Abstract
Evidence is growing at an increasing-pace that amyloid fibers are not just the result of aberrant protein folding associated with neurodegenerative diseases, but are widespread in nature for beneficial reasons. Amyloid is an attractive building material because its robust design and simple repetitive structure make for very durable and metabolically cheap material. But this requires that the production of amyloid be put under firm control. This appears to involve the use of four to five chaperones that are expressed under the control of the same promoter as the amyloid proteins. Significant progress has been made in deciphering this process in E. coli's csg operon, also found in Salmonella. Recently, we have discovered a new and unrelated operon (fap) responsible for amyloid production in Pseudomonas, which also confers biofilm-forming properties to E. coli. Intriguingly, this operon shares a number of features with csg, namely two homologous proteins (one of which, FapC, has been shown to be directly involved in amyloid build-up) and a small number of auxiliary proteins. However, FapC seems to be less economically structured than its E. coli counterpart, with a smaller number of repeats and very large and variable linker regions. Furthermore, the putative chaperones are not homologous to their csg counterparts and have intriguing homologies to proteins with other functions. These findings suggest that controlled amyloid production has arisen on many independent occasions due to the usefulness of the product and offers the potential for intriguing insights into how nature disarms and reconstructs a potentially very dangerous weapon.
Key words: Pseudomonas, Fap, curli, chaperone, recycling, detection, amyloid operation
When proteins make a move, of which we don't approve
What is it that always intervenes?
Urea and SDS, they have their place I guess,
But first: send chaperones!
Let strand meet friend,
And till their pleats at hand
They've got to be protected,
All their folds respected
Till a state we like can be selected!
Fondly adapted from Tom Lehrer “Send the Marines.”
The Problem of Protein Aggregation
Protein aggregation and the accumulation of characteristic β-sheet rich amyloid structures is a widespread problem, giving rise to neurodegenerative diseases such as Alzheimer and Parkinson in humans. These amyloid structures are typically long and thin needle-like structures, formed by the stacking of parallel or antiparallel β-strands into an indefinitely long β-sheet orthogonal to the fibril axis and often stabilized by the lateral association of several different sheets1 (Fig. 1). Essentially all proteins can be induced to fibrillate under extreme conditions,2 typically due to the formation of partially unfolded states which can satisfy hydrogen bonding requirements by forming intermolecular β-sheet structures. Until recently it was thought that aggregation under physiological conditions was limited to a small number of individual proteins. In fact, aggregation appears to be a generic problem of aging. A recent proteomics study identified several hundred proteins that become less soluble as C. elegans ages. The sequences of these proteins were characteristically enriched for aliphatic residues favoring β-sheet structures.3 These aggregates are usually disposed of through the formation of aggresomes,4 but this regulation becomes less efficient with age. Age-triggered aggregation problems are thus a combination of an age-related decline in the cell's ability to regulate protein homeostasis (proteostasis) and a rising propensity to aggregate, e.g., due to changes in the cellular environment or the accumulation of chemical insults.
Figure 1.
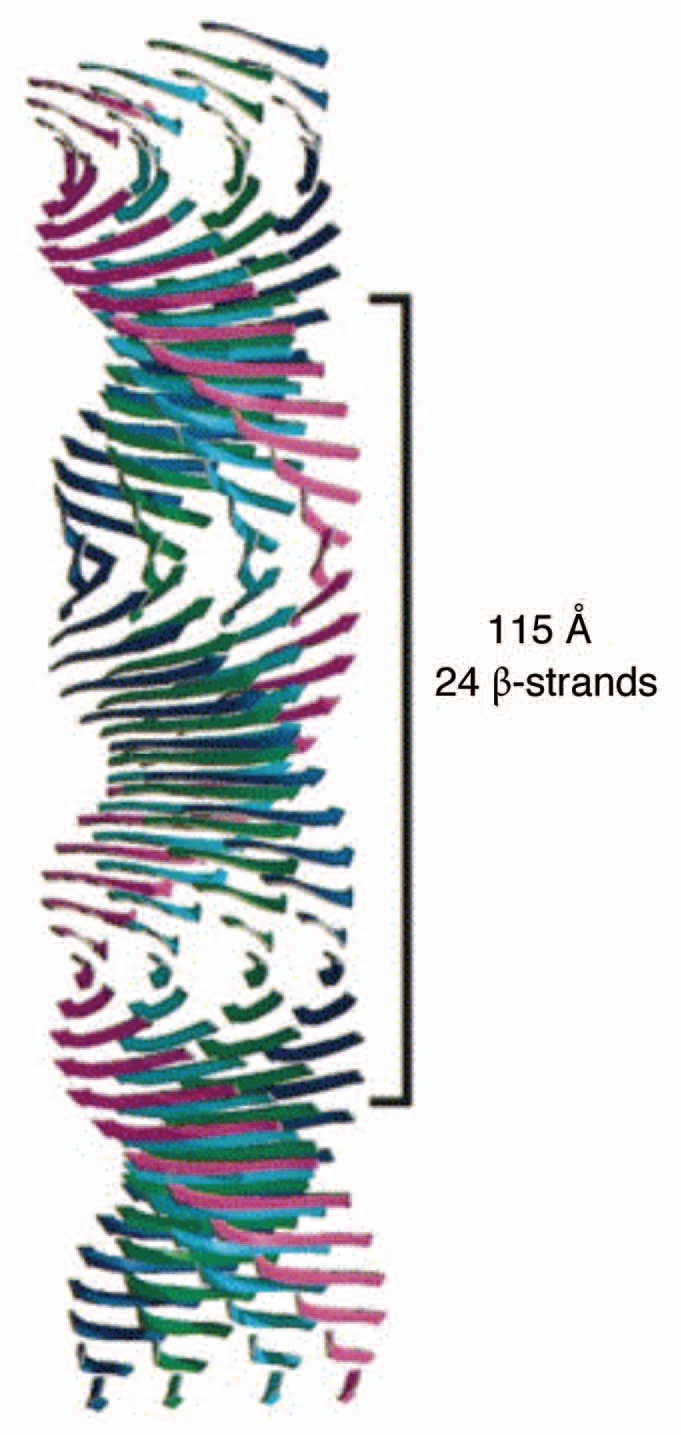
Model of the generic amyloid fibril structure, based on X-ray fiber diffraction data. Here four β-sheets (separated by a distance of around 10–12 Å) make up the protofilament structure, running parallel to the fibril axis with β-strands (separated by 4.8 Å) perpendicular to the fibril axis. Reprinted with permission from Sunde M, et al.1
Although these problems are most intensely studied in long-lived eukaryotic multicellular organisms, similar problems can occur in rapidly dividing unicellular organisms such as bacteria. If the bacterial cellular machinery is unable to fold a particular protein to its proper native conformation, the protein will typically precipitate within inclusion bodies. Interestingly, these bodies are not mere amorphous graveyards. The proteins in inclusion bodies show amyloid properties such as binding of the fibril-specific dye Thioflavin T, the ability to seed growth of new amyloid structures5–7 and sequence specificity, meaning that different co-expressed aggregating proteins do not localize to the same inclusion body.8 Heterologously expressed proteins tend to retain their original conformational properties in the bacterial environment. Thus prion proteins from yeast strains, which can “infect” properly folded versions of the same protein to engender a new fold, retain this property when expressed in E. coli.9,10 In fact there are bona fide bacterial models of amyloid proteinopathy wherein aggregation leads to aging, rather than vice versa. The bacterial plasmid-encoded protein RepA can assemble into amyloid fibers upon binding to DNA in vitro and an exacerbation of this propensity in the hyper-amyloidogenic mutant RepA-WH1 leads to the accumulation of cytoplasmic amyloid inclusions, which increases the bacterial generation time 5-fold11 provided they are retained in the bacterial cell. This is surely the fastest-reproducing and most economical animal model of amyloid deposition disease!
From Pathology to Functionality: Useful Amyloid and Their Widespread Prevalence
However, all is not doom and gloom with regards to amyloid. There is a growing number of examples of amyloid structures performing beneficial functions in nature as functional amyloid.5,12–17 In humans, the Pmel17 protein deposits as insoluble structures which help form melanosomes with covalently linked melanin.18 The Pmel17 protein is incredibly aggregation-prone, even aggregating within seconds in molar concentrations of the powerful chemical denaturant guanidinium chloride.18 However, this potentially devastating potential is kept in check by having Pmel17 attached to a membrane, from which it is released controllably through protease cleavage and subsequent dissociation. Amyloid is also found in silk moth oocytes and embryos,19 spider silks20 and fungi such as Aspergillus where they help hyphae penetrate the air-water interface interface.21
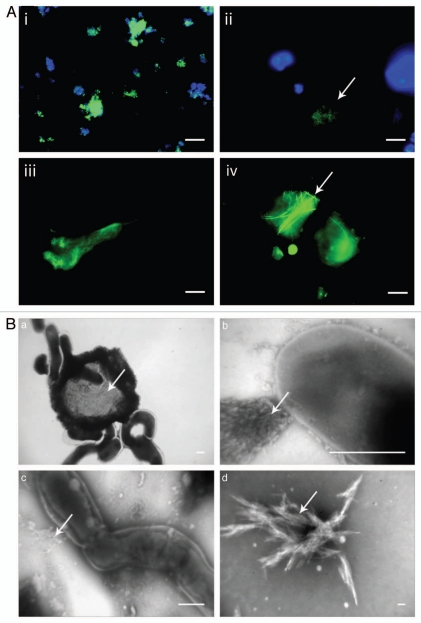
Perhaps the most widespread occurrence of amyloid is observed in the bacterial realm. We have combined fluorescence in situ hybridization with Thioflavin T staining and binding of conformationally specific antibodies targeting amyloid to investigate the prevalence of amyloid in bacterial biofilm communities. Our results show that 5–40% of all bacterial species in habitats from lakes, seawater, drinking water and waste water treatment systems harbor amyloid.22,23 The key to these observations is the diversity of location and function. We find amyloids in a wide range of bacterial species, including Proteobacteria (Alpha-, Beta-, Gamma- and Delta-proteobacteria), Bacteriodetes, Chloroflexi and Actinobacteria, as well as others not yet identified. Amyloid can localize on the cell surfaces and in the extracellular matrix between the cells, both as diffuse halos and as long distinct fibers (Fig. 2A). Some filamentous bacteria, e.g., from the phylum Chloroflexi, also express amyloids as a part of the sheath or close to the septum between the individual cells in the filaments. In the Gram-negative Mycolata family, it was necessary to strip off the lipid top layer by harsh treatment with alkaline alcohol to reveal the underlying amyloid structure, indicating that the amyloid, shown to consist of one to two protein components, was deeply integrated into the cellular envelope where it may perform essential structural functions24 (Fig. 2B).
Figure 2.
(A) Functional amyloid occurs in different species in different shapes and sizes. Antibody labeling using the amyloid-specific antibody WO1 is shown in green and nuclear counterstaining (DAPI) in blue. (i) C. glutamicum. FuBA is present around all cells. (ii) G. obscurus. FuBA occurs in large extracellular aggregates. The arrows indicate extracellular material with a high level of amyloid but low cell density. Bars 10 µm. (iii) M. avium. Velvet-like substances strongly bind WO2. (iv) T. spumae. Long (up to 50 µm) fibrils (arrows) are present. Bars 10 µm. (B) Saponification of G. amarae at increasing temperatures reveals gradual liberation of fibril-like substances: TEM micrographs with 1% phosphotungstic acid staining of (a) nonsaponified G. amarae, (b) bacteria saponified for 4 days at 37°C, (c) bacteria saponified for 4 days at 60°C and (d) bacteria saponified for 4 days at 80°C. Bars in (a–c) represent 0.5 µm; the bar in d represents 100 µm. The arrows indicate the positions of (a) a dense extracellular matrix and (b to d) fibrillar material. Reprinted with permission from Jordal PB, et al.24
Why Functional Amyloid?
Why is amyloid so widespread? This is not difficult to answer: amyloid is excellent building material. The β-strands are stacked with near-perfect design, and it has been estimated that there is only one misalignment per 30,000 strands in a typical amyloid structure.25 Unlike pathological amyloid, which often can be dissolved by denaturants, SDS or even pure water,26 functional amyloid generally resists boiling SDS (though exceptions do occur), thus providing a convenient selection criterion for its isolation24,27 and is only dissolved by high concentrations of formic acid.24,27,28 Furthermore, functional amyloid has typically been evolved to fibrillate, meaning that this process can occur over a broad range of conditions rather than a small “window of opportunity” engendered by the fortuitous accumulation of structural flexibility under specific structure-promoting conditions. The archetypal CsgA, which is the major component of the E. coli amyloid structure called curli,28 fibrillates to the same amyloid structure in vitro under a broad range of pH, temperature, concentration and ionic strength, according to fiber diffraction and Fourier Transform Infrared Spectroscopy (Dueholm MS and Otzen DE, unpublished observations). Note that unphysiological extremes of pH may still lead to different structures, as seen by recent solid state NMR studies of the fungal HET-s protein at pH 3 versus pH 7 29. This evolutionarily optimized robustness of design means that amyloid can be used as reliable building material in materials ranging from the cement of barnacle adhesive plaques30 to templates for gold nanowires31 and biomimetic silks in materials and medical applications.32 The amyloid state has also been shown to constitute a storage state for peptide hormones normally secreted in secretory granules of the endocrine system,33 though in this case extreme stability is not desired; rather, the amyloid here has to constitute a rapidly mobilizable source of material and therefore the amyloid can probably dissociate quite simply by a switch in pH.
Interestingly, functional amyloid may possess a unique structural fold rather than simply being the conventional cross-β structure observed in pathological amyloid by the combined efforts of fiber diffraction1,34 and X-ray crystallography.35,36 Although CsgA fibrils give rise to the classical diffraction distances at 5 and 10 Å characteristic of cross-β structures and representing inter-strand and inter-sheet distances, respectively, solid state NMR and electron microscopy fail to demonstrate the expected in-register parallel β-sheet architecture.37 It is speculated that CsgA may instead form β-helix structures.37 This is consistent with the reported β-solenoid structure of the fungal HET-s protein38 and our own observations that Pseudomonas amyloid gives rise to an additional diffraction distance around 6.3 Å, in addition to those corresponding to inter-strand and inter-sheet distances.27 It is worth noting that similar β-helix structures have been proposed for the passenger domains of bacterial autotransporters such as Ag43,39 which we have shown to generate Thioflavin-T positive structures in vivo.40 Future high-resolution structures of functional amyloid may resolve whether this unusual structure is formed in and contributes to the high stability of functional amyloid.
Going Green: The Challenge of Recycling Functional Amyloid
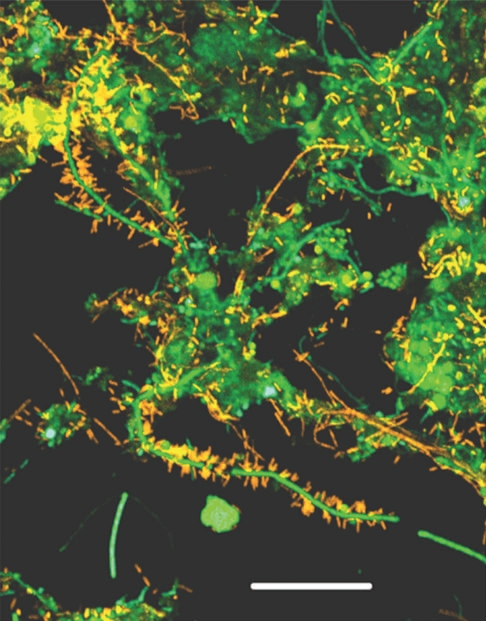
The heavy-duty durability of functional amyloid raises the question of its recycling. Clearly the large quantities of amyloid produced would lead to an insurmountable waste problem if they could not be degraded properly. Degradation obviously does occur. For example, the sulfide-oxidizing Thiothrix is encased in an amyloid-containing sheath, but the amyloid disappears in places where the absence of elemental sulfur grains indicates a loss of metabolic activity and cell death.23 There is as yet no clear understanding of how this recycling might occur, though there are tantalizing clues. The amyloid-producing filamentous bacterium Chloroflexi is often covered by multiple copies of epiphytic Candidatus Epiflobacter species (Saprospiraceae, Bacteroidetes) (Fig. 3).41 This epiphyte is specialized in protein degradation, excreting high levels of proteases and consuming amino acids but not acetate or glucose.42 It is tempting to speculate that these proteases feast on the accessible amyloid. Despite its robustness, CsgA is also susceptible to degradation by proteases (Yde A, Nielsen PH and Otzen DE, unpublished observations). It is likely that different protease-based recycling schemes will be uncovered as we learn more about the biology of these structures.
Figure 3.
Filamentous Chloroflexi (green) are often covered by epiphytic “Candidatus epiflobacter sp.” (Saprospiraceae, Bacteroidetes, colored yellow) which are specialized in protein degradation.41 Figure provided courtesy of Per Halkjær Nielsen.
Very recently it has been shown that something as simple as D-amino acids can disassemble biofilms in different bacteria (B. subtilis, S. aureus and P. aeruginosa)43 by releasing the TasA amyloids from the cell surface, with a mixture of four different amino acids (Tyr, Met, Trp and Leu) being particularly effective (IC50 ∼10 nM). This appears to occur when incorporation of D-amino acids into the cell wall somehow persuades the YqxM protein, which is involved in associating TasA with cells, to cast off the amyloid fibrils. However, in this case, amyloid is released rather than actually degraded, so this intriguing mechanism merely postpones the problem of recycling.
The Hows of Functional Amyloid: Policing the Masses
While the question of why functional amyloid is produced is easy and rather trivial to address, it is much more interesting to focus on the how aspect. This aspect has extremely important consequences for our understanding and appreciation of the mechanism of pathological amyloid formation. Given the functional amyloids' extremely strong drive towards aggregation, it is clear that nature has put a premium on a tight regulation of the process. Thus an amyloid that is suitably “constrained” within one organism due to a strong regulatory network can wreak havoc when unleashed on another organism where these safeguards are not in place. A good example is once again provided by CsgA, whose regulation is described below. When CsgA fibrils are injected into mice, they exacerbate murine amyloid protein A amyloidosis, presumably by either directly seeding the amyloid deposition or by boosting the physiological responses that lead to the production of this protein.44
We have a great deal of understanding of how biophysical factors modulate protein aggregation, e.g., the impact of sequence variation, protein stability and structure. Overall fibrillation requires a loss of native densely packed structure (facilitated e.g., by mutant destabilization, loss of co-factors or proteolytic cleavage) in order to form flexible dynamic regions, which can undergo the significant conformational changes required for amyloid formation. Aggregation of these flexible regions to amyloid structures is promoted by a certain degree of hydrophobicity, a modest amount of charge and a propensity to form β-sheet rather than α-helix.45–47 The challenge is to use this knowledge to predict how aggregation-prone sequences fare in a cellular environment, where other protein factors play a crucial role in controlling the spatial and temporal aspects of aggregation. There are already attempts to model these processes in silico48 but they are currently hampered by lack of precise quantitative data. Insight may be provided by a better understanding of the way in which functional amyloid formation is regulated in the cell. While we still lack many details, a picture of how these processes are regulated inside the cell, is gradually emerging, mainly based on the extensive work of curli formation in E. coli spearheaded by Normark, Hultgren and Chapman and others.
Curli are biologically important amyloid fibers which contribute to biofilm formation, host cell adhesion and invasion and immune system activation.49 In E. coli, the final curli fibrils consist of two subunits, CsgA and CsgB, found as major and minor components, respectively. Both contain 5 imperfect repeats, which are rich in Gln/Asn residues and show the tinctorial and structural properties of amyloid.28 In vitro CsgA polymerizes spontaneously with a classic nucleation-polymerization time profile, in which a lag phase is followed by a steep growth phase until all monomer has been incorporated into the fibrils. This happens over a wide variety of conditions (pH, protein concentration, ionic strength) (Dueholm MS and Otzen DE, unpublished results; Smith DR and Chapman M, personal communication), indicating that amyloid formation is robustly encoded.
This strong drive towards aggregation cannot go unchecked in vivo, however. Several additional chaperoning factors are involved, found within the Csg operon. In vivo curli formation occurs by a process of extracellular nucleation-precipitation (ENP),49,50 in which CsgA and CsgB are exported to the surface of the cell. Here CsgB somehow anchors on the membrane surface and provides the nucleation site for CsgA to attach and initiate growth of the curli fibril.51 CsgA and CsgB are exported to the surface of the cell via CsgG, an oligomeric lipoprotein in the outer membrane that appears to contain a central pore52 and from which the curli appear to emanate on the bacterial cell surface.53 CsgG is also needed to stabilize the proteins in the cytosol, since CsgA and CsgB do not accumulate in the cell in its absence,54 possibly because they are proteolytically degraded. This process of stabilization and secretion requires the N-terminal 22 residues of the mature CsgA, a sequence that is not part of the five imperfect repeats.52
The whole process of controlled secretion and amyloid formation is rigorously orchestrated, involving tight coordination between several different chaperones. The two chaperones CsgE and CsgG interact with CsgG at the outer membrane,52 and expression of CsgF is severely compromised in E. coli strains lacking CsgE and abolished by the absence of CsgG.55 Conversely, clustering of the CsgG pores around the curli strains require the presence of CsgE and CsgF as well as CsgA and CsgB.53 Strains lacking CsgF secrete CsgA in a soluble state that only assembles to curli to a small extent.28 This is because CsgF mediates CsgB cell-association and protease-resistance and thus provides the impetus for the anchoring and seeding of the curli fibrils.55 It remains unclear as yet whether CsgF directly binds CsgB to the membrane surface or promotes its folding, allowing it to assume a state that is both anchored to the membrane (via the C-terminal repeat) and folded. These two phenomena are not mutually exclusive. Like CsgA and CsgB, CsgF contains a high proportion of Gln and Asn which suggests that CsgF may have amyloid-forming or -stabilizing properties of its own.55
Thus, while aggregation is generic to proteins, functional amyloid has two distinguishing features: a simple composition (an abnormally high percentage of Asn, Gln, Gly and Ala) and a whole family of anxious auxiliary proteins working in an incredibly tightly choreographed pas-de-deux. Aggregation is still driven by the protein's intrinsic aggregation propensity but this only takes place in the right environment. The available evidence suggests that CsgA starts out in the unfolded state and folds directly to the amyloid state, thus avoiding potentially cytotoxic intermediates and providing a fast track to the right structure.56 Presumably the existence of five repeats within the protein provides for an intramolecular initation of the cross-β fold, reducing the need for formation of oligomeric but not yet fibrillar species. Such oligomeric species, formed by proteins involved in amyloid diseases such as Aβ and α-synuclein and many others, are able to permeabilize membranes and synthetic phospholipid and this has been suggested to provide the basis for aggregate toxicity.57–59
Gratifyingly, it is becoming clear that changes thay increase amyloidogenicity in vitro can have the same effect in vivo. We introduced the term gatekeeper residues to describe residues whose replacement by mutagenesis enhanced in vitro misfolding60 and aggregation,61 and these types of residues have been shown to be overrepresented in potential hot-spots of aggregation.62 Gatekeeping residues (glycines and asparagines) have been identified in CsgA; mutants lacking these gatekeepers polymerize in vitro significantly faster than wild-type CsgA and polymerized in vivo in the absence of the nucleation machinery, resulting in mislocalized fibers and cytotoxicity.63 Conversely, compounds that prevent CsgA fibrillation in vitro also inhibit CsgA biogenesis and formation of biofilms of uropathogenic E. coli.64
From the Specific to the General: Diversity of Mechanisms Regulating Functional Amyloid Formation
The csg operon provides a simple but elegant solution to the problem of spatio-temporal control of amyloid formation. Yet, given the enormous prevalence of functional amyloid in many different walks of life, some diversity in the details of regulation is to be expected. A way forward is to identify and compare the proteins involved in functional amyloid production from different diverse species. This is a bit of a fishing expedition where persistence and flexibility are both required—not all functional amyloid bites the investigative hook. In this context, the extreme stability of these functional amyloid structures is both a blessing and a curse, since they provide for a method and a criterion for purification but are also typically recalcitrant towards mass spectrometric identification methods. Our approach has been to take samples from ecological niches where amyloid might be expected to play a role, such as biofilm growing in non-chlorinated water reservoirs.22 By growing these samples on agar plates containing Congo Red, initial hints about amyloid production can be obtained, since amyloid-producing colonies will be dark-red or even brown (Fig. 4). Subsequent amyloid production can be verified using conformationally specific antibodies that recognize the amyloid fold.65 This presupposes, however, that the amyloid is accessible to the large antibody molecules, and this cannot be expected to be the case if the amyloid is firmly embedded in the cell membrane, as appears to be the case for various Gram-positive Mycolata species (cfr. Fig. 2B and discussion above).24 Subsequently 16S rRNA sequencing identifies the bacterial species. Many amyloid-producing species can be identified at this stage, but so far it has only been possible to purify amyloid to single- (or double-) band purity for a very small number of cases. The process is a variation over the theme of cell disruption and degradation of the cell wall to release the extracellular amyloid, followed by preparation SDS-PAGE in which only the functional amyloid resists solubilization in boiling SDS-PAGE.27 Finally, the lyophilized amyloid can be solubilized in high concentrations (typically ≥80%) formic acid, which allows us to visualize it by conventional SDS-PAGE.
Figure 4.
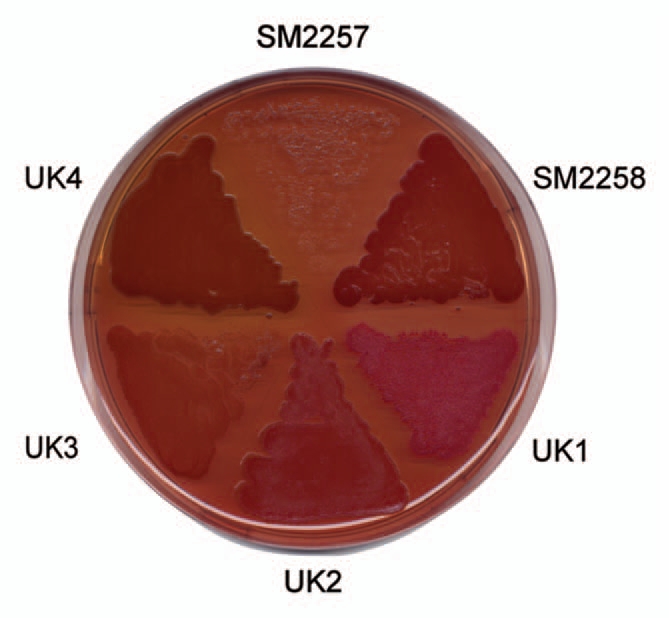
Colony morphotypes of bacterial strains when grown on Congo red agar plates. Bacteria were grown for 48 h at 26°C. It was subsequently possible to purify and identify amyloid from the strain designated UK4.
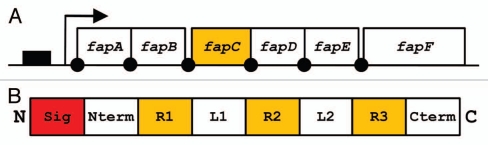
We have so far had most success with a Pseudomonas strain designated UK4, whose 16S rRNA showed 99.8% similarity with that of P. gessardii.27 It was possible to purify this protein to two bands, which in size corresponded to a 25 kDa monomer and a 50 kDa dimer. Such dimers are not unexpected in view of amyloid proteins' tendency to aggregate, and are seen for CsgA from both E. coli and Salmonella. Although the protein was clearly amyloid according to electron microscopy, secondary structure analyses and Thioflavin T fluorescence, identifying the protein proved remarkably difficult. A combination of chemically assisted fragmentation mass spectrometry and N-terminal sequencing yielded enough information to clone part of the associated gene sequence, but full identification required genomic sequencing. However, an additional advantage of this approach was that it allowed us to identify not only the amyloid in question, which we designate FapC, but also the other five proteins (FapA–B and FapD–F) under the control of the same promoter (Fig. 5A). Gratifyingly, when transplanted into E. coli, this operon was able to induce the production of a very large amount of biofilm, indicating that this is a functional entity which can be recognized across the bacterial spectrum.27 Several important insights were obtained by a closer analysis of the operon sequences.
Figure 5.
(A) Organization of the fap operon. Promoter region indicated as solid box, protein coding sequences indicated by open boxes. Arrow indicates transcription start site and solid dots are potential ribosomal binding sites. (B) Schematic representation of FapC. The N-terminal signal sequence (Sig) is cleaved off during translocation to the outer membrane, an N-terminal domain (Nterm), three repeats (R1–3) separated by two linker regions (L1–2) and a C-terminal tail (Cterm).
The Amyloid Forming Protein FapC Has Multiple Repeats with Variable Linkers
Like CsgA, FapC contains a signal sequence, an N-terminal sequence and several imperfect repeats (Fig. 5B). These features are also found for a number of homologs found in other Pseudomonas species.27 However, there are some notable differences. The N-terminal sequence is 37 residues long and not homologous to CsgA's 22-residue sequence. Given that CsgA's N-terminal sequence is likely involved in cytosolic stabilization and export through CsgG, this challenge will presumably be tackled in a different way for FapC. FapC only has three repeats, unlike CsgA's five, though they are longer (30 residues versus CsgA's 22–23 residues). These repeats are rich in Gln and Asn (a total of 38%, which compares well with the 19% in CsgA), and also contain large amounts of Gly and Ala (33%, compared to 27% in CsgA). The complete absence of aromatic residues in the repeats emphasizes that aromatic residues play no role in these functional amyloids, as seen also for CsgA 66 but in contrast to other modes of stacking β-strands in amyloid67,68 and prions.69 Rather the composition is indicative of a very economical use of simple amino acids, which presumably involves minimal metabolic cost.74 An additional intriguing distinction is the presence of large linkers between FapC's variable repeats. In contrast, CsgA has no linkers between its five repeats, only a few residues to execute a tight β-turn. While the linker between the first two repeats is relatively tightly conserved in length between the different species (35–38 amino acids), the linker between the second and third repeat varies between 36 and 275 residues and shows a distinct predilection for small and generally hydrophilic amino acids (Ser, Gly, Ala, Val, Thr and to some extent Asn and Gln). It remains entirely speculative at present what the possible role of this linker could be. Some possibilities include: (1) modulation of the bacterial cell surface, rendering it more hydrophilic, (2) structured domains with actual function (unlikely in view of the paucity of aliphatic and aromatic residues usually found in the core of globular proteins), (3) modulation of the aggregation propensity of the FapC molecule. We are currently analyzing the behavior of different FapC analogs to address these questions.
The presence of these linkers highlights an important aspect with regards to the extent of sequence incorporation into the final amyloid structure. Pathogenic aggregates are formed by proteins that can assume a wide variety of structures in their native state. Thus they have to undergo a very fundamental conformational change in order to access the amyloid state. This need not involve the entire polypeptide chain. As little as ten contiguous Gln residues can provide an amyloid zipper that can be used to construct an amyloid backbone on which the rest of the protein can hang.70 Even for amyloids whose aggregation plays a biological role such as the yeast prion protein Sup35, only a fraction of the protein is actually involved in amyloid formation.71 Thus only a subsection of the protein need assume a flexible state that can be packed into the amyloid structure.72 Obviously the most efficient use of the protein as amyloid building material requires that as much as possible of the protein be incorporated. In practice this means that the protein should start out in the unfolded state so that all the protein is structurally accessible. CsgA and its accessory amyloid component CsgB both assume a natively unfolded state in vitro under physiological conditions prior to amyloid formation,56 and the entire sequence (apart from the N-terminal part required for secretion) is incorporated into the amyloid structure. This is very unlikely to be the case for FapC. Linkers may in different ways affect the aggregation process by collapsing part of the structure in the non-aggregated state or instead keeping it extended, thus, for example, obstructing formation of unwanted β-strand registers.
The Role of the Other Operon Members
FapB: Like CsgA, FapC is undoubtedly heavily dependent on the other Fap products for its correct aggregation. An immediate parallel to the csg operon is suggested by the fact that FapC also has an internal homolog, namely FapB. However, the three repeats in this protein, while very rich in Asn, Gln, Gly and Ala (which together constitute 63% of the entire sequence), are only 17 residues long, and it is unclear whether this is sufficient to trigger the seeded nucleation that CsgB can confer on CsgA. Complementation studies using appropriate knock-out strains51 would be a simple way to address this, and this will be aided by the successful heterologous expression of the fap operon in E. coli. It will also be intriguing to see whether CsgB can complement FapC and FapB ditto CsgA, though the differences in repeat lengths suggest to us that this will not be the case.
Of the four remaining fap gene products, FapD turns out to have a very high degree of homology with a putative C39 peptidase. This peptidase facilitates secretion of bacteriocin and is involved in quorum sensing.73 Remarkably, its substrates typically contain a double glycine motif, which is also found in FapF. This opens up for a scenario in which the biofilm induced by FapC aggregation (and exported by the action of FapA and FapE?) can be regulated by the action of FapD and FapF. Note also that the six fap genes all lie downstream of the fap promoter, in contrast to the csg promoter, which lies between CsgA and CsgB on the one side and CsgCDEF on the other. This points at possible differences in the regulation of their translation.
Conclusion
Functional amyloid provides a fascinating new angle on the conundrum of why proteins have such a pronounced tendency to aggregate. Rather than being a malicious ploy to frustrate the protein scientist, this can actually serve a purpose or rather many different purposes. As a simple comparison between amyloid-producing operons from E. coli and Pseudomonas indicates, amyloid protein can be produced in many different ways and using different motifs. In teasing out the details of how nature manages to control and harness this potentially chaotic process to produce the grand architecture of cellular amyloid extensions by a suite of wonderfully adapted chaperone proteins, one is invariably reminded of the homage to a Greco-Egyptian queen that could just as well be penned to functional amyloid: Age cannot wither them nor custom stale their infinite variety.…
Acknowledgements
I am very grateful to Per Halkjær Nielsen for a long-standing and fruitful collaboration on bacterial amyloid and for providing Figure 3. This work would not have been possible without the dedicated efforts of in particular Morten S. Dueholm, Poul Larsen and Peter Lüttge Jensen. In addition, I acknowledge sterling support from Kåre Lehmann Nielsen, Mads Sønderkær, Jeppe Lund Nielsen, Jan J. Enghild, Steen Vang Petersen and Gunna Christiansen and insightful bioinformatic analysis by Janus Schatz-Jakobsen and Emil Laust Kristoffersen. I also appreciate inspiring discussions with Matt Chapman. Our work has been generously supported by Villum Kann Rasmussen (BioNET) and the Lundbeck Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/13676
References
- 1.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:12–17. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 5.Sabaté R, De Groot NS, Ventura S. Protein folding and aggregation in bacteria. Cell Mol Life Sci. 2010;67:2695–2715. doi: 10.1007/s00018-010-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrió M, González-Montalbán N, Vera A, Villaverde A, Ventura S. Amyloid-like properties of bacterial inclusion bodies. J Mol Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6:195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart RA, Rinas U, Bailey JE. Protein composition of Vitreoscilla hemoglobin inclusion bodies produced in Escherichia coli. J Biol Chem. 1990;265:12728–12733. [PubMed] [Google Scholar]
- 9.Sabaté R, Espargero A, Saupe SJ, Ventura S. Characterization of the amyloid bacterial inclusion bodies of the HET-s fungal prion. Microb Cell Fact. 2009;8:56. doi: 10.1186/1475-2859-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasmer C, Benkemoun L, Sabaté R, Steinmetz MO, Coulary-Salin B, Wang L, et al. Solid state NMR spectroscopy reveals that E. coli inclusion bodies of HET-s(218–289) are amyloids. Angew Chem Int Ed. 2009;48:4858–4860. doi: 10.1002/anie.200806100. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Tresguerres ME, De La Espina SM, Gasset-Rosa F, Giraldo R. A DNA-promoted amyloid proteinopathy in Escherichia coli. Mol Microbiol. 2010;77:1456–1469. doi: 10.1111/j.1365-2958.2010.07299.x. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen PH, Dueholm MS, Thomsen TR, Nielsen JL, Otzen DE. Functional bacterial amyloids in biofilms. In: Flemming HC, Szwezyk U, Wingender J, editors. Annual Biofilm Highlights. 2010. [Google Scholar]
- 13.Otzen DE, Nielsen PH. We find them here, we find them there: Functional bacterial amyloid. Cell Mol Life Sci. 2008;65:910–927. doi: 10.1007/s00018-007-7404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maury CP. The emerging concept of functional amyloid. J Internal Med. 2009;265:329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith AM, Scheibel T. Functional amyloids used by organisms: a lesson in controlling assembly. Macromol Chem Phys. 2010;211:127–135. [Google Scholar]
- 16.Badtke MP, Hammer ND, Chapman MR. Functional amyloids signal their arrival. Sci Signal. 2009;2:43. doi: 10.1126/scisignal.280pe43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. TIBS. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2005;4:1–8. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iconomidou VA, Chryssikos GD, Gionis V, Galanis AS, Cordopatis P, Hoenger A, et al. Amyloid fibril formation propensity is inherent into the hexapeptide tandemly repeating sequence of the central domain of silkmoth chorion proteins of the A-family. J Struct Biol. 2006;156:480–488. doi: 10.1016/j.jsb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Kenney JM, Knight D, Wise MJ, Vollrath F. Amyloidogenic nature of spider silk. Eur J Biochem. 2002;269:4159–4163. doi: 10.1046/j.1432-1033.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 21.Wösten HAB, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 22.Larsen P, Dueholm M, Christiansen G, Nielsen JL, Otzen DE, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Env Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 23.Larsen P, Nielsen JL, Otzen DE, Nielsen PH. Amyloid-like adhesins in floc-forming and filamentous bacteria in activated sludge. Appl Env Microbiol. 2008;74:1517–1526. doi: 10.1128/AEM.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordal PB, Dueholm M, Larsen P, Pedersen SV, Enghild JJ, Christiansen G, et al. Widespread abundance of functional bacterial amyloid in mycolata and other gram-positive bacteria. Appl Env Microbiol. 2009;75:4101–4110. doi: 10.1128/AEM.02107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles TPJ, Smith JF, Craig A, Dobson CM, Welland ME. Spatial persistence of angular correlations in amyloid fibrils. Phys Rev Lett. 2006;96:238301. doi: 10.1103/PhysRevLett.96.238301. [DOI] [PubMed] [Google Scholar]
- 26.Tang L, Li HT, Du HN, Zhang F, Hu XF, Hu HY. Study of the disassembly-assembly process of alpha-synuclein fibrils by in situ atomic force microscopy. Micron. 2006;37:675–679. doi: 10.1016/j.micron.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Dueholm MS, Petersen SV, Sønderkær M, Larsen P, Christiansen G, Hein KL, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07269.x. In press. [DOI] [PubMed] [Google Scholar]
- 28.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Eschericia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasmer C, Soragni A, Sabaté R, Lange A, Riek R, Meier BH. Infectious and noninfectious amyloids of the HET-s(218–289) prion have different NMR spectra. Angew Chem Int Ed. 2008;47:5839–5841. doi: 10.1002/anie.200704896. [DOI] [PubMed] [Google Scholar]
- 30.Barlow DE, Dickinson GH, Orihuela B, Kulp JL, Rittschof D, Wahl KJ. Characterization of the adhesive plaque of the barnacle Balanus amphitrite: amyloid-like nanofirils are a major component. Langmuir. 2010;26:6549–6556. doi: 10.1021/la9041309. [DOI] [PubMed] [Google Scholar]
- 31.Scheibel T, Parthasarathy R, Sawicki G, Lin XM, Jaeger H, Lindquist SL. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci USA. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heim M, Keerl D, Scheibel T. Spider silk: from soluble protein to extraordinary fiber. Angew Chem Int Ed. 2009;48:3584–3596. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- 33.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, et al. The common architecture of cross-β amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 37.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, et al. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2009;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 39.Klemm P, Schembri MA. Bacterial adhesins: function and structure. Int J Med Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 40.Knudsen SK, Westergaard UB, Franzmann M, Stensballe A, Otzen DE. Effect of glycosylation on biophysical and flocculative properties of the extracellular domain of Ag43. Biochem J. 2008;412:563–577. doi: 10.1042/BJ20071497. [DOI] [PubMed] [Google Scholar]
- 41.Xia Y, Kong Y, Thomsen TR, Nielsen PH. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing saprospiraceae (“Candidatus epiflobacter” spp.) in activated sludge. Appl Env Microbiol. 2008;74:2229–2238. doi: 10.1128/AEM.02502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y, Kong Y, Nielsen PH. In situ detection of protein-hydrolysing microorganisms in activated sludge. FEMS Microbiol Ecol. 2007;60:156–165. doi: 10.1111/j.1574-6941.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- 43.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundmark K, Westermark G, Olsén A, Westermark P. Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: cross-seeding as a disease mechanism. Proc Natl Acad Sci USA. 2005;102:6098–6102. doi: 10.1073/pnas.0501814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 46.Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and protein. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 48.Sneppen K, Lizana L, Jensen MH, Pigolotti S, Otzen DE. Modeling proteasome dynamics in Parkinson's disease. Phys Biol. 2009;6:36005. doi: 10.1088/1478-3975/6/3/036005. [DOI] [PubMed] [Google Scholar]
- 49.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bact. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci USA. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epstein EA, Reizian MA, Chapman MR. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol. 2009;191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 55.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci USA. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, et al. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volles MJ, Lee SJ, Rochet JC, Shtilerman MD, Ding TT, Kessler JC, et al. Vesicle permeabilization by protofibrillar α-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry. 2001;40:7812–7819. doi: 10.1021/bi0102398. [DOI] [PubMed] [Google Scholar]
- 59.Lashuel HA, Hartley D, Petre BM, Weals T, Lansbury PT. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 60.Otzen DE, Oliveberg M. Salt-induced detour through compact regions of the protein folding landscape. Proc Natl Acad Sci USA. 1999;96:11746–11751. doi: 10.1073/pnas.96.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otzen DE, Kristensen P, Oliveberg M. Designed protein tetramer zipped together with an Alzheimer sequence: a structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rousseau F, Serrano L, Schymkowitz J. How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci USA. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nature Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc Natl Acad Sci USA. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 68.Azriel R, Gazit E. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. An experimental support for the key role of the phenylalanine residue in amyloid formation. J Biol Chem. 2001;276:34156–34161. doi: 10.1074/jbc.M102883200. [DOI] [PubMed] [Google Scholar]
- 69.Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. The formation of Escherichia coli curli amyloid fibrils is mediated by prion-like peptide repeats. J Mol Biol. 2005;352:245–252. doi: 10.1016/j.jmb.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 70.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 71.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedersen JS, Christiansen G, Otzen DE. Modulation of S6 fibrillation by unfolding rates and gatekeeper residues. J Mol Biol. 2004;341:575–588. doi: 10.1016/j.jmb.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 73.Dirix G, Monsieurs P, Dombrecht B, Daniels R, Marchal K, Vanderleyden J, et al. Peptide signal molecules and bacteriocins in gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides. 2004;25:1425–1440. doi: 10.1016/j.peptides.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 74.Smith DR, Chapman MR. Economical Evolution: Microbes reduce the synthetic cost of extracellular proteins. mBio. 2010;1:e00131–e00110. doi: 10.1128/mBio.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]