Abstract
Different patterns of strand asymmetry have been documented in a variety of prokaryotic genomes as well as mitochondrial genomes. Because different replication mechanisms often lead to different patterns of strand asymmetry, much can be learned of replication mechanisms by examining strand asymmetry. Here I summarize the diverse patterns of strand asymmetry among different taxonomic groups to suggest that (1) the single-origin replication may not be universal among bacterial species as the endosymbionts Wigglesworthia glossinidia, Wolbachia species, cyanobacterium Synechocystis 6803 and Mycoplasma pulmonis genomes all exhibit strand asymmetry patterns consistent with the multiple origins of replication, (2) different replication origins in some archaeal genomes leave quite different patterns of strand asymmetry, suggesting that different replication origins in the same genome may be differentially used, (3) mitochondrial genomes from representative vertebrate species share one strand asymmetry pattern consistent with the strand-displacement replication documented in mammalian mtDNA, suggesting that the mtDNA replication mechanism in mammals may be shared among all vertebrate species, and (4) mitochondrial genomes from primitive forms of metazoans such as the sponge and hydra (representing Porifera and Cnidaria, respectively), as well as those from plants, have strand asymmetry patterns similar to single-origin or multi-origin replications observed in prokaryotes and are drastically different from mitochondrial genomes from other metazoans. This may explain why sponge and hydra mitochondrial genomes, as well as plant mitochondrial genomes, evolves much slower than those from other metazoans.
Keywords: Archaea, DNA replication, deamination, GC skew, mitochondria, mutation, origin of replication, selection.
INTRODUCTION
DNA strand asymmetry refers to the differential distribution of nucleotides between the two DNA strands, e.g., one has more A or C than the other. This implies a violation of Chargaff’s parity rule 2 [1], i.e., A = T and C = G within each strand. Consequently, strand asymmetry is typically measured by nucleotide skews such as GC skew and AT skew [2-9], referred hereafter as SG and SA:
| (1) |
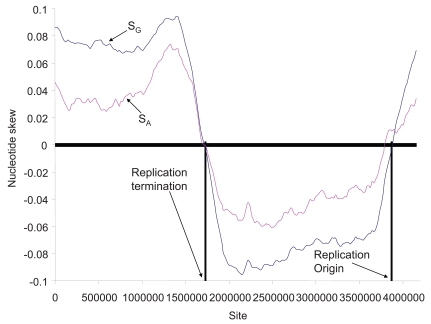
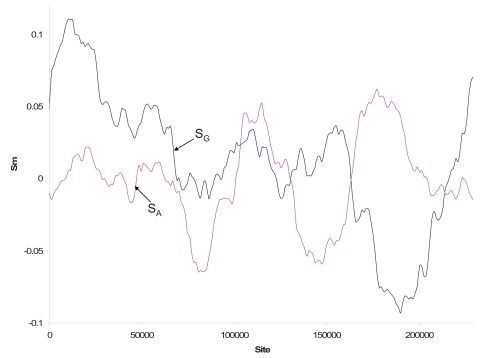
Chargaff’s parity rule 2 may be satisfied at the genomic level in spite of strong local strand asymmetry. For example, Bacillus subtilis studied by Chargaff and his colleagues [1] has its genomic nucleotide frequencies being 28.18%, 21.81%, 21.71%, and 28.30% for A, C, G and T, according to the genomic sequence deposited in GenBank (NC_000964). Thus, both SG and SA are close to 0 (SG = - 0.0021, SA = 0.0023). However, B. subtilis genomic DNA exhibits strong local asymmetry (Fig. 1). The asymmetry differs between the leading and the lagging strands, with the leading strand having more G than C and the lagging strand more C than G [2]. The strand compositional asymmetry is strong enough to allow the identification of the bacterial origin of replication (Fig. 1) whose flanking sequences change direction in GC skew [2, 10-14] or in the components of the Z-curve [11, 15-17]. For this reason, strand asymmetry is often computed locally instead of globally, with the nucleotide skews computed with a sliding window. The validity and effectiveness of the in silico methods using strand asymmetry to identify the origin of replication in prokaryotic species are well established by many experimental verifications of the predicted replication origins [18] and the utility of these methods in practice has been demonstrated by many recent studies on prokaryotic genomes [15, 17, 19-24], mitochondrial genomes [25-28] and plasmid genomes [16].
Fig. (1).
Nucleotide skew plot for the Bacillus subtilis genome (NC_000964), with window size = 537179 and step size = 21078. Each data point is at the beginning of its sliding window. The replication origin is identified as the genomic site where the GC skew (SG) changes from negative to positive and the replication termination is the site where SG changes from positive to negative.
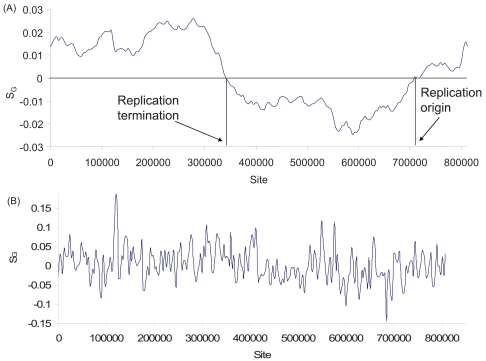
The nucleotide skews in Eq. (1) were extended in two ways. The first leads to the cumulative skew [8] which is based on summation of adjacent skew values and is equivalent to nucleotide skews with a wider sliding window than what is used to compute individual skew values. For example, the Mycoplasma pneumoniae genome has been used to illustrate the advantage of the cumulative skew method which detects the replication origin while the original GC skew method does not [8]. The real difference is not really between the two methods, but between two different widths of the sliding window (Fig. 2). A wide sliding window detects a clear change in polarity of strand asymmetry (Fig. 2A), but a narrow window fails to (Fig. 2B). In this review, the window size for skew plots is optimized with the criterion that the autocorrelation between the GC skew values of neighboring sliding windows is maximized. This method is implemented in DAMBE [29, 30].
Fig. (2).
SG plot for the Mycoplasma pneumoniae genome (NC_000912), with window size = 136694 and step size = 4081 (A), and with window size = 4000 and step size = 3000 (B).
The second extension of the nucleotide skews is to the word or motif skew [31] which is defined as
| (2) |
where m is either a nucleotide (e.g., G or A) or a motif (e.g., ACG), mrc is the reverse complement of m (mrc = C if m = G, or mrc = CGT if m = ACG), and Nx is the number of x in the sliding window (where x is either m or mrc). GC skew and AT skew are special cases of Sm when m is equal to either G or A, respectively, i.e., GC Skew is SG and AT skew is SA. It is for this reason that I have denoted GC Skew and AT Skew by SG and SA, respectively, in Eq. (1).
While transcription is known to contribute to strand asymmetry [32, 33], the most important contributor to strand asymmetry is DNA replication associated with differential strand-specific mutation bias [21, 22, 34], which is confirmed by a study that assesses the contribution of both transcription and replication to strand asymmetry [23]. Because different replication mechanisms often lead to different patterns of strand asymmetry, much can be learned of replication mechanisms by examining strand asymmetry. In this review I will summarize the different patterns of strand asymmetry in different prokaryotic and mitochondrial genomes as a basis to infer the mechanism of DNA replication that gives rise to the diversity of strand asymmetry patterns. Based on the empirical evidence, I argue that (1) the common assumption of the single-origin DNA replication in bacterial species may not be valid because bacterial genomes from the endosymbionts Wigglesworthia glossinidia and Wolbachia (from Drosophila melanogaster) exhibit patterns of strand asymmetry strongly indicative of multiple origins of replication, (2) different replication origins in some archaeal genomes leave quite different patterns of strand asymmetry, suggesting that different replication origins in the same genome may be differentially used, (3) the pattern of strand asymmetry from mammalian mitochondrial genomes is consistent with the strand-displacement model of replication well documented in mammalian mitochondria [35-40], and this pattern is shared among mitochondrial genomes from representative vertebrate species, suggesting a similar DNA replication mechanism among vertebrate mitochondrial genomes, and (4) primitive forms of metazoans such as sponge and hydra, as well as plants, have mitochondrial strand asymmetry patterns similar to prokaryotes and drastically different from higher metazoans, suggesting that mitochondrial genomes in plants and in primitive invertebrate such as sponge and hydra share the a similar replication mechanism as their bacterial ancestor with a much lower replication error rate than that in mammalian mitochondrial genomes whose strand-displacement replication is highly error-prone. This sheds light on why mitochondrial genomes from mammals evolve much faster than those from sponge, hydra and plants.
DNA REPLICATION AND STRAND ASYMMETRY IN PROKARYOTIC GENOMES
It is generally assumed that bacterial genomes have a single origin of replication [41, 42] whereas archaeal genomes tend to have multiple origins of replication [43, 44]. However, experimental verification of the exact number of replication origins is difficult and only a handful of prokaryotic species have their replication origins experimentally verified. Comparison of strand asymmetry patterns can shed lights on different replication mechanisms because different types of DNA replication typically lead to different patterns of strand asymmetry.
BACTERIAL GENOMES
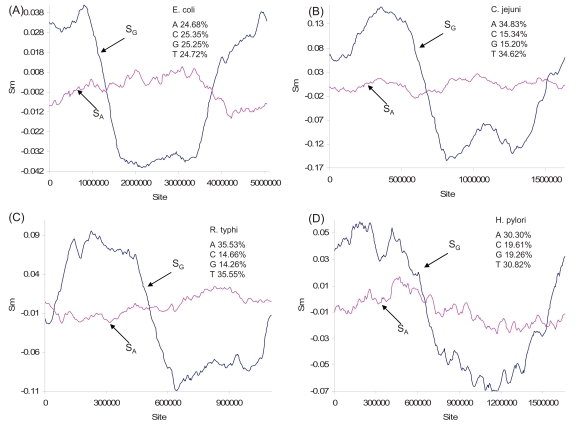
Many studies have documented strand asymmetry in eubacterial genomes associated with their single-origin mode of genome replication [2, 9, 45-47]. In general, there is an excess of G in the leading strand in many prokaryotic genomes examined [8, 17, 48-51], with the bias generally attributed to strand-biased deamination of C to U or m5C to T [9, 45, 52-54]. However, the distributions of nucleotides A and T along the leading and the lagging strands are much less consistent (Fig. 3) as has been documented before [17]. For this reason, SG has been used much more frequently in in silico identification of the replication origin and termination than SA.
Fig. (3).
Nucleotide skew plots for the genomes of (A) Escherichia coli UTI89 (NC_007946, window size = 773338 and step size = 25328), (B) Campylobacter jejuni (NC_002163, window size = 251018 and step size = 8207), (C) Rickettsia typhi wilmington (NC_006142, window size = 191456 and step size = 5557) and (D) Helicobacter pylori (NC_000915, window size = 296433 and step size = 8339). Genomic nucleotide frequencies are shown for each species.
In general, the pattern of SG is highly consistent with the single-origin replication across a diverse array of bacterial species. This has led to the common assumption that all bacterial genomes replicate with a single origin. The assumption is reinforced by the strong conservation of the molecular machinery for bacterial DNA replication. For example, the DNA replication initiation factor DnaA protein from a marine cyanobacterium (Prochlorococcus marinus CCMP1375) can specifically recognize the chromosomal origin of replication (oriC) of both E. coli and B. subtilis [55]. Thus, given that many bacterial genomes are known to replicate with a single origin of replication, and that all bacterial genomes may be replicated the same way, it is natural for us to assume that all bacterial genomes replicate with a single origin of replication.
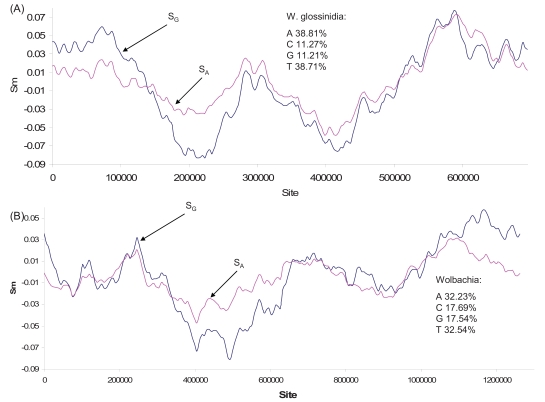
The pattern of strand asymmetry in Fig. (3), however, is not universal among bacterial species (Fig. 4). The possibility of multiple origins of replication is particularly strong in the AT-rich genome of two endosymbionts: Wigglesworthia glossinidia in tse-tse flies (Glossina brevipalpis) and Wolbachia in Drosophila melanogaster (Fig. 4). The nucleotide skew plots with multiple changes of polarity are similar to that for the yeast (Saccharomyces cerevisiae) chromosome 1 replicated with multiple origins of replication (Fig. 5). Thus, the assumption of single-origin replication in bacteria [41, 42] may be questionable.
Fig. (4).
Nucleotide skew plots for the genome of (A) Wigglesworthia glossinidia (NC_004344), with window size = 89186 and step size = 3488, and (B) Wolbachia endosymbiont (NC_002978) of Drosophila melanogaster, with window size = 167632 and step size = 6338.
Fig. (5).
Nucleotide skew plot for the yeast (Saccharomyces cerevisiae) chromosome 1 (NC_001133), with window size = 29463 and step size = 1151.
There is no strong theoretical reason against some bacterial species having multiple origins of replication, other than the probably far-fetched possibility that daughter genomes arising from multiple origins of replication may fail to segregate properly into the two daughter cells. Escherichia coli genomes with an additional oriC inserted about 1 Mb apart from the regular oriC position seem to replicate normally, with both replication origins functioning identically and with no detectable difference in generation time or cell morphology from the wild-type cells [56]. This implies that, if mutation leads to the creation of an additional ectopic replication origin in an E. coli cell, there may be no strong selection against the mutant.
While multiple origins of replication typically would lead to multiple changes in polarity in the nucleotide skew plot, one should be careful in inferring multiple origins of replication based only on the observation of multiple changes in polarity in the nucleotide skew plots, because multiple changes in polarity can result from a variety of factors. For example, horizontal gene transfer is frequent in bacterial species, and a horizontally transferred sequence segment is likely to have quite different strand asymmetry patterns from the host genome, leading to additional changes in polarity in the skew plots. In other words, multiple changes in polarity in the skew plots may not result from multiple origins, but may instead result in the recent incorporation of multiple horizontally transferred genes. Similarly, there might be heterogeneity in strand asymmetry among different genes. For example, RNA genes typically form extensive secondary structure in which stems are double stranded and requires A=T and C=G (except for cases of U/G pairs in RNA). This implies that RNA genes should have different strand asymmetry patterns than the rest of the genomes, leading to additional changes in polarity in the skew plot. Also, if an rRNA gene cluster is duplicated in the opposite strand (which is the case for Wigglesworthia glossinidia), and if the rRNA is highly conserved (which is also true in W. glossinidia), then the recipient strand will have an irregular skew value at the position of the new rRNA genes.
To alleviate these potential problems, I have generated the skew plots that included or excluded the protein-coding and rRNA genes. Such treatments do not alter the pattern of nucleotide skews in Fig. (4). While the pattern in SG is indicative of multiple origins of replication (Fig. 4), it is difficult to exclude alternative explanations. If genes switch strands frequently, then the strand asymmetry will be weak with multiple shallow peaks/valleys. This problem is particularly relevant to Wolbachia because of its mosaic genomic structure resulting from extensive recombination. My point is to highlight what is unresolved for future studies.
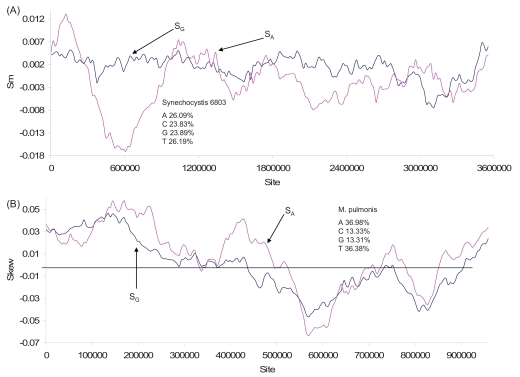
In the cyanobacterium Synechocystis sp. 6803, SG exhibits no recognizable change of polarity for any width of the sliding window. Its dnaA gene is located at sites 1350236..1351579 where no change in polarity of the strand asymmetry was observed in nearby sequence regions (Fig. 6A). While SA decreases and increases dramatically (Fig. 6A), its change is typically not indicative of the origin of replication. The nucleotide skew plot in Fig. (6A) does not favor the hypothesis that the Synechcystis sp. 6803 genome has a single origin of replication that is fired consistently in all genome replications.
Fig. (6).
Nucleotide skew plots for the genome of (A) the cyanobacterium Synechocystis 6803 (NC_000908), with window size = 436362 and step size = 17867 and (B) Mycoplasma pulmonis (NC_002771), with window size = 142301 and step size = 4819.
The nucleotide skew plots for the AT-rich Mycoplasma pulmonis genome (Fig. 6B) also do not suggest a single origin of replication because of multiple SG changes in polarity. Instead of a sharp change in polarity, there is a long stretch of the genome with SG values hovering above and below the zero line (Fig. 6B). The genome contains many putative DnaA boxes [57], which is expected given the AT-richness of the genome. The genome is also peculiar in that a plasmid carrying an oriC would, after only a few passages, integrate into the predicted genomic oriC region [57]. This could lead to multiple origins of replication clustered together, with each having the potential to fire during genome replication. Such a hypothesis would potentially explain why there is a long stretch of genomic DNA with SG values close to zero (Fig. 6B), i.e., no strand asymmetry can be established within genomic regions with closely spaced multiple replication origins.
The bacterial oriC is AT-rich and is expected to occur more frequently in AT-rich genomes. This suggests that AT-rich genomes have a greater tendency to harbor multiple origins of replication than GC-rich genomes. In this context, it is interesting to note that the bacterial species with a strong multi-origin replication signature in their strand asymmetry patterns, i.e., Mycoplasma pulmonis, Wigglesworthia glossinidia and Wolbachia are highly AT-rich genomes.
What bacterial genome would benefit from having multiple origins? If the genome is extraordinarily long, if the replication process is slow, or if the replication machinery (DNA-replication initiation and elongation proteins and enzymes) can be produced cheaply in multiple copies, then multiple replication origins would seem beneficial. Genomic data are available to address such a question.
Another point worth making in bacterial nucleotide skew plots is the diversity in the relationship between SG and SA (Figs. 1-4, 6). This diversity is unexpected given the common proposal that the main contributor to strand asymmetry is the strand-biased deamination of C to U or m5C to T during DNA replication [9, 45, 52-54]. If the strand asymmetry is maintained mainly by the C→U/T mutations, then we expect a negative relationship between SG and SA, because reductions in C and increases in T will cause both an increase in SG and a decrease in SA. Such a negative correlation is indeed observed in Buchnera aphidicola genome (not shown), but a strong positive correlation between SG and SA is also observed (e.g., all genomes in the genus Bacillus). Such a positive correlation cannot be explained by the pure C→U/T mutation bias [24, 58].
ARCHAEAL GENOMES
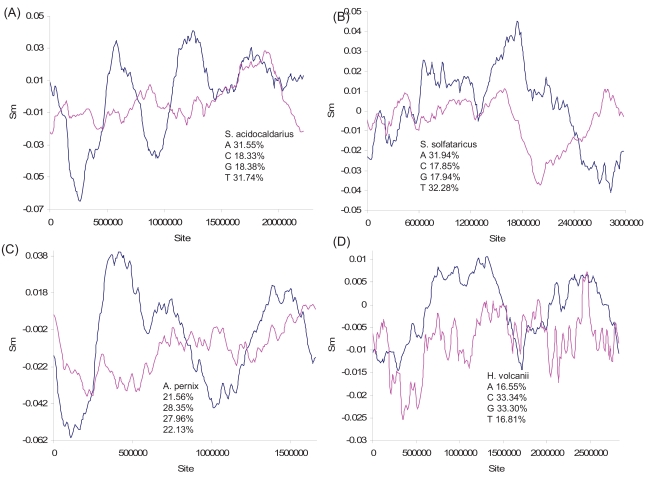
Multiple replication origins are typically assumed for archaeal genome replication [43, 44, 59]. Multiple origins of replication implies multiple changes in polarity in nucleotide skew plots, which is well exemplified by several archaeal species with experimentally verified multiple origins of replication (Fig. 7). Sulfolocus salfataricus and S. acidocaldarius both have three origins of replication [60, 61]. It is noteworthy that the SG curve in S. acidocaldarius (Fig. 7A) has valleys of different depths, similar to that for the yeast chromosome 1 (Fig. 5). These valleys of different depths suggest that some replication origins are fired more frequently than others, leading to stronger strand asymmetry than other replication origins. In eukaryotes, different replication origins are not used synchronously or equally frequently [62]. This may also be true for archaeal replication origins. Differential usage of different replication origins has been documented in Haloferax volcanii [63]. In any case, the SG pattern in Fig. (7A) casts doubt on the claim that the three replication origins in Sulfolocus species fire synchronously in each cell cycle [61].
Fig. (7).
Nucleotide skew plots for genomes of (A) Sulfolobus acidocaldarius (NC_007181, window size = 317575, step size = 11129), (B) Sulfolobus solfataricus (NC_002754), window size = 413369, step size = 14961), (C) Aeropyrum pernix (NC_000854, window size = 238220, step size = 8348), and (D) Haloferax volcanii (NC_013967), window size = 405353 and step size = 14238. The species also contains three smaller replicons whose nucleotide skew plots are not shown).
The genome of Aeropyrum pernix contains two verified origins of replication, which is consistent with the SG plot (Fig. 7C). The different peaks and valleys again suggest different firing frequencies of different origins of replication. The two origins share some homology with two of the three replication origins in Sulfolocus species [42]. This raises the question of how Sulfolocus species acquired their third replication origin, i.e., whether it arose by accumulated mutations in the genome or whether it is acquired by capturing extrachromosomal element. The finding of a viral integrase element near the replication origins lends support for the latter [42].
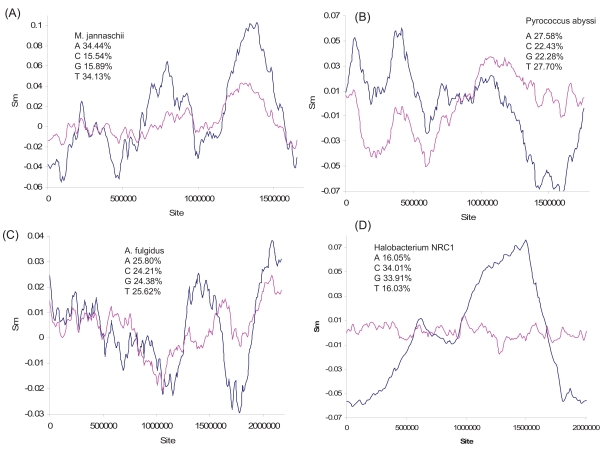
The main chromosome of the halophilic archaeon Haloferax volcanii (which also has three smaller replicons) contains two origins of replication [63], which is also suggested by the two major changes in polarity in the SG plot (Fig. 7D). The origin of replication has not been identified in the Methanococcus jannaschii genome, but the multiple changes in polarity in the SG plot (Fig. 8A) from the genome strongly suggest multiple origins of replication. The genome also exhibits multiple peaks and valleys in marker frequency distributions [64], consistent with the interpretation of multiple origins of replication. The shared feature of multiple replication origins among these taxonomically diverse archaeal species suggests that multi-origin replication is the norm in Archaea.
Fig. (8).
Nucleotide skew plot for genomes of (A) Methanococcus jannaschii (NC_000909, window size = 213047 and step size = 8324), (B) Pyrococcus abyssi (NC_000868, window size = 225116, step size = 8825), (C) Archaeoglobus fulgidus (NC_000917, window size = 299976, step size = 10892), and (D) Halobacterium NRC1 (NC_001133, window size = 307668 and step size = 10071).
Previous studies suggest only a single origin of replication in the genomes of three archaeal species: Pyrococcus abyssi [65, 66], Archaeoglobus fulgidus [64], and Halobacterium NRC1 [67]. While the SG plot of Halobacterium NRC1 is consistent with a single-origin replication (Fig. 8D), the SG plot for A. fulgidus has two peaks, suggesting two putative replication origins.
DNA REPLICATION AND STRAND ASYMMETRY IN MITOCHONDRIAL GENOMES
Mitochondrial DNA (mtDNA) replication has been studied most thoroughly in mammals. Mammalian mtDNA has two strands of different buoyant densities and consequently named the H-strand and the L-strand. The two strands have different nucleotide frequencies, with the H-strand rich in G and T and the L-strand rich in A and C, which strongly affects the codon usage of genes on the two strands [28]. This strand asymmetry can be well explained by the strand-displacement model of mtDNA replication [35-40].
During mtDNA replication, the L-strand is first used as a template to replicate the daughter H-strand, starting at the origin of replication OH, while the parental H-strand was left single-stranded for an extended period because the complete replication of mtDNA takes nearly two hours [35-37]. After about 2/3 of the daughter H-strand has been synthesized and the second origin of replication (OL) is exposed, the parental H-strand is used as a template to synthesize the daughter L-strand. Thus, different parts of the H-strands are in single-stranded form for different periods of times.
Spontaneous deamination of both A and C [52, 53] occurs frequently in human mtDNA [68]. Deamination of A leads to hypoxanthine that pairs with C, generating an A/T→G/C mutation. Deamination of C leads to U, generating C/G→U/A mutations. Among these two types of spontaneous deamination, the C→U mutation occurs more frequently than the A→G mutation [53]. In particular, the C→U mutation mediated by the spontaneous deamination occurs in single-stranded DNA more than 100 times as frequent as double-stranded DNA [54]. Note that these C→U sites will immediately be used as template to replicate the daughter L-strand, leading to a G→A mutation in the L-strand after one round of DNA duplication. Such mutation patterns are expected to leave their footprints on different parts of the H-strands left single-stranded for different periods of time.
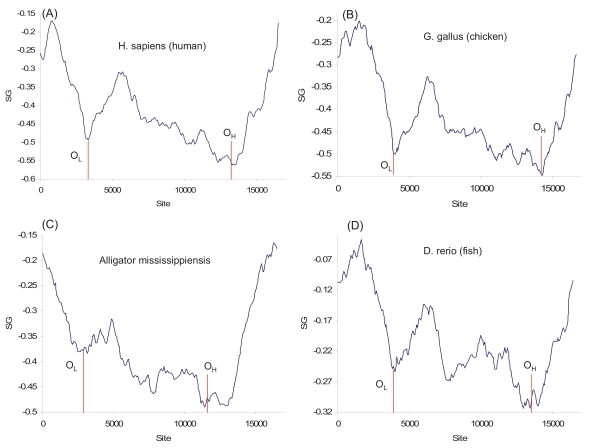
While experimental evidence for the strand-displacement model is limited to mammalian species, the nearly identical pattern of strand asymmetry among representative vertebrate species (Fig. 9) suggests that the replication mechanism is most likely shared. The reduction in SG correspond to the reduction of C in the H strand (and the associated G in the L strand), allowing us to infer the location of replication origins OH and OL (Fig. 9).
Fig. (9).
SG plots for the L-strand of the mitochondrial genomes of (A) Homo sapiens (NC_012920), (B) Gallus gallus (NC_001323), (C) Alligator mississippiensis (NC_001922), and (D) Danio rerio (NC_002333). Inferred locations of the two replication origins (OH and OL) are indicated.
The pattern of strand asymmetry among mitochondrial genomes in vertebrate species is dramatically different from those of prokaryotic species or the yeast (Figs. 1-8). In particular, the SG values for the vertebrate species are all negative (and would be all positive for the complementary strand), in contrast to the SG values of prokaryotic species which fluctuate above and below the zero line. This suggests not only local strand asymmetry, but also global strand asymmetry in vertebrate mitochondrial genomes. This is confirmed by the genomic SG, computed from genomic C and G frequencies from representative vertebrate mitochondrial genomes (Table 1). Invertebrate mitochondrial genomes also exhibit consistent and strong global strand asymmetry (Table 1), except for the most primitive ones such as the sponge (Oscarella lobularis) and the hydra (Hydra oligactis), representing Porifera and Cnidaria, respectively. The sponge and hydra mtDNAs have SG values similar to those in plant mtDNA. The two animal groups they represent are also similar to plants in having slower evolutionary rates in their mtDNA than in their nuclear genomes [69], in contrast to other metazoans whose mtDNA evolves much faster than their nuclear genomes. As evolutionary rate is largely determined by mutations introduced during DNA replication, one would expect that mtDNA in plants and in primitive invertebrates such as Porifera and Cnidaria should have DNA replication different from the strand-displacement model established for mammalian mtDNA. The nucleotide skew plots (Figs. 9, 10) are consistent with this suggestion.
Table 1.
Nucleotide Frequencies (PA, PC, PG and PT) and GC bias (SG) for Representative Metazoans and Plants. Note that SG of the Complementary Strand has the Same Value but a Different Sign
| Species | Accession | Length | PA | PC | PG | PT | SG |
|---|---|---|---|---|---|---|---|
| Oscarella lobularis | NC_014863 | 20260 | 0.333 | 0.176 | 0.173 | 0.318 | -0.006 |
| Hydra oligactis | NC_010214 | 16314 | 0.348 | 0.114 | 0.124 | 0.414 | 0.039 |
| Caenorhabditis elegans | NC_001328 | 13794 | 0.314 | 0.089 | 0.149 | 0.448 | 0.253 |
| Schistosoma japonicum | NC_002544 | 14085 | 0.249 | 0.084 | 0.206 | 0.462 | 0.422 |
| Drosophila melanogaster | NC_001709 | 19517 | 0.418 | 0.103 | 0.076 | 0.404 | -0.150 |
| Ciona intestinalis | NC_004447 | 14790 | 0.342 | 0.095 | 0.119 | 0.444 | 0.116 |
| Branchiostoma lanceolatum | NC_001912 | 15076 | 0.269 | 0.159 | 0.214 | 0.358 | 0.148 |
| Eptatretus burgeri | NC_002807 | 17168 | 0.328 | 0.229 | 0.106 | 0.337 | -0.366 |
| Mitsukurina owstoni | NC_011825 | 17743 | 0.323 | 0.254 | 0.134 | 0.290 | -0.309 |
| Danio rerio | NC_002333 | 16596 | 0.319 | 0.239 | 0.160 | 0.281 | -0.198 |
| Xenopus laevis | NC_001573 | 17553 | 0.331 | 0.235 | 0.135 | 0.300 | -0.270 |
| Alligator mississippiensis | NC_001922 | 16646 | 0.312 | 0.295 | 0.135 | 0.257 | -0.371 |
| Gallus gallus | NC_001323 | 16775 | 0.303 | 0.325 | 0.135 | 0.238 | -0.412 |
| Mus musculus | NC_005089 | 16299 | 0.345 | 0.244 | 0.124 | 0.287 | -0.328 |
| Marchantia polymorpha | NC_001660 | 186609 | 0.285 | 0.210 | 0.214 | 0.291 | 0.009 |
| Cycas taitungensis | NC_010303 | 414903 | 0.264 | 0.235 | 0.235 | 0.266 | 0.000 |
| Arabidopsis thaliana | NC_001284 | 366924 | 0.279 | 0.225 | 0.222 | 0.273 | -0.006 |
| Oryza sativa indica | NC_007886 | 491515 | 0.279 | 0.219 | 0.220 | 0.283 | 0.002 |
| Sorghum bicolor | NC_008360 | 468628 | 0.281 | 0.220 | 0.217 | 0.282 | -0.008 |
| Triticum aestivum | NC_007579 | 452528 | 0.279 | 0.221 | 0.222 | 0.278 | 0.002 |
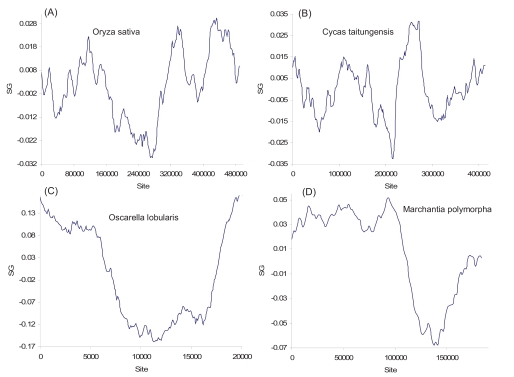
Fig. (10).
SG plots for mitochondrial genomes of (A) Oryza sativa (NC_007886), (B) Cycas taitungensis (NC_010303), (C) the sponge Oscarella lobularis (NC_014863), and (D) the liverwort Marchantia polymorpha (NC_001660).
The pattern of mtDNA strand asymmetry in higher plants (e.g., Oryza sativa and Cycas taitungensis), as characterized by the SG plots (Fig. 10A-B), suggests multiple origins of replication with the SG curve sharply crossing the zero line multiple times. This is similar to those observed in eukaryotic nuclear genomes or in archaeal genomes with multiple replication origins. Interestingly, for primitive forms of plants such as the liverwort Marchantia polymorpha, or primitive forms of metazoans such as the sponge Oscarella lobularis, the pattern of strand asymmetry (Fig. 10 C-D) is indistinguishable from what is typically seen in bacterial genomes with a single origin of replication. The SG plot of the Hydra oligactis mitochondrial genome is similar to that of Oscarella lobularis except for a slightly more pronounced secondary peak. All these patterns of strand asymmetry is dramatically different from those observed in vertebrate mtDNA (Fig. 9) and may explain the extremely slow rate of evolution between plants/sponge and higher metazoans. In other words, mitochondrial genomes in plants and primitive invertebrates may maintain the high-fidelity replication in their bacterial ancestor, whereas the error-prone strand-displacement replication evolved, likely as a secondary consequence of some advantageous traits, in a lineage leading to vertebrate mitochondrial genomes. The diversification of mtDNA replication mechanisms has not been thoroughly explored in the context of evolution.
In summary, patterns of strand asymmetry are diverse among different taxonomic groups and can tell us much about the molecular mechanism of DNA replication. The single-origin replication may not be universal among bacterial species as the endosymbionts (Wigglesworthia glossinidia, and Wolbachia species), the cyanobacterium Synechocystis 6803, and Mycoplasma pulmonis all have their genomes exhibiting strand asymmetry patterns consistent with the multi-origin mode of replication. Different replication origins in some archaeal genomes leave quite different patterns of strand asymmetry, suggesting that different replication origins in the same genome may be differentially used. Vertebrate species share one strand asymmetry pattern consistent with the strand-displacement replication documented in mammalian mtDNA, suggesting that the mtDNA replication in mammals may be universal among vertebrates. Mitochondrial genomes from primitive forms of metazoans such as the sponge and hydra, as well as those from plants have strand asymmetry patterns similar to the single-origin or multi-origin types of DNA replication observed in prokaryotes. This may explain why sponge and hydra mtDNA, as well as plant mtDNA, evolves much slower than other metazoan mtDNA.
I should finally emphasize the importance of using statistical criteria when referring to peaks or changes in polarity in the skew plots. Take SG for example, the standard deviation has been formulated as [2]:
| (3) |
A peak in the SG plot therefore refers specifically to a peak that protrude above the line of mean SG+1.96s, and a valley below the line of mean SG-1.96s, assuming the 0.05 significance level and that the window is sufficiently wide for the distribution of SG approximating the normal distribution. I encourage all programmers to include the 95% confidence intervals for nucleotide or word skew plots.
ACKNOWLEDGEMENT
This study is supported by NSERC’s Discovery Grants and the CAS/SAFEA International Partnership Program for Creative Research Teams. This project was completed when I was on sabbatical in Prof. C. Primmer’s laboratory in University of Turku.
APPENDIX 1
How to generate nucleotide skew plots in DAMBE
Download and install DAMBE which is freely available at http://dambe.bio.uottawa.ca/dambe.asp
Download any genomic sequence that you wish to generate nucleotide skew plots from, e.g., E. coli K12 genome NC_010473 from GenBank and save to your computer, say C:\data\EcoliK12.gbk. Alternatively, you can use sequence files already on your computer.
Start DAMBE, click ‘File|Open standard sequence file’. Browse to C:\data and open the ‘EcoliK12.gbk’ file.
In the ensuing ‘Process GenBank File’ dialog, the default is ‘Whole sequence’. Keep the default and click the ‘OK’ button.
In the next dialog, the default is ‘Non-protein nuc. seq’. Keep the default and click the ‘Go’ button. The sequence will be displayed
Click ‘Seq.Analysis|Genome|GC Skew’. In the ensuing dialog, check the ‘Circular genome’ checkbox and click ‘Go’ button.
Two plots will be generated, one for SG and one for SA. The window-specific data underlying the plots are also displayed.
REFERENCES
- 1.Rudner R, Karkas J D, Chargaff E. Separation of B. subtilis DNA into complementary strands. III. Direct Analysis. Proc. Natl. Acad. Sci. USA. 1968;60:921–922. doi: 10.1073/pnas.60.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobry J R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13(5):660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 3.Morton R A, Morton B R. Separating the effects of mutation and selection in producing DNA skew in bacterial chromosomes. BMC Genomics. 2007;8:369. doi: 10.1186/1471-2164-8-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimori S, Washio T, Tomita M. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genomics. 2005;6(1):26. doi: 10.1186/1471-2164-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Chambaud I, Heilig R, Ferris S, Barbe V, Samson D, Galisson F, Moszer I, Dybvig K, Wroblewski H, Viari A, Rocha E P, Blanchard A. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29(10):2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contursi P, Pisani F M, Grigoriev A, Cannio R, Bartolucci S, Rossi M. Identification and autonomous replication capability of a chromosomal replication origin from the archaeon Sulfolobus solfataricus. Extremophiles. 2004;8(5):385–391. doi: 10.1007/s00792-004-0399-y. [DOI] [PubMed] [Google Scholar]
- 8.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26(10):2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lobry J R, Sueoka N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002;3(10):1–14. doi: 10.1186/gb-2002-3-10-research0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worning P, Jensen L J, Hallin P F, Staerfeldt H H, Ussery D W. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 2006;8(2):353–361. doi: 10.1111/j.1462-2920.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Zhang C T. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 2003;302(4):728–734. doi: 10.1016/s0006-291x(03)00252-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Li K. Single-base discrimination mediated by proofreading 3' phosphorothioate-modified primers. Mol. Biotechnol. 2003;25(3):223–228. doi: 10.1385/MB:25:3:223. [DOI] [PubMed] [Google Scholar]
- 13.Frank A C, Lobry J R. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics. 2000;16(6):560–561. doi: 10.1093/bioinformatics/16.6.560. [DOI] [PubMed] [Google Scholar]
- 14.Green P, Ewing B, Miller W, Thomas P J, Green E D. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003;33(4):514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 15.Guo F B, Ou H Y, Zhang C T. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31(6):1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo F B, Yuan J B. Codon usages of genes on chromosome, and surprisingly, genes in plasmid are primarily affected by strand-specific mutational biases in Lawsonia intracellularis. DNA Res. 2009;16:91–104. doi: 10.1093/dnares/dsp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo F B, Ning L W. Strand-specific Composition Bias in Bacterial Genomes. In: Seligmann H, editor. DNA Replication-Current Advances. InTech; 2011. [Google Scholar]
- 18.Sernova N V, Gelfand M S. Identification of replication origins in prokaryotic genomes. Brief Bioinform. 2008;9(5):376–391. doi: 10.1093/bib/bbn031. [DOI] [PubMed] [Google Scholar]
- 19.Guo F B, Yu X J. Separate base usages of genes located on the leading and lagging strands in Chlamydia muridarum revealed by the Z curve method. BMC Genomics. 2007;8:366. doi: 10.1186/1471-2164-8-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C T, Zhang R, Ou H Y. The Z curve database: a graphic representation of genome sequences. Bioinformatics. 2003;19(5):593–599. doi: 10.1093/bioinformatics/btg041. [DOI] [PubMed] [Google Scholar]
- 21.Chen C L, Duquenne L, Audit B, Guilbaud G, Rappailles A, Baker A, Huvet M, d'Aubenton-Carafa Y, Hyrien O, Arneodo A, Thermes C. Replication-associated mutational asymmetry in the human genome. Mol. Biol. Evol. 2011;28(8):2327–2337. doi: 10.1093/molbev/msr056. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa K, Suzuki H, Tomita M. Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index. BMC Genomics. 2009;10:640. doi: 10.1186/1471-2164-10-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Necsulea A, Lobry J R. A new method for assessing the effect of replication on DNA base composition asymmetry. Mol. Biol. Evol. 2007;24(10):2169–2179. doi: 10.1093/molbev/msm148. [DOI] [PubMed] [Google Scholar]
- 24.Marín A, Xia X. GC skew in protein-coding genes between the leading and lagging strands in bacterial genomes: New substitution models incorporating strand bias. J. Theor. Biol. 2008;253(3):508–513. doi: 10.1016/j.jtbi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Nikolaou C, Almirantis Y. Deviations from Chargaff's second parity rule in organellar DNA Insights into the evolution of organellar genomes. Gene. 2006;381:34–41. doi: 10.1016/j.gene.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Krishnan N M, Seligmann H, Raina S Z, Pollock D D. Detecting gradients of asymmetry in site-specific substitutions in mitochondrial genomes. DNA Cell Biol. 2004;23(10):707–714. doi: 10.1089/1044549042476901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnan N M, Seligmann H, Stewart C B, De Koning A P, Pollock D D. Ancestral sequence reconstruction in primate mitochondrial DNA: compositional bias and effect on functional inference. Mol. Biol. Evol. 2004;21(10):1871–1883. doi: 10.1093/molbev/msh198. [DOI] [PubMed] [Google Scholar]
- 28.Xia X. Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes. Gene. 2005;345(1):13–20. doi: 10.1016/j.gene.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Xia X. Data analysis in molecular biology and evolution. Boston: Kluwer Academic Publishers; 2001. p. 277. [Google Scholar]
- 30.Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001;92(4):371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 31.Lopez P, Philippe H, Myllykallio H, Forterre P. Identification of putative chromosomal origins of replication in Archaea. Mol. Microbiol. 1999;32(4):883–886. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- 32.Mugal C F, von Grunberg H H, Peifer M. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol. Biol. Evol. 2009;26(1):131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- 33.Touchon M, Nicolay S, Arneodo A, d'Aubenton-Carafa Y, Thermes C. Transcription-coupled TA and GC strand asymmetries in the human genome. FEBS Lett. 2003;555(3):579–582. doi: 10.1016/s0014-5793(03)01306-1. [DOI] [PubMed] [Google Scholar]
- 34.Audit B, Nicolay S, Huvet M, Touchon M, d'Aubenton-Carafa Y, Thermes C, Arneodo A. DNA replication timing data corroborate in silico human replication origin predictions. Phys. Rev. Lett. 2007;99(24):248102. doi: 10.1103/PhysRevLett.99.248102. [DOI] [PubMed] [Google Scholar]
- 35.Clayton D A. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 36.Shadel G S, Clayton D A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 37.Clayton D A. Transcription and replication of mitochondrial DNA. Hum. Reprod. 2000;15(Suppl 2):11–17. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 38.Bogenhagen D F, Clayton D A. The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 2003;28(7):357–360. doi: 10.1016/S0968-0004(03)00132-4. [DOI] [PubMed] [Google Scholar]
- 39.Brown W M, Aiken S P. Felbamate: clinical and molecular aspects of a unique antiepileptic drug. Crit. Rev. Neurobiol. 1998;12(3):205–222. doi: 10.1615/critrevneurobiol.v12.i3.30. [DOI] [PubMed] [Google Scholar]
- 40.Brown T A, Cecconi C, Tkachuk A N, Bustamante C, Clayton D A. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19(20):2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mott M L, Berger J M. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5(5):343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 42.Robinson N P, Bell S D. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl. Acad. Sci. USA. 2007;104(14):5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelman L M, Kelman Z. Multiple origins of replication in archaea. Trends Microbiol. 2004;12(9):399–401. doi: 10.1016/j.tim.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Barry E R, Bell S D. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006;70(4):876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank A C, Lobry J R. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238(1):65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- 46.Karlin S. Bacterial DNA strand compositional asymmetry. Trends Microbiol. 1999;7(8):305–308. doi: 10.1016/s0966-842x(99)01541-3. [DOI] [PubMed] [Google Scholar]
- 47.Rocha E P, Danchin A, Viari A. Universal replication biases in bacteria. Mol. Microbiol. 1999;32(1):11–16. doi: 10.1046/j.1365-2958.1999.01334.x. [DOI] [PubMed] [Google Scholar]
- 48.McLean M J, Wolfe K H, Devine K M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998;47(6):691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 49.Freeman J M, Plasterer T N, Smith T F, Mohr S C. Patterns of Genome Organization in Bacteria. Science. 1998;279(5358):1827. [Google Scholar]
- 50.Francino M P, Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13(6):240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 51.Perriere G, Lobry J R, Thioulouse J. Correspondence discriminant analysis: a multivariate method for comparing classes of protein and nucleic acid sequences. Comput. Appl. Biosci. 1996;12(6):519–524. doi: 10.1093/bioinformatics/12.6.519. [DOI] [PubMed] [Google Scholar]
- 52.Sancar A, Sancar G B. DNA repair enzymes. Annu. Rev. Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- 53.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 54.Frederico L A, Kunkel T A, Shaw B R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry (Mosc) 1990;29(10):2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 55.Richter S, Hess W R, Krause M, Messer W. Unique organization of the dnaA region from Prochlorococcus marinus CCMP1375, a marine cyanobacterium. Mol. Gen. Genet. 1998;257(5):534–541. doi: 10.1007/s004380050679. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt D J. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. USA. 2011;108(26):E243–50. doi: 10.1073/pnas.1100874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordova C M, Lartigue C, Sirand-Pugnet P, Renaudin J, Cunha R A, Blanchard A. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J. Bacteriol. 2002;184(19):5426–5435. doi: 10.1128/JB.184.19.5426-5435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia X, Wang H C, Xie Z, Carullo M, Huang H, Hickey D A. Cytosine usage modulates the correlation between CDS length and CG content in prokaryotic genomes. Mol. Biol. Evol. 2006;23(7):1450–1454. doi: 10.1093/molbev/msl012. [DOI] [PubMed] [Google Scholar]
- 59.Kelman L M, Kelman Z. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 2003;48(3):605–615. doi: 10.1046/j.1365-2958.2003.03369.x. [DOI] [PubMed] [Google Scholar]
- 60.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. U S A. 2004;101(18):7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson N P, Dionne I, Lundgren M, Marsh V L, Bernander R, Bell S D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116(1):25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 62.Robinson N P, Bell S D. Origins of DNA replication in the three domains of life. FEBS J. 2005;272(15):3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 63.Norais C, Hawkins M, Hartman A L, Eisen J A, Myllykallio H, Allers T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007;3(5):e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maisnier-Patin S, Malandrin L, Birkeland N K, Bernander R. Chromosome replication patterns in the hyperthermophilic euryarchaea Archaeoglobus fulgidus and Methanocaldococcus (Methanococcus) jannaschii. Mol. Microbiol. 2002;45(5):1443–1450. doi: 10.1046/j.1365-2958.2002.03111.x. [DOI] [PubMed] [Google Scholar]
- 65.Matsunaga F, Forterre P, Ishino Y, Myllykallio H. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA. 2001;98(20):11152–11157. doi: 10.1073/pnas.191387498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288(5474):2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 67.Berquist B R, DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 2003;185(20):5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka M, Ozawa T. Strand asymmetry in human mitochondrial DNA mutations. Genomics. 1994;22(2):327–335. doi: 10.1006/geno.1994.1391. [DOI] [PubMed] [Google Scholar]
- 69.Shearer T L, Van Oppen M J, Romano S L, Worheide G. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria) Mol. Ecol. 2002;11(12):2475–2487. doi: 10.1046/j.1365-294x.2002.01652.x. [DOI] [PubMed] [Google Scholar]