Abstract
Colorectal cancer (CRC) is a leading cause of cancer death worldwide. Epidemiological risk factors for CRC included dietary fat intake; consequently, the role of genes in the fatty acid biosynthesis and metabolism pathways is of particular interest. Moreover, hyperlipidaemia has been associated with different type of cancer and serum lipid levels could be affected by genetic factors, including polymorphisms in the lipid metabolism pathway. The aim of this study is to assess the association between single-nucleotide polymorphisms (SNPs) in fatty acid metabolism genes, serum lipid levels, body mass index (BMI) and dietary fat intake and CRC risk; 30 SNPs from 8 candidate genes included in fatty acid biosynthesis and metabolism pathways were genotyped in 1780 CRC cases and 1864 matched controls from the Molecular Epidemiology of Colorectal Cancer study. Information on clinicopathological characteristics, lifestyle and dietary habits were also obtained. Logistic regression and association analysis were conducted. Several LIPC (lipase, hepatic) polymorphisms were found to be associated with CRC risk, although no particular haplotype was related to CRC. The SNP rs12299484 showed an association with CRC risk after Bonferroni correction. We replicate the association between the T allele of the LIPC SNP rs1800588 and higher serum high-density lipoprotein levels. Weak associations between selected polymorphism in the LIPC and PPARG genes and BMI were observed. A path analysis based on structural equation modelling showed a direct effect of LIPC gene polymorphisms on colorectal carcinogenesis as well as an indirect effect mediated through serum lipid levels. Genetic polymorphisms in the hepatic lipase gene have a potential role in colorectal carcinogenesis, perhaps though the regulation of serum lipid levels.
Introduction
Colorectal cancer (CRC) is a leading cause of death worldwide, with over 1 million of new cases and a half a million of deaths around the world every year (1,2). Risk factors for CRC include advanced age, medical history of benign adenomatous polyps and inflammatory bowel diseases, family history of CRC, low intake of vegetables and fruits and high intake of dietary fat (particularly animal fat) and processed meat (3–6). Chronic consumption of non-steroidal anti-inflammatory drugs, hormone replacement therapy and statins are protective (7,8). The role of other lifestyle factors such as tobacco smoking or alcohol consumption remains inconclusive (9–11).
Dietary fat is a recognized risk factor for CRC. Polymorphisms in genes involved in the regulation of lipid biosynthesis, transport and metabolism are potential susceptibility factors but the effects of these genes in CRC risk may be mediated and modified through their implication in energy balance, obesity or physical activity (12–17).
The aim of the present study is to assess the association between single-nucleotide polymorphisms (SNPs) in lipid metabolism genes, in particular in genes related to fatty acid biosynthesis and metabolism and CRC risk. The potential mediation of serum lipid levels, body mass index (BMI) and dietary fat intake using a comprehensive approach based on structural equation modelling in relation to CRC has been assessed as well. The analyses have been performed within a large population-based case control study conducted in Israel, the Molecular Epidemiology of Colorectal Cancer (MECC) study.
Methods
Patients
The MECC study has already been described in detail (8,18). In brief, it is a population-based case-control study of incident, pathologically confirmed, invasive CRC diagnosed between May 31, 1998, and March 31, 2004, and who lived in a geographically defined area of Northern Israel.
Controls with no prior history of CRC were identified from the same source population using the Clalit Health Services (CHS) database, covering ∼70% of the older population. Controls were matched to cases by year of birth, gender, primary clinic location and Jewish or Arab ethnicity. Participants provided written informed consent at the time of enrolment. The Institutional Review Boards at the Carmel Medical Center and the University of Michigan approved all procedures. The study population included 2100 matched pairs. For this analysis, only 1780 cases and 1864 controls with available DNA for genetic analysis will be used. The demographic characteristics of the subjects not included do not differ from those analysed (data not shown).
Life-style information
Participants were interviewed face-to-face by trained monitors with a comprehensive epidemiological questionnaire that assessed demographic information, personal and family history of cancer, reproductive history and medical history, medication use, health habits and a food frequency questionnaire. Patient’s weight was recorded by self-report, as estimated 1 year before diagnosis for cases and at the time of interview for controls. BMI was estimated 1 year before the diagnosis from self-reported weight and height.
Comprehensive dietary habits were obtained with the use of a validated food-frequency questionnaire modified for the Israeli diet. Total fat, saturated fat, monounsaturated fat and polyunsaturated fat intakes were estimated from the questionnaire using local food composition tables. Similar estimates provided total energy consumption. Serum lipid levels at the time to CRC diagnosis were available from electronic medical records of CHS for a subset of the study population. Physical activity was recorded for the longest occupation and also recreational physical exercise. These were combined into an ordinal index ranging from sedentary to intense physical activity.
SNP selection and genotyping
Eight genes (APOE, LIPC, LPL, CYP1A2, ACSL5, PPARG, PTGS2) related to lipid metabolism pathways, mainly fatty acid metabolism, were selected for analysis within a larger exploratory project including other hypothesis. Basically, candidate genes were selected from fatty acid biosynthesis and metabolism pathways of the KEGG pathway database (19). Moreover, a literature review was conducted to identify molecular epidemiological studies that analysed the relationship between SNPs in fatty acid metabolism and CRC risk. We searched in Pubmed citation database combinations of the following terms: ‘polymorphism or SNP’ and ‘lipid metabolism or fatty acid metabolism’ and ‘colon or colorectal cancer’. A total of 30 SNPs in these 8 candidate genes (Table II) were selected as maximally informative htSNPs when the regions were analysed with HaploTagger. SNPs were genotyped using Illumina Beadstation and BeadExpress platforms. Data were analysed using unsupervised Illumina BeadStation SNP calling automated routines, and the distribution of genotypes was compared to those expected from Hardy–Weinberg equilibrium. Genotypes were also confirmed with a second platform in at least 1% of samples.
Table II.
Association (P-valuesa) between polymorphisms in fatty acid metabolism genes and CRC risk, serum lipid levels, BMI and fat intake
| SNP | CRC | HDL 1 year before | LDL 1 year before | BMI | Fat densityf | Energy (Kcal) |
| APOE_rs405509 | 0.44 | 0.16 | 0.62 | 0.67 | 0.50 | 0.0024 |
| APOE_rs439401 | 0.24 | 0.49 | 0.63 | 0.71 | 0.034 | 0.048 |
| APOE_rs7259620 | 0.66 | 0.15 | 0.30 | 0.30 | 0.52 | 0.067 |
| LIPC_rs12913969 | 0.44 | 0.29 | 0.78 | 0.98 | 0.48 | 0.26 |
| LIPC_rs16940302 | 0.66 | 0.87 | 0.69 | 0.060 | 0.86 | 0.31 |
| LIPC_rs16940372 | 0.0086 | 0.14 | 0.76 | 0.63 | 0.56 | 0.86 |
| LIPC_rs17190510 | 0.70 | 0.29 | 0.45 | 0.42 | 0.36 | 0.24 |
| LIPC_rs1800588 | 0.51 | 0.025 | 0.18 | 0.92 | 0.99 | 0.71 |
| LIPC_rs2099190 | 0.17 | 0.36 | 0.19 | 0.81 | 0.57 | 0.84 |
| LIPC_rs3751542 | 0.85 | 0.58 | 0.82 | 0.73 | 0.20 | 0.68 |
| LIPC_rs4774302 | 0.14 | 0.40 | 0.92 | 0.18 | 0.20 | 0.30 |
| LIPC_rs4775053 | 0.0099 | 0.92 | 0.88 | 0.05 | 0.84 | 0.77 |
| LIPC_rs4775072 | 0.0028 | 0.49 | 0.11 | 0.023 | 0.19 | 0.64 |
| LIPC_rs6083 | 0.65 | 0.87 | 0.92 | 0.10 | 0.48 | 0.22 |
| LIPC_rs634746 | 0.54 | 0.75 | 0.18 | 0.10 | 0.90 | 0.36 |
| LIPC_rs7166788 | 0.65 | 0.87 | 0.80 | 0.35 | 0.30 | 0.63 |
| LIPC_rs7174210 | 0.17 | 0.14 | 0.99 | 0.017b | 0.50 | 0.97 |
| LIPC_rs8035006 | 0.20 | 0.87 | 0.33 | 0.68 | 0.31 | 0.35 |
| LIPC_rs9652472 | 0.0005 | 0.86 | 0.087 | 0.070 | 0.55 | 0.52 |
| LPL_rs268 | 0.034c | — | — | 0.72 | 0.090c | — |
| LPL_rs328 | 0.97 | 0.15 | 0.61 | 0.55 | 0.49 | 0.99 |
| CYP1A2_rs2470890 | 0.76 | 0.13 | 0.42 | 0.014d | 0.75 | 0.34 |
| CYP1A2_rs762551 | 0.81 | 0.10 | 1.00 | 0.71 | 0.98 | 0.91 |
| CYP2C9_rs1057910e | — | 0.13 | 0.05 | — | — | — |
| ACSL5_rs17129748 | 0.10 | 0.11 | 0.63 | 0.15 | 0.91 | 0.19 |
| ACSL5_rs2419629 | 0.19 | 0.45 | 0.99 | 0.20 | 0.43 | 0.29 |
| ACSL5_rs7919710 | 0.12 | 0.27 | 0.91 | 0.19 | 0.86 | 0.20 |
| PPARG_rs1801282 | 0.0059d | 0.36 | 0.37 | 0.023b | 0.13 | 0.11 |
| PPARG_rs3856806 | 0.033c | 0.27 | 0.95 | 0.23 | 0.020 | 0.37 |
| PTGS2_rs5275 | 0.37 | 0.54 | 0.78 | 0.77 | 0.63 | 0.84 |
Nominal P-value, P < 0.05; Bonferroni correction P < 0.0017 Bold values indicate the most significant associations.
P-values for log-additive model are shown (except when indicated).
P-value for dominant model.
P-value for codominant model.
P-value for recessive model.
Genotyping rate under 50%.
Adjusted by energy intake.
Statistical methods
Unconditional logistic regression was used to assess the association between genotypes and CRC risk. All models included age and gender to account for the matching design and avoid excluding incomplete pairs due to DNA unavailability or missing values in other variables. Multivariate-adjusted odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were calculated for each genotype compared to those homozygous for the common allele. Also a log-additive model was fitted (trend test). Linear regression was used to assess the associations with the quantitative variables serum lipid levels, BMI and dietary fat intake. In these models, mean differences and 95% CI were calculated. Analysis was performed with the SNPstats (20) web software, including the estimation of haplotypes for multiple SNPs of a gene and SNPassoc library for R package (21). A panel of 300 anonymous SNPs useful for population stratification analysis was analysed in these subjects in relation to other larger genotyping project. No relevant population structure was identified that could not be explained by reported ethnicity. The potential confounding of this variable was rejected after a sensitivity analysis and was not used in the models to maximize efficiency.
A global 5% significance level was desired for the analyses of SNPs. Thus, a P-value < 0.0017 was considered statistically significant following the Bonferroni method to account for the number of tested SNPs as independent hypothesis. All reported P-values are two tailed, adjusted for age, sex and ethnicity but uncorrected for multiple comparisons.
To simultaneously study the associations between SNPs, mediator variables and CRC, we also fitted structural equation models (SEMs) to prespecified path hypothesis. SEM was used to assess the direct effect of genetic polymorphisms on CRC (unexplained by candidate variables), and also their indirect effect, mediated through lifestyle-related factors by including all variables in the same model to assess overall associations. These analyses were performed using structural equation modelling library from R package (21) and MPLUS v. 5.21 (22). SEM was used to test the conceptual model since, in contrast to traditional analytical procedures as linear regression analysis, SEM allows distinguishing between direct and indirect effects and provides information on the degree of fit for the entire model. In SEM, the covariance structure that follows from the proposed model is fitted to the observed covariance. The maximum likelihood estimate method yields estimates of the regression coefficients in the model, standard errors and a overall goodness-of-fit test (23).
Results
The study population included 3644 participants, 1780 cases and 1864 controls, with available data regarding epidemiological interviews and genetic analysis. Serum lipid levels 1 year prior to CRC diagnosis were available only for 272 cases and 625 controls. A description of the subjects’ characteristics is in Table I. Cases had larger reported weight and BMI 1 year before the diagnosis. Also had more often family history of CRC, lower physical activity and vegetables intake. Long-term regular use of aspirin and statins were also inversely associated with CRC risk.
Table I.
Baseline characteristics of cases and controls from the study population
| Characteristic | Cases | Controls | P-value |
| N | 1780 | 1864 | |
| Clinical/baseline characteristics | |||
| Age, mean (SD) | 70.21 (11.67) | 70.58 (11.67) | 0.74 |
| Sex | 0.65 | ||
| Male, N (%) | 894 (50.22) | 931 (49.95) | |
| Female, N (%) | 886 (49.78) | 933 (50.05) | |
| Height, mean (SD) | 165.78 (8.68) | 165.42 (8.90) | 0.50 |
| Weight, year before diagnosis, mean (SD) | 75.13 (14.45) | 73.71 (13.99) | 0.0093 |
| BMI, mean (SD) | 27.26 (4.63) | 26.89 (4.63) | 0.037 |
| History of CRC in first-degree relative | 0.0019 | ||
| No, N (%) | 1571 (89.98) | 1722 (92.88) | |
| Yes, N (%) | 175 (10.02) | 132 (7.12) | |
| Physical activity index N (%) | 0.0000 | ||
| Sedentary | 1231 (69.16) | 1105 (59.28) | |
| Intermediate | 311 (17.47) | 426 (22.85) | |
| Active | 238 (13.37) | 333 (17.86) | |
| Vegetable intake, mean (SD) | 7.47 (4.48) | 7.85 (3.69) | 0.0022 |
| Regular aspirin intake | |||
| No, N (%) | 1192 (69.18) | 1170 (62.84) | 0.0001 |
| Yes, N (%) | 531 (30.82) | 692 (37.16) | |
| Regular statin intake | |||
| No, N (%) | 1630 (94.17) | 1623 (87.68) | 0.0000 |
| Yes, N (%) | 101 (5.83) | 228 (12.32) | |
| Dietary fat intakea | 692 | 1854 | |
| Energy, mean (SD) | 1768.15 (1006.97) | 1814.12 (898.08) | 0.13 |
| 1710 | 1862 | ||
| Total fat, mean (SD) | 69.35 (67.48) | 66.14 (44.91) | 0.0044 |
| Fat densitya | 1687 | 1854 | |
| Total fat, mean (SD) | 0.33 (0.07) | 0.32 (0.06) | 0.00053 |
| Serum lipids (1 year prior to diagnosis) | 272 | 625 | |
| HDL, mean (SD) | 50.03 (12.80) | 50.66 (12.11) | 0.32 |
| LDL, mean (SD) | 122.06 (31.37) | 120.78 (29.40) | 0.60 |
Cases with energy >10000 Kcal have been excluded.
Though energy intake was similar for cases and controls, total fat intake and the proportion of energy derived from fat (nutrient density) was larger for cases than controls. Serum lipid levels [high-density lipoprotein (HDL) and low-density lipoprotein (LDL)] measured 1 year prior to diagnosis were similar in cases and controls.
Fatty acid SNPs and CRC risk
Thirty SNPs in eight fatty acid biosynthesis and metabolism genes were genotyped in CRC cases and controls. A detailed description of the allele and genotype frequencies for the studied SNPs is in supplementary Table I (available at Mutagenesis Online). Two SNPs (rs16940302 of LIPC and rs1057910 of CYP2C9) were not in Hardy–Weinberg equilibrium. Nevertheless, none of these two SNPs showed significant associations with the variables included in this study, CRC risk or serum lipid levels. The P-values for main-effect associations assessed between the SNPs and CRC risk, serum lipid levels, BMI and dietary fat intake are summarized in Table II. Except for those indicated in the table, the reported P-values correspond to the log-additive model and are adjusted for age and sex.
We first considered whether genetic variation in these genes was associated with CRC risk. An association between selected SNPs in LIPC gene and CRC risk was observed. These associations remained statistically significant when BMI, serum lipid levels (HDL and LDL) and dietary fat intake were also included in the model as potential confounders. Weak associations were also observed between polymorphisms in PPARG gene and CRC risk. Details for the analysis of these SNPs are shown in Table III. The association for LIPC rs9652472 with CRC risk was significant after Bonferroni correction. The G allele of this SNP was associated with a per-allele OR of 1.52 (95% CI 1.20–1.92, P-value = 0.0005).
Table III.
Association between polymorphisms in fatty acid metabolism genes and CRC risk
| SNP | Controls, N (%) | Cases, N (%) | ORa | 95% CI | P-value |
| LIPC_rs16940372 | 0.012 | ||||
| A/A | 1029 (69.7) | 621 (73.2) | 1.00 | ||
| A/G | 403 (27.3) | 215 (25.4) | 0.84 | 0.68–1.04 | |
| G/G | 45 (3) | 12 (1.4) | 0.41 | 0.20–0.84 | |
| Log-additive | 1477 (63.5) | 848 (36.5) | 0.65 | 0.64–0.94 | 0.0086 |
| LIPC_rs4775053 | 0.021 | ||||
| C/C | 1184 (80.1) | 667 (82.3) | 1.00 | ||
| T/C | 277 (18.7) | 138 (17.0) | 0.78 | 0.61–1.00 | |
| T/T | 18 (1.2) | 5 (0.6) | 0.32 | 0.09–1.09 | |
| Log-additive | 1479 (64.6) | 810 (35.4) | 0.74 | 0.59–0.93 | 0.0099 |
| LIPC_rs4775072 | 0.011 | ||||
| C/C | 1380 (93.1) | 768 (90.1) | 1.00 | ||
| T/C | 100 (6.7) | 87 (9.6) | 1.66 | 1.18–2.32 | |
| T/T | 2 (0.1) | 2 (0.2) | 2.10 | 0.29–15.22 | |
| Log-additive | 1482 (63.5) | 852 (36.5) | 1.64 | 1.19–2.25 | 0.0028 |
| LIPC_rs9652472 | 0.0018 | ||||
| A/A | 1269 (86.2) | 694 (81.8) | 1.00 | ||
| A/G | 195 (13.2) | 150 (17.7) | 1.57 | 1.22–2.02 | |
| G/G | 8 (0.5) | 4 (0.5) | 1.43 | 0.40–5.15 | |
| Log-additive | 1472 (63.4) | 848 (36.6) | 1.52 | 1.20–1.92 | 0.0005 |
| PPARG_rs1801282 | 0.017 | ||||
| C/C | 1307 (88.4) | 710 (87.4) | 1.00 | ||
| C/G | 163 (11.0) | 102 (12.6) | 1.11 | 0.84–1.48 | |
| G/G | 9 (0.6) | 0 (0.0) | 0.00 | — | |
| Log-additive | 1479 (64.6) | 812 (35.4) | 0.99 | 0.75–1.29 | 0.92 |
| PPARG_rs3856806 | 0.033 | ||||
| C/C | 1285 (86.8) | 682 (83.9) | 1.00 | ||
| T/C | 182 (12.3) | 128 (15.7) | 1.31 | 1.01–1.70 | |
| T/T | 13 (0.9) | 3 (0.4) | 0.34 | 0.07–1.51 | |
| Log-additive | 1480 (64.5) | 813 (35.5) | 1.16 | 0.91–1.48 | 0.22 |
Age, sex, BMI 1 year before diagnosis, serum lipid levels and total dietary fat intake adjusted OR. Only significative associations are shown.
Since multiple SNPs were assessed for LIPC and three of them showed significant associations, a haplotype analysis was performed. However, this was not more informative than the analysis of individual SNPs since they were chosen as haplotype tagging SNPs. This large gene contains four haplotype blocks (supplementary Figure 1, available at Mutagenesis Online). The frequencies were under 0.15 and no associations between LIPC haplotypes and CRC risk were observed (data not shown).
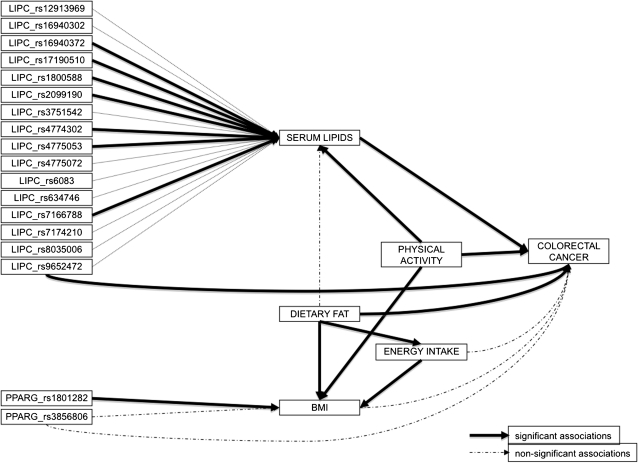
Fig. 1.
Path analysis using SEM in R. The continuous arrows show the associations that remain significant in the SEM model (P > 0.02). Discontinuous arrows show associations that were no longer significant and could be considered indirect effects.
Fatty acid SNPs and serum lipid levels
Serum lipid levels 1 year before diagnosis were only available for a limited number of subjects (272 cases and 625 controls). Thus, the power to detect associations with SNPs was reduced. Only the SNP rs1800588 in LIPC was marginally associated to HDL levels in MECC study participants (Table IV). The T allele of this SNP was associated with higher HDL levels; mean differences were increased in 1.61 points per allele (95% CI 0.20–3.01, P-value = 0.025). No SNP was related to LDL serum levels.
Table IV.
Association between polymorphisms in fatty acid metabolism genes and serum lipid levels 1 year before CRC diagnosis.
| SNP | N | Mean | SE | Differencea | 95% CI | P-value |
| HDL | ||||||
| LIPC_rs1800588 | 0.057 | |||||
| C/C | 555 | 49.85 | 0.53 | 0.00 | ||
| T/C | 243 | 50.68 | 0.71 | 1.17 | −0.56 to 2.89 | |
| T/T | 31 | 55.35 | 2.29 | 4.54 | 0.39 to 8.68 | |
| Log-additive | 829 | 1.61 | 0.20 to 3.01 | 0.025 |
Age, sex, BMI 1 year before diagnosis and total dietary fat intake adjusted OR. Only significative associations are shown.
Fatty acid SNPs and BMI
Selected polymorphism in the LIPC gene, specifically the SNPs rs16940302, rs7174210, rs4775053, and rs4775072, were found to be associated with changes in the median of BMI of the MECC study participants. Moreover, an association between rs2470890 in CYP1A2 gene and rs1801282 in PPARG gene and BMI were also observed (Table II, for details see Table V). None of these associations remained statistically significant after Bonferroni correction.
Table V.
Association between polymorphisms in fatty acid metabolism genes and BMI
| SNP | N | Mean | SE | Differencea | 95% CI | P-value |
| LIPC_rs4775072 | 0.074 | |||||
| C/C | 1959 | 27.12 | 0.11 | 0.00 | ||
| T/C | 162 | 27.98 | 0.43 | 0.84 | 0.08 to 1.60 | |
| T/T | 4 | 29.02 | 0.56 | 1.61 | −2.89 to 6.12 | |
| Log-additive | 2125 | 0.84 | 0.12 to 1.56 | 0.023 | ||
| LIPC_rs7174210 | 0.032 | |||||
| G/G | 1704 | 27.06 | 0.11 | 0.00 | ||
| T/G | 391 | 27.71 | 0.27 | 0.68 | 0.16 to 1.19 | |
| T/T | 25 | 26.69 | 0.95 | −0.40 | −2.50 to 1.45 | |
| Log-additive | 2120 | 0.48 | 0.02 to 0.95 | 0.042 | ||
| CYP1A2_rs2470890 | 0.036 | |||||
| T/T | 598 | 27.20 | 0.20 | 0.00 | ||
| T/C | 1079 | 27.36 | 0.15 | 0.18 | −0.28 to 0.65 | |
| C/C | 431 | 26.73 | 0.23 | −0.51 | −1.09 to 0.07 | |
| Log-additive | 2108 | −0.22 | −0.50 to 0.07 | 0.15 | ||
| PPARG_rs1801282 | 0.066 | |||||
| C/C | 1871 | 27.10 | 0.11 | 0.00 | ||
| C/G | 243 | 27.86 | 0.31 | 0.74 | 0.12 to 1.37 | |
| G/G | 9 | 27.20 | 0.15 | −0.03 | −3.03 to 2.98 | |
| Log-additive | 2123 | 0.64 | 0.05 to 1.22 | 0.032 |
Age, sex, serum HDL and LDL levels and total dietary fat intake adjusted OR. Only significative associations are shown.
Fatty acid SNPs and diet
The potential association of the SNPs with energy intake, fat intake and physical activity was also explored to exclude unexpected findings. None of the SNPs analysed was related to these behavioural variables (Table II).
Path analysis of SNPs, dietary fat intake, lipid levels, BMI, physical activity, energy and CRC
The main aim of this analysis was to perform a combined analysis of all players in the lipid metabolism. Path analysis using SEMs provide an interesting tool for this since a simultaneous estimate of associations can be done for an a priori proposed model. We built the model from the observed associations in the multiple univariate analyses previously described and tried to fit SEMs to identify the associations resistant to adjustment for indirect effects.
Figure 1 shows the proposed model reduced to relevant associations. The continuous arrows show the associations that remain significant in the SEM model. Discontinuous arrows show associations that were no longer significant and could be considered as indirect effects. This analysis confirms the direct effect of LIPC rs9652472 polymorphism on CRC and the indirect effect of several other LIPC polymorphisms through serum lipid levels. It is remarkable that some of these associations were weak and not significant in the univariate analysis, but the path analysis that accounts for global confounding has revealed the associations. Other behavioural variables like total energy intake, fat intake and physical exercise also remain related among each other and associated to colon cancer, showing the relevance of these factors as mediator variables. SNPs of PPARG in the path analysis lose the direct effect on CRC risk, but rs1801282 maintains its association to BMI. Supplementary Table II (available at Mutagenesis Online) shows the coefficients and P-values with the final model of the path analysis.
Discussion
We have examined the association between polymorphisms in fatty acid metabolism genes and the risk of CRC in the context of energy balance accounting for behavioural variables: dietary intake and physical exercise and mediator variables: BMI and serum lipid levels. Among the genes studied, LIPC and PPARG have shown a significant association to CRC.
We have focused this analysis in fatty acid metabolism as an extension of our previous study on cholesterol metabolism (18). Though both pathways could be interrelated, the postulated carcinogenic mechanism for fatty acids would be more related to energy balance and possibly to insulin that has not been studied in the present study.
LIPC, serum lipids and CRC risk
The LIPC gene encodes a hepatic triglyceride lipase, which is highly expressed in the liver, and has the dual function of triglyceride, phospholipids and lipoproteins hydrolase and binding factor for receptor-mediated lipoprotein uptake (24–26). This gene has been extensively studied in relation to cardiovascular diseases (27–36). Hyperlipidaemia, characterized by increased total cholesterol (TC), triglycerides, and LDL and decreased HDL, has also been associated with cancer risk (37). Both genetic and lifestyle factors (such as diet or body weight) affect serum lipid levels. Hyperlipidaemia has been associated with several genetic polymorphisms in the lipid metabolism pathway (28–31,38). Thus, the potential role of the LIPC gene in fatty acid metabolism is of particular interest in colorectal carcinogenesis, possibly mediated through hyperlipidaemia.
In this study, several polymorphisms in LIPC have been found to be associated with CRC risk, although no particular haplotype was related to CRC. The most prominent was the association of the G allele of LIPC rs9652472 with increased risk of CRC. This effect is significant after Bonferroni correction and remains in a SEM that includes all the observed significant associations among intervening variables.
We also replicate the already described associations between the T allele of the LIPC SNP rs1800588 and higher serum HDL levels (28,29,31). It is assumed that this association is due to the change of the rate of transcription of the LIPC gene for the carriers of the T allele of this promoter polymorphism (32,39,40). Other studies also have reported that the T allele is associated with higher TC and triglycerides (41).
It is interesting that the LIPC SNP most associated to CRC, rs9652472, is different than that associated to HDL levels. Though there is a degree of linkage disequilibrium between both SNPs, the LIPC gene is large and complex, with 14 exons that have evolved in different haplotype blocks (supplementary Figure 1, available at Mutagenesis Online). Our results showed a direct effect of LIPC gene polymorphisms on colorectal carcinogenesis and also an indirect effect mediated through serum lipid levels, even including dietary fat intake in the models as potential confounders. This leaves open the possibility that this gene has an effect on CRC by an additional mechanism independent of lipid metabolism.
Two previous studies have analysed the association between polymorphisms in the hepatic lipase gene (LIPC) and CRC risk, but with conflicting results. While the study by Webb et al. (42) reported an association between 10 polymorphisms and CRC risk, including the SNP rs3829462 in the LIPC gene, Frank et al. (43) did not replicate these associations. The SNP most associated in our study, rs9652472, is in linkage disequilibrium with rs3829462 (D’ = 1), though due to the low allele frequency of both SNPs (below 3% in CEU population) the concordance of alleles is not high (r2 = 0.001).
Additional evidence on the role of polymorphisms in lipid metabolism genes in CRC is scant. The recent study from Hoeft et al. (44), after the examination of 43 fatty acid metabolism-related genes (from 392 tagged SNPs) in 1225 CRC cases and 2032 controls of the EPIC cohort, reported associations between polymorphisms in HPGD, PLA2G6, TRPV3 and PTGER2 genes and the risk of CRC. This study selected a different set of genes than ours (only one SNP in PPARG was coincident and LIPC was not analysed at all) and was focused exclusively on main genetic effects, ignoring other mediator variables as we have analysed.
PPARG, BMI and CRC risk
The PPARG gene encodes the gamma subunit of the peroxisome proliferator-activated receptor subfamily of nuclear receptors. This protein plays an important role in the regulation of adipocyte differentiation, glucose and lipid homeostasis, and intracellular insulin-signalling events. Consequently, PPARG has been implicated in the pathology of numerous diseases including obesity, diabetes, atherosclerosis and cancer (45–52). Moreover, experimental evidence has suggested that activation of PPARG in the colon results in growth inhibition and differentiation and reduces the malignant potential of CRC cells (53,54).
We observed that the T allele of the PPARG SNP rs1801282, which results in a proline-to-alanine substitution in codon 12 with a clear functional effect, was weakly associated with a CRC risk reduction. This is in accordance with other studies (14,51,52,54–58).
The path analysis has revealed that the effect of PPARG gene polymorphisms on colorectal carcinogenesis is mediated through BMI. The univariate analysis showed significant direct associations between PPARG and CRC, but the model that considered other mediators such as dietary fat intake, energy and physical activity only retained as significant the association between PPARG rs1801282 and BMI.
Strengths and limitations
This study is one of the largest conducted so far in which polymorphisms in lipid metabolism have been studied in the global context of diet, physical activity and serum lipid levels in relation to CRC risk. The population-based case-control design, though restrospective, is capable to identify a series of associations that allow a global picture of the key players. Some of the reported associations had been previously reported in studies of specific factors, which reinforces the strength of the proposed model.
Probably the study’s most important limitation is that the data on blood lipids were only available for lipids related with cholesterol (HDL, LDL and TC) and in a limited subset of the study. The estimates for the associations with these variables were less precise, but could be incorporated into the comprehensive path analysis since this statistical technique only requires the matrix of correlation coefficients. The analyses performed in this study only used HDL and LDL, which predominantly reflect the cholesterol metabolism rather that fatty acid metabolism which we were interested in. The availability of serum triglyceride would have been interesting, but this is not routinely requested in blood tests and could not be retrieved. Anyway this is, as far as we know, the only study on CRC that explores simultaneously the effects of polymorphisms in lipid metabolism genes with diet, BMI and blood lipid levels.
The sample size of this study, though large, is not powered enough to assess gene–gene or gene–environment interactions. The focus of our study has been the main effects, but further investigations should be done in larger or pool studies on the putative role of gene–gene interactions in this pathway.
Conclusion
Although our findings requires validation in other independent populations, further characterization of LIPC and PPARG polymorphisms may provide new insights into their contribution to incidence and progression of CRC.
Supplementary data
Supplementary Figure 1 and Table 1 are available at Mutagenesis Online.
Funding
National Cancer Institute (N01-CN43308 to S.M.L. and NCI R01-CA81488 to S.B.G.); University of Michigan's Cancer Center Support Grant (5 P30 CA46592); the Catalan Institute of Oncology and the Private Foundation of the Biomedical Research Institute of Bellvitge (IDIBELL); the Instituto de Salud Carlos III (grants PI08-1635, PI08-1359, PS09-1037); CIBERESP (CB06/02/2005); the ‘Acción Transversal del Cancer’; the Catalan Government DURSI (grant 2009SGR1489); the AECC (Spanish Association Against Cancer) Scientific Foundation.
Supplementary Material
Acknowledgments
Conflict of interest statement: None declared.
References
- 1.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 4.van Duijnhoven FJ, Bueno-De-Mesquita HB, Ferrari P, et al. Fruit, vegetables, and colorectal cancer risk: the European Prospective Investigation into Cancer and Nutrition. Am J Clin Nutr. 2009;89:1441–1452. doi: 10.3945/ajcn.2008.27120. [DOI] [PubMed] [Google Scholar]
- 5.Gerber M. Background review paper on total fat, fatty acid intake and cancers. Ann Nutr Metab. 2009;55:140–161. doi: 10.1159/000229000. [DOI] [PubMed] [Google Scholar]
- 6.Potter JD, Hunter D. Colorectal cancer. In: Adami HO, Hunter D, Trichopoulos D, editors. Textbook of Cancer Epidemiology. New York: Oxford University Press; 2008. , pp. 275–307. [Google Scholar]
- 7.Clevers H. Colon cancer—understanding how NSAIDs work. N Engl J Med. 2006;354:761–763. doi: 10.1056/NEJMcibr055457. [DOI] [PubMed] [Google Scholar]
- 8.Poynter JN, Gruber SB, Higgins PD, Almog R, Bonner JD, Rennert HS, Low M, Greenson JK, Rennert G. Statins and the risk of colorectal cancer. N Engl J Med. 2005;352:2184–2192. doi: 10.1056/NEJMoa043792. [DOI] [PubMed] [Google Scholar]
- 9.Liang PS, Chen TY, Giovannucci E. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. Int J Cancer. 2009;124:2406–2415. doi: 10.1002/ijc.24191. [DOI] [PubMed] [Google Scholar]
- 10.Park JY, Mitrou PN, Dahm CC, Luben RN, Wareham NJ, Khaw KT, Rodwell SA. Baseline alcohol consumption, type of alcoholic beverage and risk of colorectal cancer in the European Prospective Investigation into Cancer and Nutrition-Norfolk study. Cancer Epidemiol. 2009;33:347–354. doi: 10.1016/j.canep.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Poynter JN, Haile RW, Siegmund KD, et al. Associations between smoking, alcohol consumption, and colorectal cancer, overall and by tumor microsatellite instability status. Cancer Epidemiol Biomarkers Prev. 2009;18:2745–2750. doi: 10.1158/1055-9965.EPI-09-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox DG, Pontes C, Guino E, Navarro M, Osorio A, Canzian F, Moreno V. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer. 2004;91:339–343. doi: 10.1038/sj.bjc.6601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landi S, Gemignani F, Bottari F, et al. Polymorphisms within inflammatory genes and colorectal cancer. J Negat Results Biomed. 2006;5:15. doi: 10.1186/1477-5751-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, Capella G, Canzian F. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–3566. [PubMed] [Google Scholar]
- 15.Jones R, Adel-Alvarez LA, Alvarez OR, Broaddus R, Das S. Arachidonic acid and colorectal carcinogenesis. Mol Cell Biochem. 2003;253:141–149. doi: 10.1023/a:1026060426569. [DOI] [PubMed] [Google Scholar]
- 16.Gong Z, Hebert JR, Bostick RM, Deng Z, Hurley TG, Dixon DA, Nitcheva D, Xie D. Common polymorphisms in 5-lipoxygenase and 12-lipoxygenase genes and the risk of incident, sporadic colorectal adenoma. Cancer. 2007;109:849–857. doi: 10.1002/cncr.22469. [DOI] [PubMed] [Google Scholar]
- 17.Tan W, Wu J, Zhang X, et al. Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis. 2007;28:1197–1201. doi: 10.1093/carcin/bgl242. [DOI] [PubMed] [Google Scholar]
- 18.Lipkin M, Chao L, Moreno V, et al. (2010) Genetic variation in 3-Hydroxy-3-Methylglutaryl CoA reductase modifies the chemopreventive activity of statins for colorectal cancer. Cancer Prevention Research. doi: 10.1158/1940-6207.CAPR-10-0007. (in press) [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:2. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 21.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 22.Muthén LK. Mplus User’s Guide. 5th edn. Los Angeles, CA: Muthén & Muthén; 1998–2009. M.B. [Google Scholar]
- 23.Joreskog KG. Modeling development: using covariance structure models in longitudinal research. Eur Child Adolesc Psychiatry. 1996;5:8–10. doi: 10.1007/BF00538536. [DOI] [PubMed] [Google Scholar]
- 24.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 25. Enrez Gene. NCBI, NLM, NIH. http://www.ncbi.nlm.nih.gov/gene (accessed January 2010) [Google Scholar]
- 26. The GeneCards Human Gene Database. Weizmann Institute of Science. http://www.genecards.org (accessed January 2010) [Google Scholar]
- 27.Chen SN, Cilingiroglu M, Todd J, Lombardi R, Willerson JT, Gotto AM, Jr., Ballantyne CM, Marian AJ. Candidate genetic analysis of plasma high-density lipoprotein-cholesterol and severity of coronary atherosclerosis. BMC Med Genet. 2009;10:111. doi: 10.1186/1471-2350-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathiresan S, Musunuru K, Orho-Melander M. Defining the spectrum of alleles that contribute to blood lipid concentrations in humans. Curr Opin Lipidol. 2008;19:122–127. doi: 10.1097/MOL.0b013e3282f70296. [DOI] [PubMed] [Google Scholar]
- 31.Yamada Y, Matsuo H, Warita S, et al. Prediction of genetic risk for dyslipidemia. Genomics. 2007;90:551–558. doi: 10.1016/j.ygeno.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Fan YM, Raitakari OT, Kahonen M, Hutri-Kahonen N, Juonala M, Marniemi J, Viikari J, Lehtimaki T. Hepatic lipase promoter C-480T polymorphism is associated with serum lipids levels, but not subclinical atherosclerosis: the Cardiovascular Risk in Young Finns Study. Clin Genet. 2009;76:46–53. doi: 10.1111/j.1399-0004.2009.01180.x. [DOI] [PubMed] [Google Scholar]
- 33.Ghatrehsamani K, Darabi M, Rahbani M, Hashemzadeh Chaleshtory M, Farrokhi E, Noori M. Combined hepatic lipase -514C/T and cholesteryl ester transfer protein I405V polymorphisms are associated with the risk of coronary artery disease. Genet Test Mol Biomarkers. 2009;13:809–815. doi: 10.1089/gtmb.2009.0080. [DOI] [PubMed] [Google Scholar]
- 34.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visvikis-Siest S, Marteau JB. Genetic variants predisposing to cardiovascular disease. Curr Opin Lipidol. 2006;17:139–151. doi: 10.1097/01.mol.0000217895.67444.de. [DOI] [PubMed] [Google Scholar]
- 36.Zambon A, Deeb SS, Pauletto P, Crepaldi G, Brunzell JD. Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. Curr Opin Lipidol. 2003;14:179–189. doi: 10.1097/00041433-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 37.Herbey II, Ivankova NV, Katkoori VR, Mamaeva OA. Colorectal cancer and hypercholesterolemia: review of current research. Exp Oncol. 2005;27:166–178. [PubMed] [Google Scholar]
- 38.Wang J, Ban MR, Zou GY, et al. Polygenic determinants of severe hypertriglyceridemia. Hum Mol Genet. 2008;17:2894–2899. doi: 10.1093/hmg/ddn188. [DOI] [PubMed] [Google Scholar]
- 39.Su Z, Zhang S, Nebert DW, Zhang L, Huang D, Hou Y, Liao L, Xiao C. A novel allele in the promoter of the hepatic lipase is associated with increased concentration of HDL-C and decreased promoter activity. J Lipid Res. 2002;43:1595–1601. doi: 10.1194/jlr.m200046-jlr200. [DOI] [PubMed] [Google Scholar]
- 40.Isaacs A, Sayed-Tabatabaei FA, Njajou OT, Witteman JC, van Duijn CM. The -514 C->T hepatic lipase promoter region polymorphism and plasma lipids: a meta-analysis. J Clin Endocrinol Metab. 2004;89:3858–3863. doi: 10.1210/jc.2004-0188. [DOI] [PubMed] [Google Scholar]
- 41.Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb EL, Rudd MF, Sellick GS, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum Mol Genet. 2006;15:3263–3271. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- 43.Frank B, Burwinkel B, Bermejo JL, et al. Ten recently identified associations between nsSNPs and colorectal cancer could not be replicated in German families. Cancer Lett. 2008;271:153–157. doi: 10.1016/j.canlet.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 44.Hoeft B, Linseisen J, Beckmann L, et al. Polymorphisms in fatty acid metabolism-related genes are associated with colorectal cancer risk. Carcinogenesis. 2010;3:466–472. doi: 10.1093/carcin/bgp325. [DOI] [PubMed] [Google Scholar]
- 45.Barroso I, Gurnell M, Crowley VE, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 46.Gurnell M, Wentworth JM, Agostini M, et al. A dominant-negative peroxisome proliferator-activated receptor gamma (PPARgamma) mutant is a constitutive repressor and inhibits PPARgamma-mediated adipogenesis. J Biol Chem. 2000;275:5754–5759. doi: 10.1074/jbc.275.8.5754. [DOI] [PubMed] [Google Scholar]
- 47.Guan Y, Zhang Y, Breyer MD. The Role of PPARs in the Transcriptional Control of Cellular Processes. Drug News Perspect. 2002;15:147–154. doi: 10.1358/dnp.2002.15.3.840011. [DOI] [PubMed] [Google Scholar]
- 48.Tamori Y, Masugi J, Nishino N, Kasuga M. Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes. 2002;51:2045–2055. doi: 10.2337/diabetes.51.7.2045. [DOI] [PubMed] [Google Scholar]
- 49.He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voutsadakis IA. Peroxisome proliferator-activated receptor gamma (PPARgamma) and colorectal carcinogenesis. J Cancer Res Clin Oncol. 2007;133:917–928. doi: 10.1007/s00432-007-0277-y. [DOI] [PubMed] [Google Scholar]
- 51.Theodoropoulos G, Papaconstantinou I, Felekouras E, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–5043. doi: 10.3748/wjg.v12.i31.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slattery ML, Curtin K, Wolff R, Ma KN, Sweeney C, Murtaugh M, Potter JD, Levin TR, Samowitz W. PPARgamma and colon and rectal cancer: associations with specific tumor mutations, aspirin, ibuprofen and insulin-related genes (United States) Cancer Causes Control. 2006;17:239–249. doi: 10.1007/s10552-005-0411-6. [DOI] [PubMed] [Google Scholar]
- 53.Brockman JA, Gupta RA, Dubois RN. Activation of PPARgamma leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology. 1998;115:1049–1055. doi: 10.1016/s0016-5085(98)70072-1. [DOI] [PubMed] [Google Scholar]
- 54.Koh WP, Yuan JM, Van Den Berg D, Ingles SA, Yu MC. Peroxisome proliferator-activated receptor (PPAR) gamma gene polymorphisms and colorectal cancer risk among Chinese in Singapore. Carcinogenesis. 2006;27:1797–1802. doi: 10.1093/carcin/bgl001. [DOI] [PubMed] [Google Scholar]
- 55.Jiang J, Gajalakshmi V, Wang J, Kuriki K, Suzuki S, Nakamura S, Akasaka S, Ishikawa H, Tokudome S. Influence of the C161T but not Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma on colorectal cancer in an Indian population. Cancer Sci. 2005;96:507–512. doi: 10.1111/j.1349-7006.2005.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murtaugh MA, Ma KN, Caan BJ, Sweeney C, Wolff R, Samowitz WS, Potter JD, Slattery ML. Interactions of peroxisome proliferator-activated receptor {gamma} and diet in etiology of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1224–1229. doi: 10.1158/1055-9965.EPI-04-0681. [DOI] [PubMed] [Google Scholar]
- 57.Slattery ML, Murtaugh MA, Sweeney C, Ma KN, Potter JD, Caan BJ, Samowitz W. PPARgamma, energy balance, and associations with colon and rectal cancer. Nutr Cancer. 2005;51:155–161. doi: 10.1207/s15327914nc5102_5. [DOI] [PubMed] [Google Scholar]
- 58.Gong Z, Xie D, Deng Z, Bostick RM, Muga SJ, Hurley TG, Hebert JR. The PPAR{gamma} Pro12Ala polymorphism and risk for incident sporadic colorectal adenomas. Carcinogenesis. 2005;26:579–585. doi: 10.1093/carcin/bgh343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.