Abstract
Cell surface receptors function to transduce signals across the cell membrane leading to a variety of biologic responses. Structurally, these integral proteins can be classified into two main families, depending on whether extracellular ligand-binding and intracellular signaling domains are located on the same protein chain (single-chain receptors, SRs) or on separate subunits (multichain receptors, MRs). Since most MRs are immune receptors, they are all commonly referred to as multi-chain immune recognition receptors (MIRRs). Recent studies reveal that, in contrast to well-structured signaling domains of SRs, those of MIRRs represent intrinsically disordered regions, the regions that lack a well-defined three-dimensional structure under physiological conditions. Why did nature separate recognition and signaling functions of MIRRs? Why for MIRRs did nature select to provide highly specific signaling through the chaos of protein disorder? What mechanisms could control this chaos in the process of transmembrane signal transduction to provide the specificity and diversity of the immune response? Here, I summarize recent findings that may not only shed light on these and other questions but also add significantly to our understanding of receptor signaling, a fundamental process that plays a critical role in health and disease.
Key words: intrinsically disordered proteins, cell receptors, receptor signaling, immune signaling, protein order, protein disorder, single-chain receptors, multichain receptors, multichain immune recognition receptors, MIRR, SCHOOL concept
Introduction
Cells communicate with each other and with their environment through an array of diverse signal-generating surface receptors that respond specifically to individual stimuli. Molecular understanding how these various receptors signal the cell to respond in different ways is of both fundamental and clinical importance. Recent findings show that while ligand binding outside the cell is mediated through well-structured protein domains, intracellular signaling domains of many receptors represent intrinsically disordered regions (IDRs), the regions that lack a well-defined three-dimensional structure under physiological conditions.1–5 Intriguingly, protein intrinsic disorder is a characteristic feature of the cytoplasmic signaling domains of those receptors, in which recognition and signaling are mediated by separate protein chains.1,4,5 Our studies of these signaling-related IDRs reveal several unusual and previously unreported biophysical phenomena1,4,6–9 that not only facilitate a rethinking process of the fundamental paradigms in protein biophysics but also open new perspectives on the molecular mechanisms of receptor signaling with multiple applications in biology and medicine.
Together, these observations raise several important questions that had been barely addressed. Why for a variety of receptors did nature separate recognition and signaling functions, thereby complicating the process of transmembrane signaling? Why for receptors with separated recognition and signaling functions did nature select to provide highly specific signaling through the chaos of protein disorder? What mechanisms could control this chaos to provide the specificity and diversity of the immune response? Here, I suggest how our new multidisciplinary knowledge about the structure, properties and function of cell surface receptors, developed over the past 10 or so years, may shed light on these and other questions.
Single- and Multichain Cell Surface Receptors
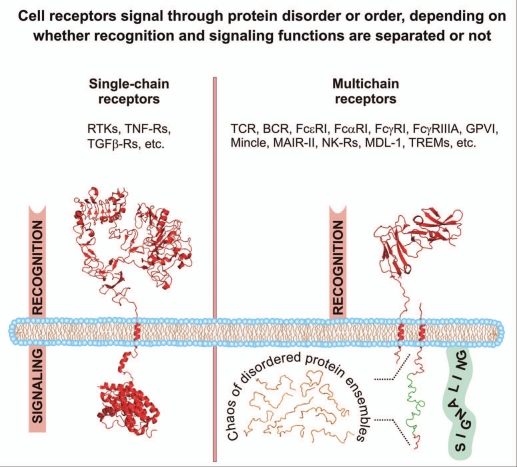
Functionally diverse and unrelated cell surface receptors can be structurally classified into two main families, depending on whether extracellular ligand-binding and intracellular signaling (effector) domains are located on the same (single-chain receptors, SRs) or separate (multichain receptors, MRs) protein chains (Fig. 1).10,11 Most multichain activating receptors are immune receptors. For this reason, they are all commonly referred to as multichain immune recognition receptors (MIRRs).10–12 MIRRs signal through receptor-associated subunits that contain in their cytoplasmic domains one or more copies of the immunoreceptor tyrosine-based activation motif (ITAM) regions13 or the YxxM motif, found in the DNAX adapter protein of 10 kD (DAP-10) cytoplasmic domain.14 Upon receptor triggering, tyrosine residues of the ITAM/YxxM regions are phosphorylated in an early and obligatory event in the signaling cascade.
Figure 1.
Intrinsic order and disorder of the cytoplasmic signaling (effector) domains of single-and multichain cell receptors. Images were created using PyMol (www.pymol.org) from Protein Data Bank entries 1NQL and 3GOP for the EGFR extracellular domain, juxtamembrane and kinase domains, respectively (shown as an exemplary structure of a single-chain receptor) and entry 1UCT for the FcαRI extracellular domain (shown as an exemplary structure of a multichain receptor recognition subunit). For illustrative purposes, the cytoplasmic domain of a multichain receptor-associated signaling subunit is shown as a monomer and using arbitrary idealized structural elements to represent the ensemble of unfolded conformations of an IDR. The immunoreceptor tyrosine-based activation motif (ITAM) is depicted in green. Abbreviations: BCR, B cell receptor; DAP-12, DNAX adapter protein of 12 kD; FcαRI, type I Fc receptor for IgA; FcγRI, type I Fc receptor for IgG; FcεRI, type I Fc receptor for IgE; FcγRIIIA, type IIIA Fc receptor for IgG; GPVI, glycoprotein VI; IDR, intrinsically disordered region; MAIR-II, myeloid-associated Ig-like receptor; MDL-1, myeloid DAP12-associating lectin 1; Mincle, a C-type lectin receptor expressed in activated phagocytes; NK-R, natural killer cell receptor; RTK, receptor tyrosine kinase; TCR, T cell receptor; TGFβ-R, transforming growth factor-beta receptor; TNF-R, tumor necrosis factor receptor; TREM, triggering receptor expressed on myeloid cells.
Why did Nature Separate Recognition and Signaling Functions of MIRRs?
Some of the resulting advantages that come with this separation are the following:
In contrast to SRs that mostly provide only a single ON/OFF signal, MIRRs often need to induce different downstream sequences and as a result, different functional outcomes to generate the diversity of the immune response.10–12 Separation of recognition and signaling functions of MIRRs makes possible the use of several different signaling subunits in one receptor (e.g., T cell receptor, TCR, that contains 4 different ITAM-containing subunits). This allows cells to diversify functional responses triggered by such MIRRs as TCR, B cell receptor (BCR) and type I Fc receptor for IgE (FcεRI).
Upon ligand binding, signaling subunits of several MIRRs are known to physically dissociate from the remaining receptor complexes, which then undergo internalization.11,15–22 Thus, separation of recognition and signaling functions allows intracellular processing of the engaged ligand-binding subunits while active signaling subunits still remain on the cell surface.
In resting cells, dissociation of ligand-binding and signaling MIRR subunits can underlie important biological processes, such as the BCR sensitization and T cell clonal anergy.10–11
The modular assembly of MIRRs (Fig. 1) permits the use of one signaling subunit for different receptors (e.g., various MIRRs signal through the ITAM-containing Fc receptor γ signaling chain, FcRγ).2,11
Intrareceptor Transmembrane Interactions: Achilles' Heel and a Promising Target for Therapy
The association of the MIRR subunits is driven mostly by the non-covalent trans-membrane interactions between recognition and signaling components (Fig. 1). These protein-protein interactions represent biochemical processes that can be influenced and controlled.23–25 Thus, advantages of separated recognition and signaling functions come at the cost of not only complicating the process of transmembrane signal transduction but also taking a risk of adding a potential point of attack.
Recent studies26–30 suggest that in order to establish a successful infection, viruses have evolved the strategy to attack this Achilles' heel of MIRRs and disrupt functional coupling between two aspects of receptor machinery: recognition and signaling. This allows viruses to cheat the immune system and enter the host cell without triggering the self-defense response.
On the other hand, intrareceptor transmembrane interactions represent a promising target for therapy of autoimmune and other MIRR-mediated diseases where there is a need to modulate receptor signaling.26,31–33 Viruses represent millions of years of evolution and the efficiency and optimization that come along with it. Thus, we can take advantage of this billion-year development process and transfer it to therapeutic strategies.26,32,33 Experimental data of both in vitro and in vivo studies31,34–38 strongly support the novel therapeutic approach.
Cell Receptors Signal through Protein Disorder or Order, Depending on Whether Recognition and Signaling Functions are Separated or Not
Cell surface receptors recognize their cognate ligands outside the cell and translate this information into an intracellular activation signal. For both receptor families, SRs and MIRRs, recognition of ligands is mediated by well-structured ligand-binding domains (Fig. 1) providing the molecular basis of recognition specificity and sensitivity. Surprisingly, a striking difference between the two families is observed in structural patterns of intracellular signal-generating regions. Recently, using a variety of biophysical techniques1,4,9 and prediction algorithms,3,5 we found that while signaling by SRs is mediated through intracellular well-defined protein structures (Fig. 1), the cytoplasmic domains of MIRR signaling subunits, including those of ζ, CD3ε, CD3δ and CD3γ chains of TCR, Igα and Igβ chains of BCR, FcRγ, DAP-10 and DNAX adapter protein of 12 kD (DAP-12), represent a novel class of intrinsically disordered proteins (IDPs).
Why for receptors with separated recognition and signaling functions, the complexes that are critical in the immune response, did nature select the chaos of protein disorder to provide intracellular signaling? MIRRs represent functionally diverse and unrelated surface receptors that are expressed on a variety of cell types. Upon binding to ligand, tyrosine residues of the ITAM/YxxM regions of MIRR signaling subunits are phosphorylated initiating a complex cascade of signaling events that results in multiple cell responses. Despite apparent similarities, the common activation motif, ITAM, provides diverse activation signals in the context of different signaling subunits of one receptor (e.g., TCR signals through ζ, CD3ε, CD3δ and CD3γ) or one subunit of different receptors (e.g., glycoprotein VI and several Fc receptors signal through FcRγ; similarly, 4 different triggering receptors expressed on myeloid cells, TREMs, generate the activation signal through DAP-12). Upon ITAM phosphorylation, the cytoplasmic domains of MIRR signaling subunits interact with multiple different binding partners such as adapters and kinases to induce different downstream cascades and, as a result, different functional outcomes.39 In this context, protein intrinsic disorder both enables and enhances the features most important for MIRR-mediated signaling: high-specificity low-affinity interactions, the multiple binding of one protein to many partners and the multiple binding of many proteins to one partner.40–41
Unusual Biophysics of Multichain Receptor-Related Protein Disorder
Our recent studies1,4,7–9 reveal several intriguing biophysical phenomena6 that are characteristic of the MIRR-related IDPs and include those that are unprecedented. Among these are the following:
Specific homodimerization of IDP molecules that is distinct from non-specific aggregation behavior seen in many systems.
Fast and slow homodimerization equilibrium, depending on the protein.
No disorder-to-order transition upon interaction with a well-folded partner protein or another IDP molecule.
Lack of significant chemical shift and peak intensity changes in the nuclear magnetic resonance (NMR) heteronuclear single quantum coherence (HSQC) spectra upon protein-protein complex formation. Note: these spectra provide a readily accessible fingerprint of proteins, where the backbone amide group of each non-proline amino acid residue contributes a single cross-peak.
Two modes of binding to model membranes: with and without folding, depending on the lipid bilayer stability.
In summary, the cytoplasmic domains of MIRR signaling subunits: (a) all represent signal-generating regions of more than 25 functionally unrelated and diverse receptors that belong to one structural family, (b) all contain similar activation motifs, ITAMs, (c) all are intrinsically disordered and (d) all feature the ability to homooligomerize, which is unusual for IDPs, and what is especially important, they all homooligomerize without disorder-to-order structural transition. This strongly suggests that MIRRs all function through similar molecular mechanisms and that their unusual biophysical properties represent the key point in the solution to the puzzle of MIRR signaling.
Cytoplasmic Homooligomerization as a Mechanism to Control Signaling through Both Protein Order and Disorder: The SCHOOL Concept
The fact that all cytoplasmic domains of MIRR signaling subunits are intrinsically disordered raises the following important question:
What mechanisms could control the chaos of intracellular MIRR disorder to provide the specificity and diversity of the immune response? The answer to this question comes from a recently proposed novel model of transmembrane signaling, the Signaling Chain HOmoOLigomerization (SCHOOL) model.2,3,10–11,42 First introduced for MIRRs,10,43 this model suggests that formation of competent signaling homooligomers in cytoplasmic milieu is the necessary and sufficient event to trigger MIRRs and induce cell activation. This dictates several important restraints on multivalent ligand binding-induced MIRR signaling: (1) sufficient interreceptor proximity in receptor dimers/oligomers, (2) correct (permissive for signaling) relative orientation of the receptors in receptor dimers/oligomers, (3) long enough duration of the receptor-ligand interaction that generally correlates with the strength (affinity/avidity) of the ligand and (4) sufficient lifetime of an individual receptor in receptor dimers/oligomers. Thus, the necessity to form specific signaling-competent homooligomers may represent a means to control the chaos of protein disorder of the ITAM-containing cytoplasmic domains of MIRR signaling subunits.
In the context of the SCHOOL concept,10,11 protein disorder of the ITAM-containing signaling regions being under control of ligand-promoted cytoplasmic homooligomerization provides a molecular basis to explain high specificity, selectivity and sensitivity of immune cells in recognition and discrimination of different antigens/ligands and how this recognition/discrimination results in different functional outcomes.
ITAMs of multichain receptors: multipliers or diversifiers of signaling? One of the members of the MIRR family, TCR, signals through 10 ITAMs present in the ζ, CD3ε, CD3γ and CD3δ invariant signaling chains of the TCR complex including 3 different ITAMs of the ζ chain. Similarly, signaling mediated by BCR is controlled by 2 ITAMs present in the Igα and Igβ signaling chains. Another MIRR, FcεRI, generates the activation signal using 2 ITAMs of the FcεRIβ and FcRγ chains. Most of experimental data in the literature reported to date strongly support the distinct rather than redundant functions for the ITAM signaling modules including those located on the different signaling chains39,44–55 and those located on the same signaling module (e.g., 3 different ITAMs on TCR ζ chain).56
Within the SCHOOL model,10,11,42,43 cytoplasmic homooligomerization controls the ITAM signaling through the different patterns of MIRR signaling subunit homooligomerization11,43 that produce distinct activation signals provided by different ITAMs. This assumes the combinatorial nature of MIRR-mediated signaling and can provide a molecular explanation for the diversity of the immune response. In this context, the diversity of cell functional outcomes in response to different ligands is higher with the more different signaling subunits (or ITAMs on the same subunit) the MIRR complex has. Thus, TCR-mediated signaling and cell activation has the highest combinatorial potential as compared to other MIRRs, explaining a high variability of distinct TCR-triggered intracellular signaling pathways and therefore distinct T cell functional responses depending on the nature of the stimulus.10,11
Cytoplasmic homooligomerization-controlled signaling mechanism: How general is the concept? Oligomerization of receptors upon ligand binding is considered to be a key regulatory factor in receptor triggering. On the other hand, structural similarities of receptors within the SR and MIRR families strongly suggest similarities in the molecular mechanisms. Within the SCHOOL concept, receptor oligomerization induced or tuned upon ligand binding outside the cell is translated across the membrane into protein homooligomerization in cytoplasmic milieu, thus providing a general platform for transmembrane signal transduction mediated by receptors of both structural families (SRs and MIRRs).2,3,11 The analysis of literature data reveal multiple experimental evidence to support the concept.2,3,11 This suggests cytoplasmic homooligomerization as a general cell mechanism to control signaling through both protein order and disorder, linking mechanistically a variety of structurally and functionally diverse receptors.
Conclusions and Perspectives
Recent progress in multidisciplinary research towards the elucidation of the structure, properties and function of cell surface receptors dramatically improves our understanding of these systems and transmembrane signal transduction at the molecular level. The complex and scientifically intriguing interplay between protein order, the chaos of protein disorder and oligomericity in the context of cell recognition and signaling—stressed throughout this article—opens new exciting perspectives for further multidisciplinary studies. Moreover, for the first time, we have an opportunity to adopt an entirely novel mind-set, not only about the nature and principles of receptor signaling, but also about the way we perform fundamental research and design therapies of receptor-mediated diseases for the foreseeable future.
References
- 1.Sigalov A, Aivazian D, Stern L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry. 2004;43:2049–2061. doi: 10.1021/bi035900h. [DOI] [PubMed] [Google Scholar]
- 2.Sigalov AB. Protein intrinsic disorder and oligomericity in cell signaling. Mol Biosyst. 2010;6:451–461. doi: 10.1039/b916030m. [DOI] [PubMed] [Google Scholar]
- 3.Sigalov AB. The SCHOOL of nature. II. Protein order, disorder and oligomericity in transmembrane signaling. Self/Nonself. 2010;1:89–102. doi: 10.4161/self.1.2.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-Binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigalov AB, Uversky VN. Differential occurrence of protein intrinsic disorder in the cytoplasmic signaling domains of cell receptors. Self/Nonself. 2011;2:55–72. doi: 10.4161/self.2.1.14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sigalov AB. Unusual biophysics of immune signaling-related intrinsically disordered proteins. Self/Nonself. 2010;1:271–281. doi: 10.4161/self.1.4.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigalov AB, Hendricks GM. Membrane binding mode of intrinsically disordered cytoplasmic domains of T cell receptor signaling subunits depends on lipid composition. Biochem Biophys Res Commun. 2009;389:388–393. doi: 10.1016/j.bbrc.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigalov AB, Kim WM, Saline M, Stern LJ. The intrinsically disordered cytoplasmic domain of the T-cell receptor zeta chain binds to the nef protein of simian immunodeficiency virus without a disorder-to-order transition. Biochemistry. 2008;47:12942–12944. doi: 10.1021/bi801602p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigalov AB. Multichain immune recognition receptor signaling: Different players, same game? Trends Immunol. 2004;25:583–589. doi: 10.1016/j.it.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Sigalov AB. The SCHOOL of nature. I. Transmembrane signaling. Self/Nonself. 2010;1:4–39. doi: 10.4161/self.1.1.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keegan AD, Paul WE. Multichain immune recognition receptors: Similarities in structure and signaling pathways. Immunol Today. 1992;13:63–68. doi: 10.1016/0167-5699(92)90136-U. [DOI] [PubMed] [Google Scholar]
- 13.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 14.Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishimoto H, Kubo RT, Yorifuji H, Nakayama T, Asano Y, Tada T. Physical dissociation of the TCR-CD3 complex accompanies receptor ligation. J Exp Med. 1995;182:1997–2006. doi: 10.1084/jem.182.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosugi A, Saitoh S, Noda S, Yasuda K, Hayashi F, Ogata M, et al. Translocation of tyrosine-phosphorylated TCRzeta chain to glycolipid-enriched membrane domains upon T cell activation. Int Immunol. 1999;11:1395–1401. doi: 10.1093/intimm/11.9.1395. [DOI] [PubMed] [Google Scholar]
- 17.La Gruta NL, Liu H, Dilioglou S, Rhodes M, Wiest DL, Vignali DA. Architectural changes in the TCR:CD3 complex induced by MHC:Peptide ligation. J Immunol. 2004;172:3662–3669. doi: 10.4049/jimmunol.172.6.3662. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Rhodes M, Wiest DL, Vignali DA. On the dynamics of TCR:CD3 complex cell surface expression and downmodulation. Immunity. 2000;13:665–675. doi: 10.1016/s1074-7613(00)00066-2. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Cramer L, Mueller H, Wilson B, Vilen BJ. Independent trafficking of Ig-alpha/Ig-beta and mu-heavy chain is facilitated by dissociation of the B cell antigen receptor complex. J Immunol. 2005;175:147–154. doi: 10.4049/jimmunol.175.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremyanskaya M, Monroe JG. Ig-independent Ig beta expression on the surface of B lymphocytes after B cell receptor aggregation. J Immunol. 2005;174:1501–1506. doi: 10.4049/jimmunol.174.3.1501. [DOI] [PubMed] [Google Scholar]
- 21.Vilen BJ, Nakamura T, Cambier JC. Antigen-stimulated dissociation of BCR mIg from Ig-alpha/Ig-beta: implications for receptor desensitization. Immunity. 1999;10:239–248. doi: 10.1016/s1074-7613(00)80024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asai K, Fujimoto K, Harazaki M, Kusunoki T, Korematsu S, Ide C, et al. Distinct aggregation of beta- and gamma-chains of the high-affinity IgE receptor on cross-linking. J Histochem Cytochem. 2000;48:1705–1716. doi: 10.1177/002215540004801213. [DOI] [PubMed] [Google Scholar]
- 23.Archakov AI, Govorun VM, Dubanov AV, Ivanov YD, Veselovsky AV, Lewi P, et al. Protein-protein interactions as a target for drugs in proteomics. Proteomics. 2003;3:380–391. doi: 10.1002/pmic.200390053. [DOI] [PubMed] [Google Scholar]
- 24.Pagliaro L, Felding J, Audouze K, Nielsen SJ, Terry RB, Krog-Jensen C, et al. Emerging classes of protein-protein interaction inhibitors and new tools for their development. Curr Opin Chem Biol. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Veselovsky AV, Archakov AI. Inhibitors of protein-protein interactions as potential drugs. Current Computer-Aided Drug Design. 2007;3:51–58. [Google Scholar]
- 26.Sigalov AB. Immune cell signaling: A novel mechanistic model reveals new therapeutic targets. Trends Pharmacol Sci. 2006;27:518–524. doi: 10.1016/j.tips.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Sigalov AB. Interaction between HIV gp41 fusion peptide and T cell receptor: Putting the puzzle pieces back together. Faseb J. 2007;21:1633–1634. doi: 10.1096/fj.07-0603ltr. [DOI] [PubMed] [Google Scholar]
- 28.Sigalov AB. Novel mechanistic insights into viral modulation of immune receptor signaling. PLoS Pathog. 2009;5:1000404. doi: 10.1371/journal.ppat.1000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigalov AB. The SCHOOL of nature. IV. Learning from viruses. Self/Nonself. 2010;1:282–298. doi: 10.4161/self.1.4.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana FJ, Gerber D, Kent SC, Cohen IR, Shai Y. HIV-1 fusion peptide targets the TCR and inhibits antigen-specific T cell activation. J Clin Invest. 2005;115:2149–2158. doi: 10.1172/JCI23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigalov AB. Novel mechanistic concept of platelet inhibition. Expert Opin Ther Targets. 2008;12:677–692. doi: 10.1517/14728222.12.6.677. [DOI] [PubMed] [Google Scholar]
- 32.Sigalov AB. New therapeutic strategies targeting transmembrane signal transduction in the immune system. Cell Adh Migr. 2010;4:255–267. doi: 10.4161/cam.4.2.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigalov AB. The SCHOOL of nature. III. From mechanistic understanding to novel therapies. Self/Nonself. 2010;1:192–224. doi: 10.4161/self.1.3.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintana FJ, Gerber D, Bloch I, Cohen IR, Shai Y. A structurally altered D, L-amino acid TCRalpha transmembrane peptide interacts with the TCRalpha and inhibits T-cell activation in vitro and in an animal model. Biochemistry. 2007;46:2317–2325. doi: 10.1021/bi061849g. [DOI] [PubMed] [Google Scholar]
- 35.Kurosaka N, Ali M, Byth K, Manolios N. The mode of anti-arthritic peptide delivery impacts on the severity and outcome of adjuvant induced arthritis. APLAR J Rheumatol. 2007;10:198–203. [Google Scholar]
- 36.Manolios N, Ali M, Amon M, Bender V. Therapeutic application of transmembrane T and natural killer cell receptor peptides. Adv Exp Med Biol. 2008;640:208–219. doi: 10.1007/978-0-387-09789-3_16. [DOI] [PubMed] [Google Scholar]
- 37.Manolios N, Ali M, Bender V. T-cell antigen receptor (TCR) transmembrane peptides: A new paradigm for the treatment of autoimmune diseases. Cell Adh Migr. 2010;4:273–283. doi: 10.4161/cam.4.2.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manolios N, Huynh NT, Collier S. Peptides in the treatment of inflammatory skin disease. Australas J Dermatol. 2002;43:226–227. doi: 10.1046/j.1440-0960.2002.00603.x. [DOI] [PubMed] [Google Scholar]
- 39.Pitcher LA, van Oers NS. T-cell receptor signal transmission: Who gives an ITAM? Trends Immunol. 2003;24:554–560. doi: 10.1016/j.it.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007;35:786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Sigalov AB. Signaling chain homooligomerization (SCHOOL) model. Adv Exp Med Biol. 2008;640:121–163. doi: 10.1007/978-0-387-09789-3_12. [DOI] [PubMed] [Google Scholar]
- 43.Sigalov A. Multi-chain immune recognition receptors: spatial organization and signal transduction. Semin. Immunol. 2005;17:51–64. doi: 10.1016/j.smim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Pike KA, Baig E, Ratcliffe MJ. The avian B-cell receptor complex: Distinct roles of Igalpha and Igbeta in B-cell development. Immunol Rev. 2004;197:10–25. doi: 10.1111/j.0105-2896.2004.0111.x. [DOI] [PubMed] [Google Scholar]
- 45.Storch B, Meixlsperger S, Jumaa H. The Ig-alpha ITAM is required for efficient differentiation but not proliferation of pre-B cells. Eur J Immunol. 2007;37:252–260. doi: 10.1002/eji.200636667. [DOI] [PubMed] [Google Scholar]
- 46.Gazumyan A, Reichlin A, Nussenzweig MC. Ig beta tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med. 2006;203:1785–1794. doi: 10.1084/jem.20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin S, Cicala C, Scharenberg AM, Kinet JP. The Fc(epsilon)RIbeta subunit functions as an amplifier of Fc(epsilon)RIgamma-mediated cell activation signals. Cell. 1996;85:985–995. doi: 10.1016/s0092-8674(00)81300-8. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Mejorada G, Rosales C. Signal transduction by immunoglobulin Fc receptors. J Leukoc Biol. 1998;63:521–533. doi: 10.1002/jlb.63.5.521. [DOI] [PubMed] [Google Scholar]
- 49.Lysechko TL, Ostergaard HL. Differential Src family kinase activity requirements for CD3 zeta phosphorylation/ZAP70 recruitment and CD3 epsilon phosphorylation. J Immunol. 2005;174:7807–7814. doi: 10.4049/jimmunol.174.12.7807. [DOI] [PubMed] [Google Scholar]
- 50.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26:357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 51.Chau LA, Bluestone JA, Madrenas J. Dissociation of intracellular signaling pathways in response to partial agonist ligands of the T cell receptor. J Exp Med. 1998;187:1699–1709. doi: 10.1084/jem.187.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen WA, Pleiman CM, Beaufils P, Wegener AM, Malissen B, Cambier JC. Qualitatively distinct signaling through T cell antigen receptor subunits. Eur J Immunol. 1997;27:707–716. doi: 10.1002/eji.1830270320. [DOI] [PubMed] [Google Scholar]
- 53.Pitcher LA, Mathis MA, Young JA, Deford LM, Purtic B, Wulfing C, et al. The CD3 gammaepsilon/deltaepsilon signaling module provides normal T cell functions in the absence of the TCR zeta immunoreceptor tyrosine-based activation motifs. Eur J Immunol. 2005;35:3643–3654. doi: 10.1002/eji.200535136. [DOI] [PubMed] [Google Scholar]
- 54.Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, et al. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3{epsilon} J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 55.Roifman CM. CD3 delta immunodeficiency. Curr Opin Allergy Clin Immunol. 2004;4:479–484. doi: 10.1097/00130832-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 56.Chae WJ, Lee HK, Han JH, Kim SW, Bothwell AL, Morio T, et al. Qualitatively differential regulation of T cell activation and apoptosis by T cell receptor zeta chain ITAMs and their tyrosine residues. Int Immunol. 2004;16:1225–1236. doi: 10.1093/intimm/dxh120. [DOI] [PubMed] [Google Scholar]