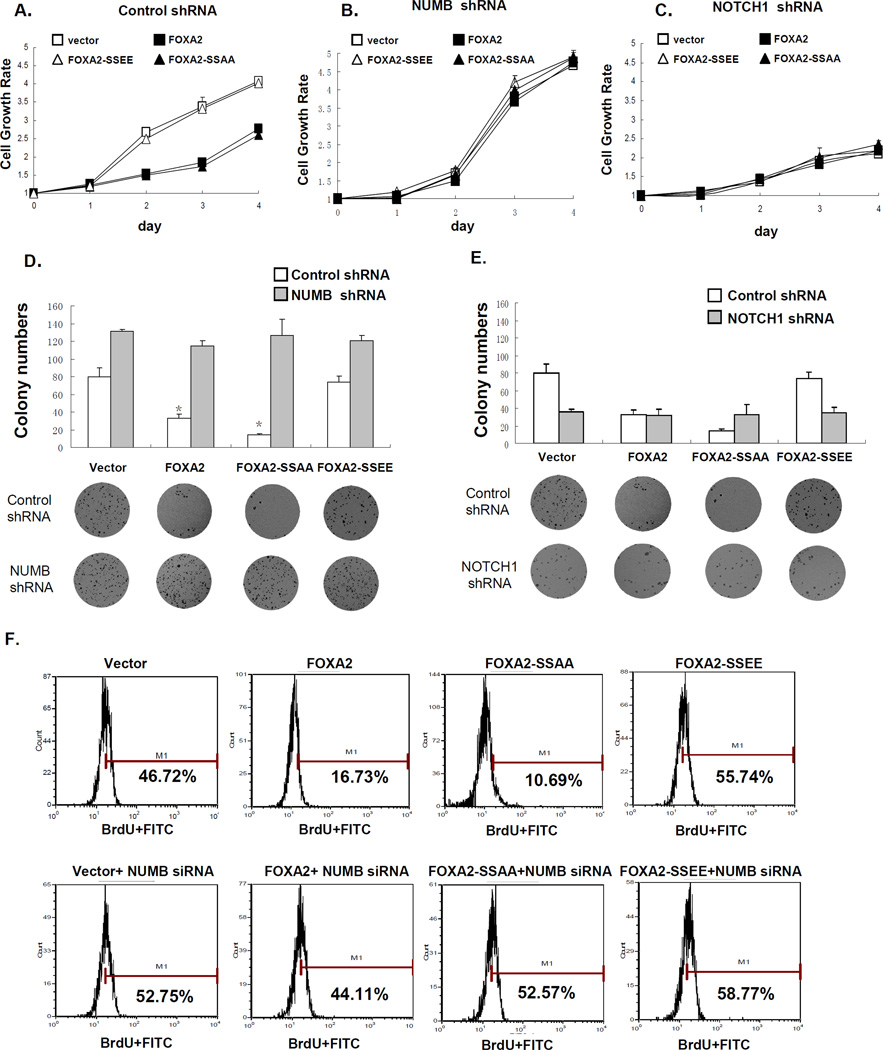
Next, to determine whether FOXA2 phosphorylation by IKKα might contribute to the biological activity of FOXA2, we examined the function of IKKα-induced FOXA2 phosphorylation in cell growth by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. We found that expression of FOXA2-SSAA, but not of FOXA2-SSEE, antagonized cell growth (Figure 5A). WT IKKα, but not KD IKKα, abrogated WT FOXA2-mediated growth suppression, presumably as a result of phosphorylation of WT FOXA2 by IKKα, and thus FOXA2 lost its ability to inhibit cell growth (Figure S5A). Knocking down NUMB reversed the cell growth inhibition by WT FOXA2 and FOXA2-SSAA, supporting our proposed suppressive role of NUMB in the TNFα/IKKα pathway (Figure 5B). In addition, when NOTCH1 was knocked down in Hep3B cells, cell growth was inhibited regardless of FOXA2 phosphorylation status, demonstrating that NOTCH1 was indeed required for cell growth (Figure 5C). When we treated nine liver cancer cell lines with two NOTCH1 pathway inhibitors, LY-411575 and γ-secretase inhibitor (GSI), cell growth decreased, further supporting that NOTCH1 activation plays an important role in liver cancer cell growth (Figure S5B). Consistent with our results above (Fig 5A), FOXA2-SSAA, but not FOXA2-SSEE, inhibited cell colony formation in an anchorage-independent growth assay (Figure 5D). Knocking down NUMB allowed colonies to form even in the presence of FOXA2-SSAA (Figure 5D). In contrast, when NOTCH1 was knocked down, colony formation was inhibited regardless of FOXA2 phosphorylation status (Figure 5E).
Figure 5. FOXA2 Blocks IKKα-Induced Cell Proliferation and Tumor Growth In Vitro.
(A, B, and C). Hep3B cells were infected with retrovirus expressing WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE in the presence of (A) control shRNA, (B) NUMB shRNA, or (C) NOTCH1 shRNA. Cells (2.5 × 103) were plated in 96-well plates, and cell growth was determined by MTT assay each day for 4 days.
(D. and E). Anchorage-independent growth assay of Hep3B cells infected with retrovirus expressing WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE in the presence of (D) NUMB shRNA or (E) NOTCH1 shRNA, compared with control shRNA. Colony numbers (mean ± SD) in week 3 are shown (n = 6). * indicates statistical significance (P < 0.05, Student’s t test). Representative images from the anchorage-independent growth assay are shown below.
(F) Hep3B cells were infected with retrovirus expressing WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE either without (top) or with (bottom) NUMB shRNA. Infected cells were subjected to BrdU assay and analyzed by flow cytometry.
To verify the status of DNA synthesis under these conditions, we also performed a bromodeoxyuridine (BrdU) incorporation assay. We found that WT FOXA2-suppressed BrdU incorporation was compromised by WT IKKα but not by KD IKKα (Figure S5C). Again, both WT FOXA2 and FOXA2-SSAA, but not FOXA2-SSEE, inhibited cell proliferation, as indicated by the loss of new DNA synthesis (Figure 5F, top). The decrease in new DNA synthesis in the presence of WT FOXA2 or FOXA2-SSAA was reversed by knocking down NUMB (Figure 5F, bottom).
FOXA2 Phosphorylation by IKKα De-Represses FOXA2-Mediated Repression of Cell Growth and Tumorigenesis In Vivo
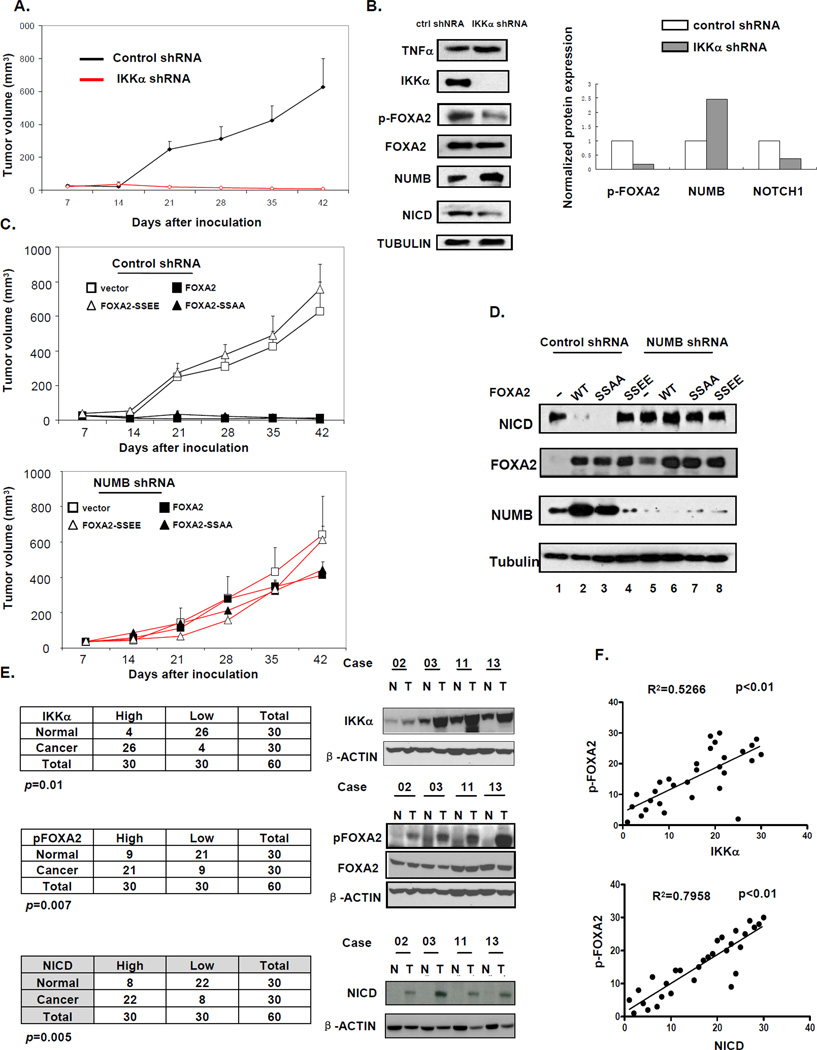
To further explore the biological function of IKKα-induced FOXA2 phosphorylation in vivo, we injected Hep3B cells into three groups of nude mice using an orthotopic liver cancer animal model (Xiong, et al., 2010). Briefly, we injected Hep3B cells into mice subcutaneously. After the tumor reached 2–3 mm in diameter, it was harvested, dissected into small pieces (1 × 1 × 1 mm3), and transplanted into the livers of new mice. We monitored the tumor volume (TV) of these mice for 6 weeks and analyzed tumor tissue samples by western blotting. To determine the role of IKKα in liver tumor growth, we injected the first group of mice with Hep3B cells infected with retrovirus expressing shRNA against IKKα. TV was much smaller in the mice harboring tumors (after transplantation) than in the control group, validating the role of IKKα in liver tumor growth (Figure 6A). Western blot analysis of these tumor tissue samples detected TNFα in tumors along with decreased FOXA2 phosphorylation, enhanced NUMB expression, and attenuated NICD expression. The second group of mice was injected with cells infected with retrovirus expressing WT or mutant FOXA2, and transplantation was performed essentially as described for the first group. We found that WT FOXA2 and FOXA2-SSAA both inhibited tumor growth, whereas FOXA2-SSEE had no effect (Figure 6C, top). In addition, NUMB expression was increased with a decrease in NICD expression in the WT FOXA2 and FOXA2-SSAA tumors (Figure 6D). Together, these results suggest that FOXA2-SSAA functions in a dominant manner to inhibit tumor growth and support the importance of IKKα in promoting FOXA2-associated HCC development.
Figure 6. FOXA2 Blocks IKKα-Induced Cell Proliferation and Tumor Growth In Vivo.
(A) Hep3B cells were infected with retrovirus expressing control or IKKα shRNA. Mice were injected subcutaneously with 1 × 106 cells. Error bars represent SD (n = 5). Please see the Experimental Procedures section for more detailed information.
(B) Tumor tissues from mice that received transplants were analyzed 10 days after transplantation and subjected to western blot analysis for indicated antibodies. Right: Expression levels of pFOXA2, NUMB, and NOTCH1 quantified with ImageJ software.
(C) Hep3B cells were infected with retrovirus expressing WT FOXA2, FOXA2-SSAA, or FOXA2-SSEE in the presence of (top) control shRNA or (bottom) NUMB shRNA. Error bars represent SD (n = 5). Please see the Experimental Procedures section for more detailed information.
(D) Tumor issues from (C) were harvested 10 days after transplantation and subjected to western blot analysis for NICD, FOXA2, and NUMB. α-Tubulin was included as control.
(E) Comparison of pFOXA2 (S107/111), IKKα, and NICD expression in tumors and their adjacent normal tissues. Protein expression was examined by immunoblotting 30 pairs (tumor and normal) of liver tissue samples and quantified with ImageJ software. Right: Four representative pairs (N, normal; T, tumor). Protein expression levels in tumor samples were normalized to those in paired normal tissues. Protein expression levels higher (or lower) than those in normal tissues were defined as “high” (or “low”).(P < 0.01, Pearson’s chi-square test.)
(F) Correlation analysis of pFOXA2 and IKKα/NICD expression levels in tumors (R2 > 0.5, Spearman’s rank correlation test).
Previously (Figure 5B and 5D), we showed that knockdown of NUMB increased cell growth rate and colony formation regardless of FOXA phosphorylation status. To demonstrate the role of NUMB in TNFα/IKKα-induced tumor growth in vivo, we knocked down NUMB by shRNA in Hep3B cells infected with retrovirus expressing WT FOXA2 and its mutants. Mice that received transplanted tumors harboring these NUMB-knockdown cells continued to show tumor growth even in the presence of FOXA2 and FOXA2-SSAA, which was probably due to restored NICD expression and is consistent with results from the in vitro assays (Figure 6C, bottom).
In addition, to further examine the aforementioned conclusion that FOXA2 phosphorylation by TNFα-induced IKKα plays a critical role in liver cancer, we examined the expression of IKKα, pFOXA2 (S107/111), and NICD in 30 freshly prepared human primary HCC tumor specimens and paired normal liver tissues by immunoblotting. We found that IKKα, pFOXA2 (S107/111), and NICD expression levels were significantly higher in tumor specimens than in the corresponding normal liver tissues (P < 0.05). In addition, pFOXA2 expression was positively correlated with expression of IKKα and NICD. These results strengthen the notion that IKKα regulates FOXA2 and activates NICD through phosphorylation at S107/111 and the physiological implication of pFOXA2 in IKKα-induced tumorigenesis (Figures 6E, 6F, and S6). Taken together, analyses of these clinical HCC tumor specimens support that FOXA2 phosphorylation by IKKα might be associated with HCC tumorigenesis.
DISCUSSION
Here, we identified an axis in the TNFα pathway that activates NOTCH1 signaling by suppressing FOXA2 transactivation activity by IKKα, leading to disruption of NUMB expression, which promotes NOTCH1-induced liver cell proliferation and growth (Figure 7).
Figure 7. Proposed Model of FOXA2 Phosphorylation by IKKα.
FOXA2 phosphorylation by IKKα leads to activation of NOTCH1 signaling through downregulation of NUMB and thereby induces tumorigenesis.
We demonstrated that TNFα stimulates the NOTCH1 pathway in liver cancer cell lines. Although NOTCH1 has been shown to play a role as an oncogene mainly in leukemia and breast cancer (Pui, 2009; Zardawi et al., 2010), one report demonstrated inhibition of HCC tumor growth through NOTCH1 signaling by induction of cell cycle arrest and apoptosis (Qi et al., 2003). Several groups that analyzed human HCC clinical tumor specimens indicated a tumor progressive role of NOTCH1 in HCC (Cantarini et al., 2006; Gramantieri et al., 2007). More recently, Ning et al. specifically showed that downregulation of NOTCH1 signaling inhibited tumor growth in human HCC in both cell lines and a mouse model (Ning et al., 2009). These studies are in line with our findings that NOTCH1 is activated by TNFα through dysregulation of FOXA2 transactivation activity to promote cell proliferation and growth.
Some FOX subfamilies, such as FOXO, have been linked to tumorigenesis and the progression of certain cancers (Yang et al., 2008). We showed here that a new FOX family member plays an important role in inflammation-induced HCC. The crystal structure of the FOXA2 DNA binding domain was published in 1993; however, the crystal structure of full-length FOXA2 is not yet available. The phosphorylation sites that we identified (S107/111) are located in the domain between TAD and WHD. It will be interesting to investigate how phosphorylation at S107/111 affects the DNA binding activity of FOXA2 and/or its interaction with other transcription factors.
In summary, the identification of FOXA2 as a downstream substrate of IKKα links the TNFα and NOTCH1 signaling pathways and provides an important new starting point for uncovering the molecular basis of TNFα-mediated human tumor growth and identifying potential targets for cancer therapy. Inhibition of FOXA2 phosphorylation or activation of NUMB could have important clinical implications for the treatment or prevention of cancer.
EXPERIMENTAL PROCEDURES
Plasmids Construction and Retroviral Infection
We constructed Myc-FOXA2- and GFP-FOXA2-expressing plasmids by inserting hFOXA2 complementary DNA (cDNA) into pcDNA6 and pCMV-EGFP vectors containing the Myc and GFP tags, respectively. We generated the GST-FOXA2-expressing plasmid by subcloning the FOXA2 fragment into the pGEX-6P-1 GST vector. Different mutant forms of the FOXA2 construct were generated by subcloning the respective FOXA2 fragment into pcDNA6 and pCMV-EGFP vectors. Mutants were generated using the QuikChange multi site-directed mutagenesis kit (Stratagene). The FOXA2 and mutant expression plasmids were constructed into retroviral pBABE-Puro and pBABE-GFP vectors for retroviral infection and expression.
siRNAs
Hep3B and Huh-7 cells were transfected with ON-TARGETplus siRNAs (all from Dharmacon RNA Technologies) by using DharmaFECT Transfection Reagents (T-2001-02).
Cell Culture and Transfection
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with 10% fetal bovine serum..
For transient transfection, cells were transfected with DNA by either Lipofectamine with Plus reagent or electroporation
In Vitro Pull-Down Assay
In vitro pull-down assays were performed as previously described (Lee, et al., 2007). Detail information is described in supplemental experimental procedure.
Immunoprecipitation, Immunoblotting, and In Vitro Kinase Assay
Immunoprecipitation and immunoblotting assays were performed as previously described (Hu et al., 2004). The in vitro IKKα kinase assay was performed as previous described (Maeda et al., 2000).
Identification of In Vivo Phosphorylation Sites by Mass Spectrometry
After protein gel electrophoresis, bands that corresponded to FOXA2 phosphorylated by IKKα in vivo were identified, excised from gels, and subjected to tryptic digestion. After being isolated by immobilized metal affinity chromatography, the enriched phosphopeptides were analyzed by micro-liquid chromatography/tandem MS.
ChIP and Re-ChIP Assay
ChIP assays were modified from the EZ-ChIP (Upstate) protocol using anti-FOXA2, anti-NOTCH1, and anti-IKKα antibodies. For sequential ChIP, the first immunoprecipitate eluted by anti-FOXA2 was further immunoprecipitated by anti-NOTCH1/anti-IKKα and IgG antibodies. The percentage of the bound DNA was quantified against the original DNA input.
Real-Time PCR
Total RNAs were extracted from cells by using the RNeasy kit (QIAGEN). RNAs were reverse-transcribed by using the SuperScript II kit (Invitrogen). Results were analyzed by using the iCycler system (Bio-Rad).
MTT Assay
Cells were plated at 5000/well for Hep3B cells in 96-well microplates. At different time points, 50 µl of MTT was added to each well, and incubation was continued for 2 h. The light absorbance was measured at 570 nm with a multiwell spectrophotometer (Labsystems).
Anchorage-Independent Growth Assay
Anchorage-independent growth of stable transfectants was determined by a previously described method (Lee, 2007). Detail information is described in supplemental information.
BrdU Incorporation Assay
Cell proliferation was assessed by using the cell proliferation enzyme-linked immunosorbent assay (Flow Cytometry kit (Roche) according to the manufacturer’s instructions.
Mouse Model for Tumorigenesis
We performed the tumorigenesis assay for retrovirus-infected cells with both liver cancer subcutaneous and orthotopic mouse model that has been described elsewhere (Xiong et al., 2010). Cells (1 × 106) were injected subcutaneously. After tumors reached 2–3 mm in diameter, tumors were harvested, dissected into small pieces (1 × 1 × 1 mm3), and transplanted into the livers of new mice (n = 5 per group). TV was calculated according to the formula described by Yaguchi et al.: TV = 0.5 × Length × Width2 (Yaguchi et al., 2006).
Immunohistochemical Staining
IHC staining was performed as described before (Hu et al., 2004). Detail information is described in supplemental information.
Statistical Analyses
Statistical analyses were performed with Student’s t test, Spearman’s rank correlation test, or Pearson’s chi-square test as indicated. P < 0.05 was considered statistically significant. R2 > 0.5 was considered statistically correlated.
Highlights.
TNFα activates NOTCH1 activity through IKKα but not IKKβ
pFOXA2 by IKKα impairs its binding to the NUMB promoter which activates NOTCH1
Orthotopic mouse model and paired HCC tissue samples validate our proposed model
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Zhang, J. Shi, B. Spohn, J.-F. Lee, and Z. Han for their technical support. This work was partially supported by NIH (R01 CA109311, P01 CA099031, and MD Anderson’s Cancer Center Support Grant, CA016672); NHRI-EX99-9603BC, DOH97-TD-I-111-TM003, and DOH98-TD-I-111-TM002 (to L.-Y.L.); The University of Texas MD Anderson Cancer Center–China Medical University and Hospital Sister Institution Fund; The Kadoorie Charitable Foundation; NSC99-2632-13-039-001-MY3; and a research assistant scholarship from the University of Texas Graduate School of Biomedical Sciences at Houston to M.L. We thank Dr. Karen Muller for scientific editing our manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and can be found with this article online.
REFERENCES
- Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Curr Opin Genet Dev. 2008;18:3–10. doi: 10.1016/j.gde.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarini MC, de la Monte SM, Pang M, Tong M, D'Errico A, Trevisani F, Wands JR. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006;44:446–457. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- Carayol N, Wang CY. IKKalpha stabilizes cytosolic beta-catenin by inhibiting both canonical and non-canonical degradation pathways. Cell Signal. 2006;18:1941–1946. doi: 10.1016/j.cellsig.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Carroll KD, Bu W, Palmeri D, Spadavecchia S, Lynch SJ, Marras SA, Tyagi S, Lukac DM. Kaposi's Sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J Virol. 2006;80:9697–9709. doi: 10.1128/JVI.00746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello G, Scala S, Palmieri G, Curley SA, Izzo F. HCV-related hepatocellular carcinoma: From chronic inflammation to cancer. Clin Immunol. 2010;134:237–250. doi: 10.1016/j.clim.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Danese S, Mantovani A. Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene. 2010;29:3313–3323. doi: 10.1038/onc.2010.109. [DOI] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Giovannini C, Lanzi A, Chieco P, Ravaioli M, Venturi A, Grazi GL, Bolondi L. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007;27:997–1007. doi: 10.1111/j.1478-3231.2007.01544.x. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Wang W, Ninomiya T, Nagano H, Ohta K, Itoh H. Liver enriched transcription factors and differentiation of hepatocellular carcinoma. Mol Pathol. 1999;52:19–24. doi: 10.1136/mp.52.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Whitsett Jeffrey A., Haitchi Hans Michael, Maeda Yutaka. Intersections between Pulmonary Development and Disease. American Journal of Respiratory and Critical Care Medicine. 2010;184:401–406. doi: 10.1164/rccm.201103-0495PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaczyn A, Bani-Yaghoub M, Tremblay R, Kubu C, Cowling R, Adams TL, Prudovsky I, Spicer D, Friesel R, Vary C, Verdi JM. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural Dev. 2010;5:31. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. The IkappaB kinase - a bridge between inflammation and cancer. Cell Res. 2008;18:334–342. doi: 10.1038/cr.2008.30. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Lee DF, Hung MC. Advances in targeting IKK and IKK-related kinases for cancer therapy. Clin Cancer Res. 2008;14:5656–5662. doi: 10.1158/1078-0432.CCR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Lee S, Andrieu C, Saltel F, Destaing O, Auclair J, Pouchkine V, Michelon J, Salaun B, Kobayashi R, Jurdic P, et al. IkappaB kinase beta phosphorylates Dok1 serines in response to IL-1, or gamma radiation. Proc Natl Acad Sci U S A. 2004;101:17416–17421. doi: 10.1073/pnas.0408061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner F, Kulik U, Klempnauer J, Borlak J. The hepatocyte nuclear factor 6 (HNF6) and FOXA2 are key regulators in colorectal liver metastases. Faseb J. 2007;21:1445–1462. doi: 10.1096/fj.06-6575com. [DOI] [PubMed] [Google Scholar]
- Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- Maeda S, Yoshida H, Ogura K, Mitsuno Y, Hirata Y, Yamaji Y, Akanuma M, Shiratori Y, Omata M. H. pylori activates NF-kappaB through a signaling pathway involving IkappaB kinases, NF-kappaB-inducing kinase, TRAF2, and TRAF6 in gastric cancer cells. Gastroenterology. 2000;119:97–108. doi: 10.1053/gast.2000.8540. [DOI] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Medyouf H, Gao X, Armstrong F, Gusscott S, Liu Q, Gedman AL, Matherly LH, Schultz KR, Pflumio F, You MJ, Weng AP. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood. 2010;115:1175–1184. doi: 10.1182/blood-2009-04-214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Ning L, Wentworth L, Chen H, Weber SM. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res. 2009;1:358–366. [PMC free article] [PubMed] [Google Scholar]
- Pui CH. T cell acute lymphoblastic leukemia: NOTCHing the way toward a better treatment outcome. Cancer Cell. 2009;15:85–87. doi: 10.1016/j.ccr.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, Guo Z, Cheng T, Cao X. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63:8323–8329. [PubMed] [Google Scholar]
- Song LL, Peng Y, Yun J, Rizzo P, Chaturvedi V, Weijzen S, Kast WM, Stone PJ, Santos L, Loredo A, et al. Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene. 2008;27:5833–5844. doi: 10.1038/onc.2008.190. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Shih DQ, Kuwajima S, Norris AW, Kahn CR, Stoffel M. Role of Foxa-2 in adipocyte metabolism and differentiation. J Clin Invest. 2003;112:345–356. doi: 10.1172/JCI18698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Ren ZG, Qiu SJ, Sun HC, Wang L, Liu BB, Li QS, Zhang W, Zhu XD, Liu L, et al. Residual hepatocellular carcinoma after oxaliplatin treatment has increased metastatic potential in a nude mouse model and is attenuated by Songyou Yin. BMC Cancer. 2010;10:219. doi: 10.1186/1471-2407-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Fukui Y, Koshimizu I, Yoshimi H, Matsuno T, Gouda H, Hirono S, Yamazaki K, Yamori T. Antitumor activity of ZSTK474, a new phosphatidylinositol 3-kinase inhibitor. J Natl Cancer Inst. 2006;98:545–556. doi: 10.1093/jnci/djj133. [DOI] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CJ, Izzo JG, Lee DF, Guha S, Wei Y, Wu TT, Chen CT, Kuo HP, Hsu JM, Sun HL, et al. Bile acid exposure up-regulates tuberous sclerosis complex 1/mammalian target of rapamycin pathway in Barrett's-associated esophageal adenocarcinoma. Cancer Res. 2008;68:2632–2640. doi: 10.1158/0008-5472.CAN-07-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardawi SJ, Zardawi I, McNeil CM, Millar EK, McLeod D, Morey AL, Crea P, Murphy NC, Pinese M, Lopez-Knowles E, et al. High Notch1 protein expression is an early event in breast cancer development and is associated with the HER-2 molecular subtype. Histopathology. 2010;56:286–296. doi: 10.1111/j.1365-2559.2009.03475.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.