Abstract
Several fatal, progressive neurodegenerative diseases, including various prion and prion-like disorders, are connected with the misfolding of specific proteins. These proteins misfold into toxic oligomeric species and a spectrum of distinct self-templating amyloid structures, termed strains. Hence, small molecules that prevent or reverse these protein-misfolding events might have therapeutic utility. Yet it is unclear whether a single small molecule can antagonize the complete repertoire of misfolded forms encompassing diverse amyloid polymorphs and soluble oligomers. We have begun to investigate this issue using the yeast prion protein Sup35 as an experimental paradigm. We have discovered that a polyphenol, (−)epigallocatechin-3-gallate (EGCG), effectively inhibited the formation of infectious amyloid forms (prions) of Sup35 and even remodeled preassembled prions. Surprisingly, EGCG selectively modulated specific prion strains and even selected for EGCG-resistant prion strains with novel structural and biological characteristics. Thus, treatment with a single small molecule antagonist of amyloidogenesis can select for novel, drug-resistant amyloid polymorphs. Importantly, combining EGCG with another small molecule, 4,5-bis-(4-methoxyanilino)phthalimide, synergistically antagonized and remodeled a wide array of Sup35 prion strains without producing any drug-resistant prions. We suggest that minimal drug cocktails, small collections of drugs that collectively antagonize all amyloid polymorphs, should be identified to besiege various neurodegenerative disorders.
Key words: amyloid, yeast prion, Sup35, prion strains, EGCG, DAPH-12
Combating Amyloidogenesis in Neurodegenerative Disorders
Protein misfolding and aggregation are common features of several of the most devastating and currently untreatable neurodegenerative diseases confronting humankind.1,2 For example, the brains of Alzheimer disease (AD) patients are characterized by extracellular protein deposits, termed plaques.3 These deposits consist of amyloid fibrils, structurally defined as fibrillar polypeptide aggregates with a cross-β conformation, in which the β-strands align orthogonal to the fiber axis to generate an intermolecular β-sheet.4 These cross-β structures are exceptionally stable (e.g., protease- and detergent-resistant) and therefore extremely difficult to clear.5 Moreover, once initiated, amyloidogenesis can cascade out of control because amyloid fibrils seed their own assembly by recruiting non-amyloid conformers to fibril ends and converting them to the cross-β form.5 Specific polypeptides that tend to populate intrinsically unfolded states, but which do not share any primary sequence homology, switch to the amyloid state in specific neurodegenerative disorders.1,6,7 For example, Aβ forms amyloid plaques in AD, tau forms neurofibrillary tangles in AD and tauopathies, a-synuclein forms amyloid inclusions called Lewy bodies in Parkinson disease (PD) and polyglutamine-expanded huntingtin forms cytosolic and nuclear inclusions in Huntington disease (HD).3
Burgeoning evidence suggests these self-templating amyloid forms can disseminate and be transmitted from cell to cell, thereby spreading neuropathology through contiguous tracts of the brain in afflicted individuals.1,8–16 Indeed, in some cases amyloid forms can be infectious.17,18 That is, they are naturally transmitted between individuals and promote phenotypic change.1,19–21 These infectious amyloid forms are termed prions.1,19–21 In mammals, infectious forms of prion protein (PrP) cause transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy in cattle, chronic wasting disease in elk and deer and variant Creutzfeldt-Jakob disease in humans.20,22–24
Despite immense experimental effort, the precise role of amyloid deposits in the pathobiology of various neurodegenerative diseases is still highly debated. While earlier work suggested that the amyloid form represents a toxic, pathogenic species, more recent studies imply that amyloid deposits could either be an epiphenomenon of protein misfolding, or even represent a neuroprotective conformation.25 Indeed, abundant evidence suggests that the soluble oligomeric intermediates that form prior to fibers during amyloidogenesis may be even more neurotoxic than amyloid fibrils. Thus, sequestration of protein into fibrillar inclusions at the expense of oligomers may be protective.26–29 However, a ubiquitous feature of amyloidogenesis is that a single polypeptide can access multiple distinct amyloid structures, termed strains.1,18,19,22,30–34 Different strains can confer different phenotypes.18,22,30–33,35,36 Some strains are highly toxic, whereas others are more benign.31,32,35 Moreover, amyloid forms represent a space-occupying lesion and due to steric constraints of the amyloid form, proteins often lose functionality in this state.37 Indeed, this loss of protein function may contribute to several neurodegenerative disorders.38–40 Hence, as a therapeutic strategy both pharma and academia continue to explore antagonists of amyloidogenesis.41–43 Encouragingly, numerous studies using diverse disease models document that the prevention or the elimination of amyloid deposits can reduce neurodegeneration.25,44
For instance, antibodies or fragments of antibodies that recognize amyloid proteins effectively inhibit the formation of amyloid deposits and can reduce the associated toxicity.45 Yet because of their size and immunological properties, antibodies can pose substantial problems as potential therapeutics.44 Another approach might involve the targeted expression of agents that prevent or reverse amyloidogenesis. For example, expression of the protein disaggregase Hsp104 can reduce protein aggregation and associated neurodegeneration in rodent models of HD and PD.46–49 However, such an approach in humans would seem to require gene therapy, which has delivered promising preclinical outcomes for several diverse disorders, including congenital blindness50,51 and PD.52–54 Yet gene therapy has not been readily translated to the clinic due to a legion of significant technical and safety issues.
As an alternative strategy, small molecules that prevent or reverse amyloidogenesis may hold more immediate therapeutic potential. Yet inhibiting amyloid formation or even remodeling pre-formed amyloids might present a daunting biophysical task for small molecules for two major reasons: (1) Small molecules lack the size and bulkiness that seem to be required to interfere with the comparatively large surfaces of protein-protein interactions within an amyloid fibril. (2) The intermolecular protein-protein contacts within amyloids are among the strongest in nature and thus may resist modulation by small molecules.5,55,56 Another challenge is found in the blood-brain barrier (BBB), which sharply limits which small molecules can enter the brain.57 Ideally, lead small molecules should efficiently cross the BBB.58 In spite of these obstacles, work by many groups has identified small molecules that are able to effectively inhibit amyloid formation and modulate, or even remodel pre-formed amyloids.19,43,59–67
The Challenge of Conformational Diversity
A colossal challenge facing any therapeutic strategy aimed at antagonizing amyloidogenesis is to cope with the conformational diversity of misfolded forms that are likely to have already accrued prior to administration of any treatment. Typically, amyloidogenic polypeptides fold spontaneously into an ensemble of distinct strains.19,30–33,68,69 Little is known about the precise atomic structures of these distinct cross-β structures in the context of full-length proteins. Yet studies with short peptides (4–12 amino acids) have yielded atomic insights into cross-β diversity.70,71 The ability of a single polypeptide to populate distinct amyloid forms is likely due to large portions of Ramachandran space being available to the main chain within the constraints of cross-β architecture.19,72 This freedom likely enables side chains to pack in different ways to achieve various unique and energy-minimized cross-β structures.19,70–72 Once each cross-β form has originated, it can then amplify by high fidelity self-templating.
The precise composition of the strain ensemble depends on the environment. For example, PrP, Aβ and polyglutamine assemble into distinct strains ensembles at 4°C versus 37°C.32,68,69 Different strain ensembles confer distinct phenotypes, including the degree of neurotoxicity.31,32,73 Precisely how distinct strains encode distinct phenotypes remains unclear, although two environmentally-sensitive kinetic properties may be important: the rate at which fibril ends self-template and the rate at which fibrils fragment to liberate new self-templating surfaces.19,36,68 Remarkably, at least 15 different prion strains have been described for mammalian PrP,22 which confer distinct transmissible neurodegenerative disorders differentiated by the rate of disease progression and pathology. This cloud of diverse misfolded forms is further elaborated by the suite of toxic, non-amyloid oligomeric species that accumulate prior to fibril formation.26,28 These oligomeric species can also take a variety of poorly defined structural forms.26,28,29
This conformational diversity severely complicates the development of potential small molecule therapeutics, particularly because small molecules are likely to be applied only after structurally diverse misfolded forms have already accumulated. Among these misfolded forms, unlike many other protein targets, there is no single well defined atomic structure immediately amenable for rational drug design.74 Thus, one approach has been to design small molecules that stabilize the natively folded state of the protein, such that it is less likely to access amyloidogenic misfolding trajectories. For example, stabilizing the tetrameric form of transthyretin (TTR) with small molecules increases the kinetic barrier associated with misfolding.67 A drug discovered using this approach is now in Phase II/III clinical trials in patients suffering from TTR amyloid polyneuropathy. However, for many amyloidogenic proteins the native state is intrinsically unfolded and it is unclear how to stabilize this form. Ideally, a small molecule therapeutic would antagonize the complete repertoire of structurally distinct misfolded forms. For example, to effectively eradicate amyloids a small molecule would have to remodel not only one specific strain but many if not all possible strains of an amyloid that might be present in a patient's brain. However, a key issue that has remained unaddressed is whether a single small molecule can directly antagonize the entire spectrum of structurally distinct misfolded forms, including diverse amyloid or prion strains.
The [PSI+] Toolbox
To address this issue for prionogenic proteins, we needed a defined system to study different prion strains comprised of pure protein. The generation of distinct mammalian prion strains with pure PrP, which infect and cause distinct transmissible diseases in wild-type mice, has proven challenging, even though several key advances have been made.17,24,35,68 Hence, we turned to Sup35, a translation termination factor, which can be readily assembled into various pure infectious amyloid strains that confer distinct heritable reductions in translation termination fidelity and comprise variants of the yeast prion [PSI+].30,33,36,61,75–77 With an array of in vitro and in vivo tools to study bona fide prion strains, Sup35 offers a powerful system to deconstruct how small molecules directly affect the folding, formation and integrity of distinct misfolded forms.
[PSI+] is one of the most prominent and studied yeast prions.21 In [psi−] cells, Sup35 is found in its soluble, non-prion form and facilitates faithful translation termination.21 In [PSI+] cells, Sup35 accesses various insoluble, prion forms.21 Consequently, Sup35 function is partially impaired, which leads to reductions in translation termination fidelity.21 The reduced function of Sup35 in [PSI+] cells uncovers many multigenic phenotypes in yeast, which depend on the particular genetic background.21,78,79 These [PSI+]-mediated phenotypes can be neutral, deleterious or occasionally even advantageous for the fitness and survival of yeast cells in a given environment.21,78,79 Distinct Sup35 prion strains manifest in the distinct strengths of the associated [PSI+] phenotypes, which are classified by the extent of loss of Sup35 function.30,36,80 In cells harboring a strong [PSI+] strain, the loss of Sup35 function in translation termination is more severe, whereas in cells harboring a weak [PSI+] strain, the loss of Sup35 function is less severe.80 These prion strain phenotypes are readily distinguished in cells that carry a premature stop codon in their ADE1 gene.80 Thus, [psi−] yeast colonies do not make functional Ade1 and accumulate a red metabolite on rich media. By contrast, on rich media, weak [PSI+] colonies are pink and strong [PSI+] colonies are white in accord with the extent of Sup35 aggregation and contingent inactivation.21,36,80 These color differences allow the straightforward quantitative assessment of the capacity of small molecules to convert [PSI+] cells into [psi−] cells in simple plating assays.60,61 Using a similar experimental approach, we can also easily assess the capacity of small molecules to modulate different strains of [PSI+] in living cells.60,61
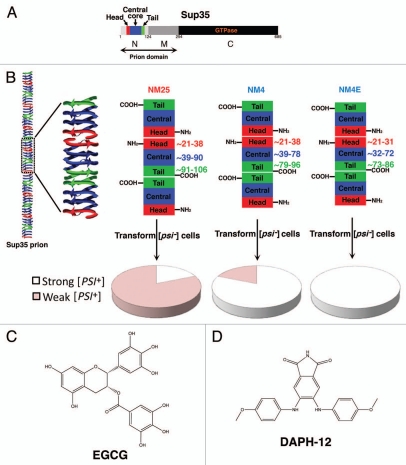
Importantly, these distinct [PSI+] strains can be readily generated beginning with pure Sup35 in the test tube (Fig. 1A).30,33,36,61,77 Sup35 is a modular protein comprised of a C-terminal GTPase domain (C, amino acids 254–685), which confers translation termination activity (Fig. 1A). Whether Sup35 adopts a prion or a non-prion state is determined by interplay between the middle domain (M, amino acids 124–253) and the N-terminal domain (N, amino acids 1–123). M is enriched in charged residues and confers solubility,81 whereas N is extremely amyloidogenic82 and of unusually low sequence complexity, composed primarily of glutamine, asparagine, glycine and tyrosine. Together N and M (NM) confer all the properties needed to form a stable prion in yeast, and are termed the prion domain.81,83 In isolation, Sup35, NM and N can all spontaneously access infectious amyloid forms.30,33,36,61,77,84 That is, if Sup35, NM or N are assembled into amyloid forms in vitro and then transformed into [psi−] cells, then a large proportion of transformants (up to 80%) become [PSI+].30,33,36,61,77,84 This change in prion phenotype occurs because the introduced amyloid forms of Sup35 convert the endogenous Sup35 to the prion state, which can then be propagated through successive generations.30,36 This transformation method of [PSI+] induction does not require cells to harbor another prion, [PIN+],30,36,61,77,84 which is usually comprised of prion conformers of the protein Rnq1.85–88 By contrast, [PSI+] can also be induced by overexpression of Sup35 or NM, but in this case [PSI+] induction depends on the presence of [PIN+]86–88 because Rnq1 prions appear to provide a template for the initial formation of Sup35 prions.86,89 However, once [PSI+] is established then [PIN+] is dispensable for [PSI+] propagation.86,87 Importantly, using pure protein, the precise strain ensemble that assembles can be controlled by altering the assembly conditions. For example, if pure NM is assembled into infectious amyloid forms at 25°C, termed NM25 and transformed into [psi−] [pin−] cells, then the majority of transformants become weak [PSI+] (Fig. 1B).30,33,36,61 By contrast, if pure NM is assembled into infectious amyloid forms at 4°C, termed NM4 and transformed into [psi−] [pin−] cells then the majority of transformants become strong [PSI+]30,33,36,61 (Fig. 1B).
Figure 1.
Sup35 prion strains and small-molecule antagonists. (A) Sup35 is a modular protein comprised of a C-terminal GTPase domain (C, amino acids 254–685, black), a highly charged middle domain (M, amino acids 124–253, dark grey) and an N-terminal domain (N, amino acids 1–123, light grey) enriched in glutamine, asparagine, tyrosine and glycine residues. Together N and M (NM) confer all the properties needed to form a stable prion in yeast. NM is termed the prion domain.83 Within N, prion recognition elements termed the “head” (red) and “tail” (green), which flank a “central core” (blue), play important roles in prion formation.33,90 (B) Sup35 prions adopt a polymeric cross-beta structure. In one proposed model (left), this amyloid structure is composed of the head (red), central core (blue) and tail (green) regions of N. The M and C domains are located on the exterior of this structure and are not depicted for clarity. If we zoom in on three adjacent monomers in the Sup35 prion polymer, we find that the prion is proposed to be maintained by an alternating sequence of head-to-head (red) and tail-to-tail (green) intermolecular contacts. The central core is sequestered in intramolecular contacts (blue). Different Sup35 prion strains assemble under different environmental conditions. Thus, NM25 assembles at 25°C or when NM is chemically crosslinked with BMB in the tail region.33,61 NM4 assembles at 4°C or when NM is chemically crosslinked with BMB in the head region.33,61 NM4E assembles in the presence of EGCG at 4°C.61 These prion strains have subtle differences in the precise residues that comprise the head, tail and central core (right). The residues that comprise the head, tail and central core are shown to the right of each central protomer. NM25, NM4 and NM4E are distinguished by their different tail-to-tail contacts and central core region.61 Moreover, NM4E has a distinct head-to-head contact.61 Infection of [psi−] [pin−] cells with NM25 yields mostly weak [PSI+] variants, whereas NM4 yields mostly strong [PSI+] variants.61 By contrast, NM4E generates purely strong [PSI+] variants61 (pie charts in lower portion). (C) Chemical structure of EGCG. (D) Chemical structure of DAPH-12.
A collection of single cysteine NM mutants, which behave like wild-type NM, have allowed the strategic placement of fluorescent probes (e.g., pyrene or acrylodan) to provide position-specific information concerning the local environment of individual residues within NM in various conformational states.33,60,61 These tools have allowed definition of the intermolecular contacts that hold Sup35 prions together, as well as the definition of the primary sequence sequestered in cross-β structure.33,60,61 Short prion recognition elements within the N-terminal domain of Sup35, termed “head” and “tail” (Fig. 1A), are proposed to make homotypic intermolecular contacts in assembled Sup35 prions (Fig. 1B). Thus, Sup35 fibers are proposed to consist of an alternating sequence of head-to-head and tail-to-tail contacts that are separated by a central core, which together comprise the cross-beta structure (Fig. 1A and B). Indeed, NM4 and NM25 strains have different intermolecular contacts and sequester overlapping but distinct portions of the N-terminal domain (N) in their amyloid core33,60,61 (Fig. 1B). The length of primary sequence sequestered in intramolecular contacts (central core) and the position of the C-terminal intermolecular contact (tail) are markedly different between NM4 and NM2533,60,61 (Fig. 1B). Although the N-terminal intermolecular contact (head) is similar in NM4 and NM25, residues N-terminal to the head are organized differently in the two strains.33,61,76 This collection of single cysteine mutants also allows the assembly of specific prion strains regardless of the assembly temperature.33,61 Thus, specific crosslinking of single cysteine NM mutants in the head with 1,4-bis-maleimidobutane (BMB) causes formation of NM4 at 4 and 25°C, whereas BMB crosslinking in the tail causes formation of NM25 at 4 and 25°C33,61 (Fig. 1B).
The atomic structures of Sup35 prion strains remain unknown and several models have been proposed.33,61,71,76,90–93 Nevertheless, the formation of bona fide Sup35 prion strain ensembles is readily controlled and these strains are distinguished at the resolution of spatial arrangements of individual amino acids.33,61,76 The combination of in vitro and in vivo approaches available to study Sup35 prions presents an unparalleled experimental platform to determine the direct effects of small molecules on different prion strains and decipher the molecular bases for these effects.
EGCG: A Strain-Selective Antagonist
We initially focused on (−)epigallocatechin-3-gallate (EGCG) (Fig. 1C), a polyphenol that is highly abundant in green tea. Several studies document that EGCG effectively inhibits the de novo amyloidogenesis of various disease-associated proteins, such as α-synuclein, tau, PrP, polyglutamine-expanded huntingtin and Aβ.56,62,63,65,94 EGCG appears to inhibit assembly by redirecting proteins into stable oligomers and monomeric forms that are not competent for amyloidogenesis.63 Importantly, these EGCG-induced conformers are much less toxic to neuroblastoma cells in culture than amyloid forms of Aβ and α-synuclein.63 EGCG also inhibited assembly of Aβ and α-synuclein that was seeded by preformed fibrils and even remodeled preformed toxic oligomers of Aβ and α-synuclein.63 Thus, EGCG can antagonize various preformed misfolded species. Yet whether EGCG could target different amyloid strains of the same polypeptide remained uncertain.
Using assembly temperature and BMB crosslinking to control the strain bias of pure NM, we demonstrated that EGCG selectively inhibited formation of NM25 by preventing the maturation of molten oligomers that is required for de novo prion formation.61,95,96 Under assembly conditions where NM4 was favored, EGCG failed to inhibit prion formation.61 We verified these findings with full-length Sup35.61 Intriguingly, however, at 4°C in the presence of EGCG, NM4 did not form.61 Instead, a new prion strain, termed NM4E, assembled with N-terminally shifted head and tail intermolecular contacts that were resistant to EGCG61 (Fig. 1B). Thus, under some environmental conditions, a single prion protein can fold into a new strain with shifted intermolecular contacts that evades an otherwise potent small molecule antagonist.61 This unexpected plasticity of prionogenesis exposes the difficulty it poses to drug development.
We corroborated these findings in vivo.61 Transformation of [psi−] [pin−] cells with NM4E yielded purely strong [PSI+], whereas NM4 generated a mixture of strains that were mostly strong [PSI+], though weak [PSI+] was also represented61 (Fig. 1B). Similarly, overexpression of NM-YFP in [psi−] [PIN+] cells in the presence of EGCG induced the formation of mostly strong [PSI+].61 [PIN+] variants can influence the stability of different [PSI+] variants,97 but upon overexpression of NM-YFP this particular [PIN+] variant facilitated the induction of ∼65% weak [PSI+] and ∼35% strong [PSI+] in the absence of EGCG.61 In the presence of EGCG, [PSI+] induction was reduced and the distribution of strains was shifted to ∼30% weak [PSI+] and ∼70% strong [PSI+].61 These data indicate that EGCG might also inhibit Rnq1 prions from cross-seeding the formation of NM25 but not NM4, a possibility that remains to be tested with pure components.89 Regardless, EGCG strongly selects against Sup35 prion strains that encode weak [PSI+] in vitro and in vivo.61 Moreover, our data suggest that the repertoire of Sup35 prion strains that can encode strong [PSI+] is more nuanced than previously suspected.33,61
EGCG also displayed strain-selective activity against preassembled Sup35 prions. Remarkably, EGCG remodeled the intra- and intermolecular contacts of NM25 to generate oligomeric species that retained some β-sheet structure, but were unable to seed the assembly of soluble NM or convert [psi−] [pin−] cells to [PSI+].61 EGCG partially remodeled NM4, but self-templating prions remained largely intact.61 At the other extreme, NM4E completely resisted remodeling by EGCG.61 This spectrum of effects was recapitulated in vivo. Thus, strong [PSI+] variants encoded by NM4E were refractory to EGCG, whereas EGCG cured strong [PSI+] strains encoded by NM4 to some extent.61 Yet, consistent with in vitro observations, EGCG showed the greatest curing activity against weak [PSI+] variants encoded by NM25.61 EGCG promoted these curing events by remodeling Sup35 prions in vivo and did not induce cellular stress responses or interfere with Hsp104 ATPase activity, that is, EGCG did not modulate cellular proteostasis.61
Typically, [PSI+] variants are stable and do not switch from weak to strong or vice versa.80,98 However, when weak [PSI+] variants were cured with EGCG, a low percentage of yeast colonies converted to a strong [PSI+] strain that could no longer be cured by EGCG.61 It is probable that some weak [PSI+] variants contained low levels of EGCG-resistant prions, which could amplify in the presence of EGCG and promote switching to strong [PSI+].61 Collectively, these in vitro and in vivo findings raise the specter that a single small molecule may be insufficient to counter prion or amyloid polymorphism, and might even select for drug-resistant forms. Indeed, this phenomenon has also become apparent in mammalian prion infections, where drug-resistant strains can arise and dominate the population after chronic exposure to a single small-molecule drug.19,99,100
Whether EGCG is strain-selective for other amyloidogenic proteins or whether Sup35 prion strains are uniquely able to escape its inhibitory effects remains uncertain. However, a rapidly assembling variant of α-synuclein can escape inhibition by EGCG, perhaps by accessing a different strain.101 While many small molecules have the capacity to inhibit amyloid formation, there are fewer examples of small molecules that remodel pre-formed amyloids. EGCG was very effective in remodeling NM25 and recent studies suggest that EGCG also remodels amyloid fibrils formed by Aβ or α-synuclein.64 In analogy to our studies using NM25, EGCG converted large Aβ or α-synuclein fibrils into smaller aggregated species.64 These smaller aggregates, unlike larger amyloid fibrils, were not toxic to neuroblastoma cells in culture.64 EGCG also remodels amyloid forms of prostatic acidic phosphatase fragments, which can potentiate HIV infection.102,103 Moreover, EGCG might help reduce amyloid burden in amyloid light chain amyloidosis.104 Thus, EGCG can target various amyloidogenic proteins although it remains unclear to what extent these effects are strain-selective.
Combinatorial Treatment with EGCG and DAPH-12
We next asked whether combining EGCG with another small molecule might help antagonize a broader spectrum of different Sup35 prion strains and prevent the formation of drug-resistant prions. We first considered 4,5-bis-(4-methoxyanilino)phthalimide (DAPH-12) (Fig. 1D). We had previously demonstrated that DAPH-12, an analog of 4, 5-dianilinophthalimide, which remodels Aβ fibrils66 but not α-synuclein, tau or mammalian prions,60 inhibits NM25 formation in vitro by disrupting early folding events in molten oligomers that nucleate prion assembly.60 DAPH-12 also antagonized assembly of NM4, but this inhibition was less pronounced than that observed with NM25.61 In contrast to EGCG, however, a new prion strain did not form in the presence of DAPH-12.61 DAPH-12 also remodeled preformed NM25, NM4 and NM4E prions, but was considerably more effective against NM25, which was converted to predominantly non-replicating conformers.60,61 Accordingly, DAPH-12 cured weak [PSI+] encoded by NM25 more efficaciously than strong [PSI+] encoded by NM4 or NM4E.61 Unlike EGCG, DAPH-12 remodeled only the intermolecular contacts of NM25 and did not affect the intramolecular contacts.60,61 Thus, although DAPH-12 also operates most potently against NM25, it modulates Sup35 prions by a distinct molecular mechanism to EGCG.60,61 Importantly, DAPH-12 never produced new drug-resistant prion strains.61
This distinct mechanistic action of DAPH-12 suggested that DAPH-12 might synergize with EGCG to antagonize Sup35 prionogenesis. Indeed, when we combined DAPH-12 and EGCG, they synergized to prevent and reverse the formation of NM4 and NM25.61 Strain-specific intra- and intermolecular contacts were most effectively disrupted by the drug combination.61 Also, the formation of the new EGCG-resistant strain, NM4E, was effectively inhibited by the additional presence of DAPH-12.61 Pre-formed NM4E was remodeled by DAPH-12 alone, and in this instance the combination was no more effective than DAPH-12 alone.61 The combination of DAPH-12 and EGCG also synergized in curing multiple distinct [PSI+] strains in vivo and, importantly, weak [PSI+] was now cured without the generation of EGCG-resistant [PSI+] strains that could escape curing.61 Overall, our findings suggest that DAPH-12 and EGCG synergize to antagonize a broader spectrum of Sup35 prion forms.61
Minimal Drug Cocktails: A Safe and Effective Strategy to Combat Neurodegeneration?
Based upon our results, we advocate the exploration of effective small molecule combination strategies for the treatment of amyloid-associated neurodegenerative diseases. It seems unlikely that a single small molecule holds the capacity to overcome the dynamics of amyloid formation, propagation and the generation and maintenance of diverse amyloid strains. The strain phenomenon has only recently been explored for non-prion amyloids31,32,69 and there are still many open questions. Yet in analogy to PrP or Sup35, it seems highly probable that other amyloidogenic proteins assemble into different strains in different individuals suffering from the same neurodegenerative disease. A single molecule might thus show positive effects only in one group of patients that harbor a particular amyloid strain but might fail in many other groups that harbor different amyloid strains. We further speculate that individual patients or even individual neurons within individual patients are likely to harbor multiple amyloid strains. In this case a single small molecule would most likely be completely ineffective. Moreover, our results indicate an even more insidious threat posed by treatment of amyloid-associated neurodegenerative diseases with a single small molecule. In analogy to the EGCG-mediated generation of a drug-resistant strain of [PSI+], the treatment with a single small molecule might produce drug-resistant and, possibly, more neurotoxic amyloid strains and thus may worsen the condition of a patient. Indeed, during mammalian prion infection continuous treatment with quinacrine can lead to the appearance of quinacrine-resistant prions that maintain the infection and neurodegeneration.100 It is already clear that small molecules can have strain-selective effects on mammalian prion infections.105 Thus, it is important to identify small-molecule scaffolds that are prone to giving rise to new amyloid polymorphs and either avoid these scaffolds altogether or incorporate them into more broad spectrum drug cocktails.
In our experimental paradigm, only the combination of EGCG and DAPH-12 cured different [PSI+] strains and precluded the emergence of a new drug resistant prion strain. Hence, we suggest that combining different small molecules that directly modulate amyloidogenesis by distinct molecular mechanisms presents a promising strategy. Ideally, a minimal collection of small molecules that together targets all conformational variants is sought to diminish any increased side effects or toxicity of each single small molecule.106 Treatments with minimal drug combinations are already successfully applied to overcome the emergence of drug resistance in microbial infections and lessons learned from these systems might also be appropriate for the treatment of neurodegenerative amyloidoses.19,107,108 Moreover, small molecule combinations that modulate proteostasis can have synergistic therapeutic effects in models of lysosomal storage disorders.109
The yeast prion [PSI+] allowed us to explore the molecular mechanism by which EGCG and DAPH-12 synergize to eradicate infectious amyloid forms. The unique and powerful experimental arsenal available for studying [PSI+] revealed the prion strain-selectivity of EGCG, the EGCG-mediated emergence of a drug-resistant prion strain and provided the mechanistic rationale to test the drug combination of EGCG and DAPH-12. While future research will be needed to explore the effects of drug cocktails in the context of other amyloidogenic proteins and other model organisms, the yeast prion system will undoubtedly continue to provide valuable insights into the interaction of small molecules with amyloidogenic proteins.
Acknowledgements
M.L.D. is supported by a research grant from the American Federation for Aging Research, from the William Wood Foundation, and the Hereditary Disease Foundation. J.S. is supported by an NIH Director's New Innovator Award (1DP2OD002177-01), an Ellison Medical Foundation New Scholar in Aging Award, an NINDS grant (1R21NS067354-0110), a Bill and Melinda Gates Foundation Grand Challenges Explorations Award and a University of Pennsylvania Diabetes and Endocrinology Research Center Pilot and Feasibility grant.
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/13597
References
- 1.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 3.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 4.Fandrich M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell Mol Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 7.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 10.Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JY, Englund E, Holton JL, Soulet D, Hagell P, Lees AJ, et al. Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 13.Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson's disease. Nat Med. 2008;14:504–506. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 14.Luk KC, Song C, O'Brien P, Stieber A, Branch JR, Brunden KR, et al. Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc Natl Acad Sci USA. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, et al. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 18.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, et al. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shorter J. Emergence and natural selection of drug-resistant prions. Mol Biosyst. 2010;6:1115–1130. doi: 10.1039/c004550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 22.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 23.Prusiner SB. The prion diseases. Brain Pathol. 1998;8:499–513. doi: 10.1111/j.1750-3639.1998.tb00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6:891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 26.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 27.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 28.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, et al. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 31.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 32.Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc Natl Acad Sci USA. 2009;106:9679–9684. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fandrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion. 2009;3:89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, et al. Protease-sensitive synthetic prions. PLoS Pathog. 2010;6:1000736. doi: 10.1371/journal.ppat.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 37.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gitler AD, Shorter J. Prime time for alpha-synuclein. J Neurosci. 2007;27:2433–2434. doi: 10.1523/JNEUROSCI.0094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorbatyuk OS, Li S, Nash K, Gorbatyuk M, Lewin AS, Sullivan LF, et al. In vivo RNAi-mediated alpha-synuclein silencing induces nigrostriatal degeneration. Mol Ther. 2010;18:1450–1457. doi: 10.1038/mt.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VM. Abnormal tau phosphorylation at Ser396 in Alzheimer's disease recapitulates development and contributes to reduced microtubule binding. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- 41.Barten DM, Albright CF. Therapeutic strategies for Alzheimer's disease. Mol Neurobiol. 2008;37:171–186. doi: 10.1007/s12035-008-8031-2. [DOI] [PubMed] [Google Scholar]
- 42.Citron M. Alzheimer's disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9:387–398. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- 43.Crowe A, Huang W, Ballatore C, Johnson RL, Hogan AM, Huang R, et al. Identification of aminothienopyridazine inhibitors of tau assembly by quantitative high-throughput screening. Biochemistry. 2009;48:7732–7745. doi: 10.1021/bi9006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schenk D. Amyloid-beta immunotherapy for Alzheimer's disease: the end of the beginning. Nat Rev Neurosci. 2002;3:824–828. doi: 10.1038/nrn938. [DOI] [PubMed] [Google Scholar]
- 45.Spencer B, Rockenstein E, Crews L, Marr R, Masliah E. Novel strategies for Alzheimer's disease treatment. Expert Opin Biol Ther. 2007;7:1853–1867. doi: 10.1517/14712598.7.12.1853. [DOI] [PubMed] [Google Scholar]
- 46.Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/o09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 48.Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, et al. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 50.Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 51.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feigin A, Kaplitt MG, Tang C, Lin T, Mattis P, Dhawan V, et al. Modulation of metabolic brain networks after subthalamic gene therapy for Parkinson's disease. Proc Natl Acad Sci USA. 2007;104:19559–19564. doi: 10.1073/pnas.0706006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 54.Stoessl AJ. Gene therapy for Parkinson's disease: early data. Lancet. 2007;369:2056–2058. doi: 10.1016/S0140-6736(07)60957-X. [DOI] [PubMed] [Google Scholar]
- 55.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 56.Roberts BE, Shorter J. Escaping amyloid fate. Nat Struct Mol Biol. 2008;15:544–546. doi: 10.1038/nsmb0608-544. [DOI] [PubMed] [Google Scholar]
- 57.Pardridge WM. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballatore C, Brunden KR, Piscitelli F, James MJ, Crowe A, Yao Y, et al. Discovery of brain-penetrant, orally bioavailable aminothienopyridazine inhibitors of tau aggregation. J Med Chem. 2010;53:3739–3747. doi: 10.1021/jm100138f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ladiwala AR, Lin JC, Bale SS, Marcelino-Cruz AM, Bhattacharya M, Dordick JS, et al. Resveratrol selectively remodels soluble oligomers and fibrils of amyloid a{beta} into off-pathway conformers. J Biol Chem. 2010;285:24228–24237. doi: 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, et al. Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc Natl Acad Sci USA. 2008;105:7159–7164. doi: 10.1073/pnas.0801934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, et al. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5:936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, et al. Green tea (−)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum Mol Genet. 2006;15:2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- 63.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 64.Bieschke J, Russ J, Friedrich RP, Ehrnhoefer DE, Wobst H, Neugebauer K, et al. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc Natl Acad Sci USA. 2010;107:7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masuda M, Suzuki N, Taniguchi S, Oikawa T, Nonaka T, Iwatsubo T, et al. Small molecule inhibitors of alpha-synuclein filament assembly. Biochemistry. 2006;45:6085–6094. doi: 10.1021/bi0600749. [DOI] [PubMed] [Google Scholar]
- 66.Blanchard BJ, Chen A, Rozeboom LM, Stafford KA, Weigele P, Ingram VM. Efficient reversal of Alzheimer's disease fibril formation and elimination of neurotoxicity by a small molecule. Proc Natl Acad Sci USA. 2004;101:14326–14332. doi: 10.1073/pnas.0405941101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 68.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, et al. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kodali R, Williams AD, Chemuru S, Wetzel R. Aβ(1–40) forms five distinct amyloid structures β-whose sheet contents and fibril stabilities correlate. J Mol Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiltzius JJ, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 72.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mobley DL, Dill KA. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shkundina IS, Ter-Avanesyan MD. Prions. Biochemistry (Mosc) 2007;72:1519–1536. doi: 10.1134/s0006297907130081. [DOI] [PubMed] [Google Scholar]
- 76.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 77.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 79.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 80.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI+] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci USA. 2002;99:16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 83.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 84.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 86.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 87.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, et al. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 89.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DL. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl Acad Sci USA. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rambold AS, Miesbauer M, Olschewski D, Seidel R, Riemer C, Smale L, et al. Green tea extracts interfere with the stress-protective activity of PrP and the formation of PrP. J Neurochem. 2008;107:218–229. doi: 10.1111/j.1471-4159.2008.05611.x. [DOI] [PubMed] [Google Scholar]
- 95.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 96.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 97.Bradley ME, Liebman SW. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kochneva-Pervukhova NV, Chechenova MB, Valouev IA, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. [PSI+] prion generation in yeast: characterization of the ‘strain’ difference. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- 99.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghaemmaghami S, Ahn M, Lessard P, Giles K, Legname G, DeArmond SJ, et al. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:1000673. doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waxman EA, Emmer KL, Giasson BI. Residue Glu83 plays a major role in negatively regulating alpha-synuclein amyloid formation. Biochem Biophys Res Commun. 2010;391:1415–1420. doi: 10.1016/j.bbrc.2009.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hauber I, Hohenberg H, Holstermann B, Hunstein W, Hauber J. The main green tea polyphenol epigallocatechin-3-gallate counteracts semen-mediated enhancement of HIV infection. Proc Natl Acad Sci USA. 2009;106:9033–9038. doi: 10.1073/pnas.0811827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Munch J, Rucker E, Standker L, Adermann K, Goffinet C, Schindler M, et al. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 104.Mereles D, Buss SJ, Hardt SE, Hunstein W, Katus HA. Effects of the main green tea polyphenol epigallocatechin-3-gallate on cardiac involvement in patients with AL amyloidosis. Clin Res Cardiol. 2010;99:483–490. doi: 10.1007/s00392-010-0142-x. [DOI] [PubMed] [Google Scholar]
- 105.Kocisko DA, Engel AL, Harbuck K, Arnold KM, Olsen EA, Raymond LD, et al. Comparison of protease-resistant prion protein inhibitors in cell cultures infected with two strains of mouse and sheep scrapie. Neurosci Lett. 2005;388:106–111. doi: 10.1016/j.neulet.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 106.Radhakrishnan ML, Tidor B. Optimal drug cocktail design: methods for targeting molecular ensembles and insights from theoretical model systems. J Chem Inf Model. 2008;48:1055–1073. doi: 10.1021/ci700452r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Staszewski S, Morales-Ramirez J, Tashima KT, Rachlis A, Skiest D, Stanford J, et al. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. Study 006 Team. N Engl J Med. 1999;341:1865–1873. doi: 10.1056/NEJM199912163412501. [DOI] [PubMed] [Google Scholar]
- 108.Cowen LE, Singh SD, Kohler JR, Collins C, Zaas AK, Schell WA, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]