Abstract
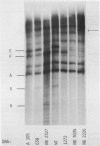
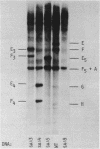
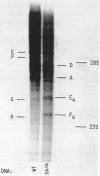
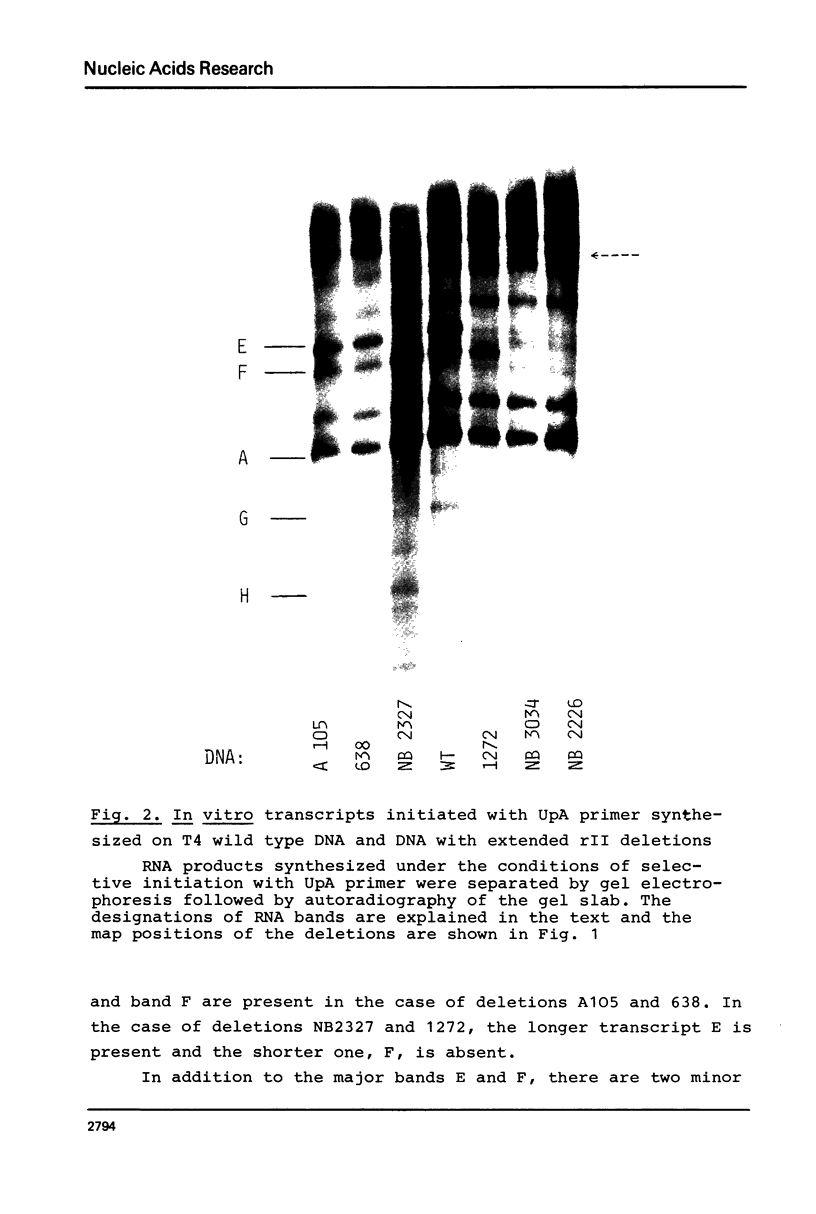
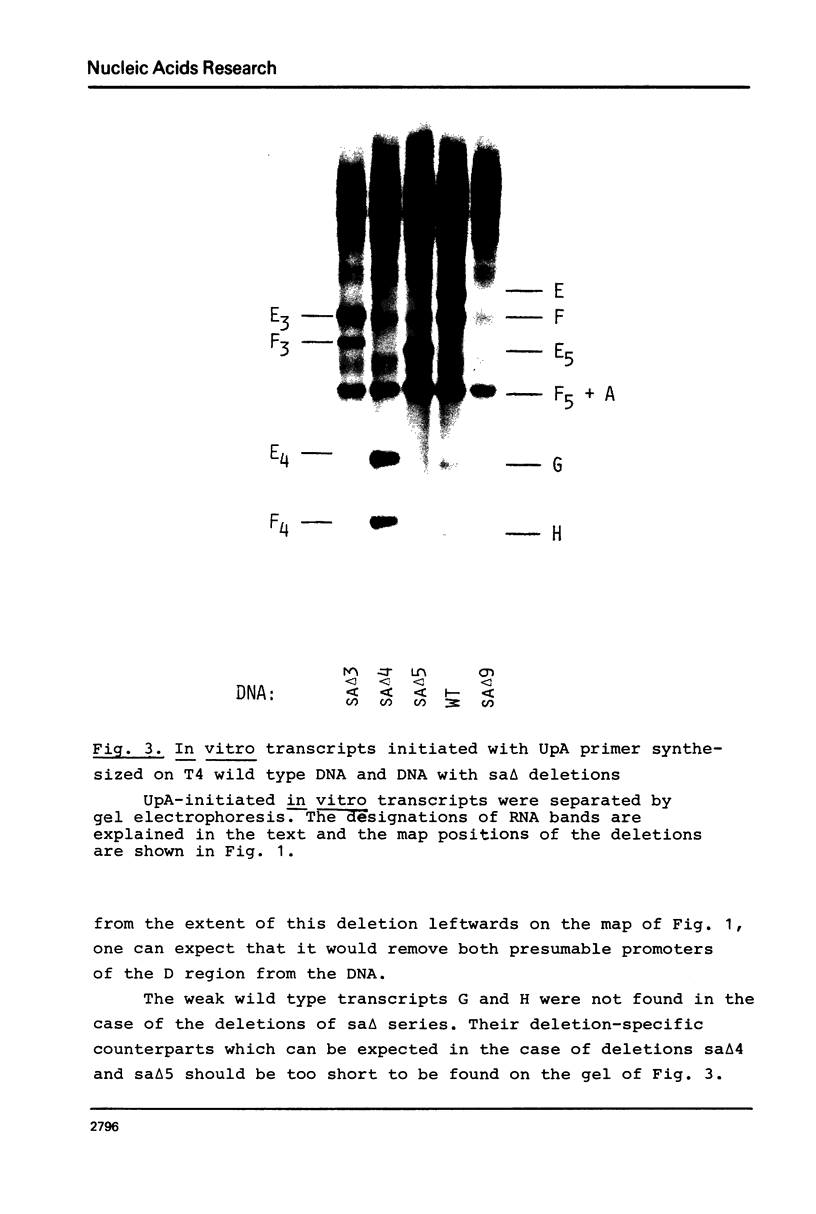
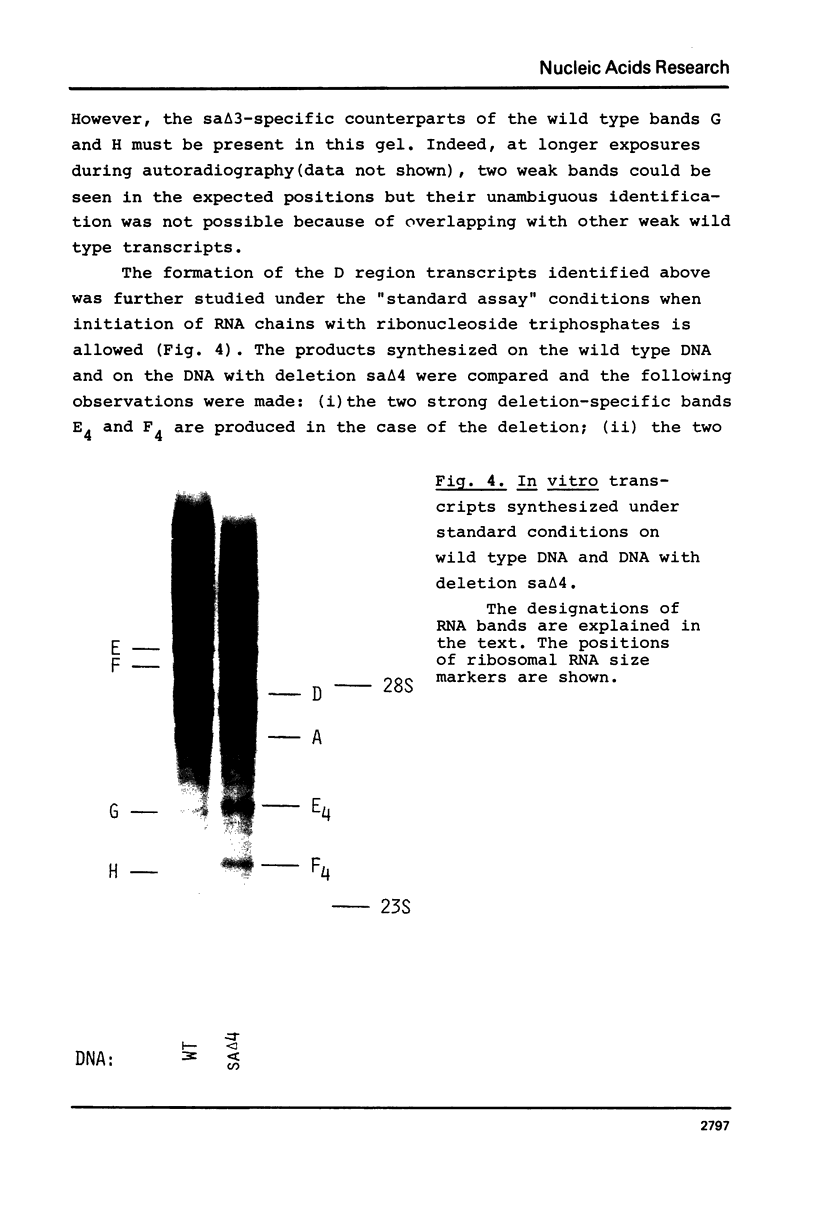
The D region of bacteriophage T4 is comprised of six closely linked genes which are situated between 161 kb and 165 kb on the T4 chromosome. We studied the transcription of these genes in vitro by using DNA templates derived from a series of deletion mutants in this region. The mixture of primary products made by Escherichia coli RNA polymerase were fractionated by gel electrophoresis into discrete RNA species. The results obtained together with the known map positions of the deletions allowed to identify four wild-type and several deletion-specific transcripts of the D region. The end points of these transcripts were approximately mapped. The results demonstrate that the D region has two promoters and two terminators, an organisation which is similar to the previously established organisation of the T4 tRNA gene cluster.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bautz F. A., Bautz E. K. Mapping of deletions in a non-essential region of the phage T4 genome. J Mol Biol. 1967 Sep 14;28(2):345–355. doi: 10.1016/s0022-2836(67)80014-7. [DOI] [PubMed] [Google Scholar]
- Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961 Mar;47(3):403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujard H., Mazaitis A. J., Bautz E. K. The size of the rII region of bacteriophage T4. Virology. 1970 Nov;42(3):717–723. doi: 10.1016/0042-6822(70)90317-x. [DOI] [PubMed] [Google Scholar]
- Daegelen P., Brody E. Early bacteriophage T4 transcription. A diffusible product controls rIIA and rIIB RNA synthesis. J Mol Biol. 1976 May 5;103(1):127–142. doi: 10.1016/0022-2836(76)90055-3. [DOI] [PubMed] [Google Scholar]
- Depew R. E., Snopek T. J., Cozzarelli N. R. Characterization of a new class of deletions of the D region of the bacteriophage T4 genome. Virology. 1975 Mar;64(1):144–145. doi: 10.1016/0042-6822(75)90086-0. [DOI] [PubMed] [Google Scholar]
- Dove W. The extent of rII deletions in phage T4. Genet Res. 1968 Apr;11(2):215–219. doi: 10.1017/s001667230001140x. [DOI] [PubMed] [Google Scholar]
- Fisher M. P., Dingman C. W. Role of molecular conformation in determining the electrophoretic properties of polynucleotides in agarose-acrylamide composite gels. Biochemistry. 1971 May 11;10(10):1895–1899. doi: 10.1021/bi00786a026. [DOI] [PubMed] [Google Scholar]
- Goldfarb A., Daniel V. Mapping of transcription units in the bacteriophage T4 tRNA gene cluster. J Mol Biol. 1981 Mar 15;146(4):393–412. doi: 10.1016/0022-2836(81)90039-5. [DOI] [PubMed] [Google Scholar]
- Goldfarb A., Daniel V. Transcriptional control of two gene subclusters in the tRNA operon of bacteriophage T4. Nature. 1980 Jul 24;286(5771):418–420. doi: 10.1038/286418a0. [DOI] [PubMed] [Google Scholar]
- Goldfarb A. IN vitro transcription of bacteriophage T4 tRNA gene cluster from two different promoters. Nucleic Acids Res. 1981 Feb 11;9(3):519–527. doi: 10.1093/nar/9.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb A., Seaman E., Daniel V. In vitro transcription and isolation of a polycistronic RNA product of the T4 tRNA operon. Nature. 1978 Jun 15;273(5663):562–564. doi: 10.1038/273562a0. [DOI] [PubMed] [Google Scholar]
- Kasai T., Bautz E. K. Regulation of gene-specific RNA synthesis in bacteriophage T4. J Mol Biol. 1969 May 14;41(3):401–417. doi: 10.1016/0022-2836(69)90284-8. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüger W. Transcription of bacteriophage T4 DNA in vitro: selective initiation with dinucleotides. Eur J Biochem. 1978 Jul 17;88(1):109–117. doi: 10.1111/j.1432-1033.1978.tb12427.x. [DOI] [PubMed] [Google Scholar]
- Schmidt D. A., Mazaitis A. J., Kasai T., Bautz E. K. Involvement of a phage T4 sigma factor and an anti-terminator protein in the transcription of early T4 genes in vivo. Nature. 1970 Mar 14;225(5237):1012–1016. doi: 10.1038/2251012a0. [DOI] [PubMed] [Google Scholar]
- Sederoff R., Bolle A., Epstein R. H. A method for the detection of specific T4 messenger RNAs by hybridization competition. Virology. 1971 Aug;45(2):440–455. doi: 10.1016/0042-6822(71)90344-8. [DOI] [PubMed] [Google Scholar]
- TESSMAN I. The induction of large deletions by nitrous acid. J Mol Biol. 1962 Oct;5:442–445. doi: 10.1016/s0022-2836(62)80033-3. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T., Mattson T., Selzer G., Van Houwe G., Bolle A., Epstein R. Bacteriophage T4 gene transcription studied by hybridization to cloned restriction fragments. J Mol Biol. 1980 Apr 15;138(3):423–445. doi: 10.1016/s0022-2836(80)80011-8. [DOI] [PubMed] [Google Scholar]