Abstract
MicroRNAs (miRNAs) are a class of short non-coding RNAs that bind mRNAs through partial base-pair complementarity with their target genes, resulting in post-transcriptional repression of gene expression. The role of miRNAs in controlling aging processes has been uncovered recently with the discovery of miRNAs that regulate lifespan in the nematode Caenorhabditis elegans through insulin and insulin-like growth factor-1 signaling and DNA damage checkpoint factors. Furthermore, numerous miRNAs are differentially expressed during aging in C. elegans, but the specific functions of many of these miRNAs are still unknown. Recently, various miRNAs have been identified that are up- or down-regulated during mammalian aging by comparing their tissue-specific expression in younger and older mice. In addition, many miRNAs have been implicated in governing senescence in a variety of human cell lines, and the precise functions of some of these miRNAs in regulating cellular senescence have helped to elucidate mechanisms underlying aging. In this Commentary, we review the various regulatory roles of miRNAs during aging processes. We highlight how certain miRNAs can regulate aging on the level of organism lifespan, tissue aging or cellular senescence. Finally, we discuss future approaches that might be used to investigate the mechanisms by which miRNAs govern aging processes.
Key words: Aging, MicroRNAs, Senescence
Introduction
Aging is known to be affected by many environmental stimuli; however, research over the past ~25 years has indicated that genetic factors also have an important role in regulating aging. After original discoveries in the nematode Caenorhabditis elegans, it has since been observed that many genetic pathways that determine lifespan do so in a manner conserved across species, from nematodes, and even yeast, to fruit flies, mice and humans. The first established aging pathway was the insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS) pathway, primarily identified through loss-of-function alleles of the genes age-1 (which encodes a phosphoinositide 3-kinase; PI3K) and daf-2 (which encodes an insulin or IGF-1 receptor) in C. elegans that lead to extended longevity (Friedman and Johnson, 1988; Kenyon et al., 1993). Other factors, such as target of rapamycin (TOR) signaling, caloric restriction and mitochondrial respiration have also been shown to have important roles during aging (Kenyon, 2010).
On the level of an individual cell, senescence contributes to overall organism aging. Senescent cells aggregate in aging tissues and organs and have been shown to be a causal factor in aging-related disorders because their removal delays the onset of such disorders (Baker et al., 2011; Campisi, 2005). Cellular senescence is characterized as irreversible growth arrest, and in response to the exhaustion of proliferative potential there are changes in cell behavior, structure and function (Campisi, 2005). Besides displaying permanent cell cycle arrest, senescent cells also have increased autofluorescence, owing to the accumulation of non-degradable macromolecules, as well as increased activity of senescence-associated (SA) β-galactosidase (Terman and Brunk, 2006). It is well established that the presence and progressive accumulation of senescent cells contributes to aging of an organism and the onset of aging-related diseases. Cellular senescence is also thought to have evolved as a tumor suppression mechanism in which preventing metastatic cellular proliferation occurs at the expense of accumulating dysfunctional senescent cells and thus limiting longevity (Campisi, 2005). Senescence can be induced in response to factors such as telomere shortening after many rounds of cell division (replicative senescence) or to stressors, including DNA and chromatin damage, oncogenes and mitogens (stress-induced premature senescence).
MicroRNAs (miRNAs) have recently emerged as important regulators of cellular senescence and aging. miRNAs are short, non-coding RNAs that regulate the expression of mRNA targets in a sequence-specific manner by inducing mRNA degradation or translational repression (Bartel, 2004). The first miRNA to be discovered was lin-4 in C. elegans, and subsequently many other miRNAs have been discovered in the nematode (Lau et al., 2001; Lee et al., 1993; Reinhart et al., 2000). Presently, thousands of miRNAs have been identified in plants and animals, with over 1400 human miRNAs reported in miRBase (http://www.mirbase.org/) (Kozomara and Griffiths-Jones, 2011). As a class of regulatory small molecules, miRNAs exhibit a wide range of biological functions, including influences on stem cell self-renewal, cell proliferation, apoptosis and metabolism (Bartel, 2004) (Box 1).
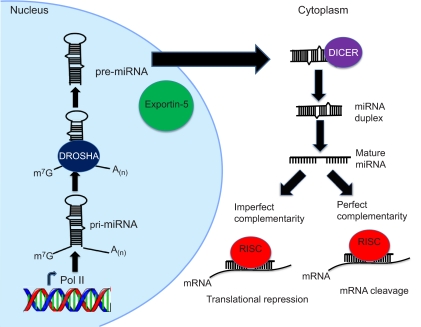
Box 1. miRNA biogenesis and post-transcriptional gene regulation
miRNAs are endogenous non-coding RNAs that post-transcriptionally repress the expression of their various target genes. miRNAs are transcribed by RNA polymerase II (RNA Pol II) and form an approximately 70-nucleotide (nt) long stem-loop primary structure or pri-miRNA, which is processed by the RNase III enzyme DROSHA into a precursor or pre-miRNA structure (see figure). The pre-miRNA is then exported from the nucleus into the cytoplasm by exportin-5 for further processing (see figure). In the cytoplasm, another RNase III enzyme, DICER, cleaves the pre-miRNA to form a linear double-stranded intermediate (see figure). The ~22-nt long mature miRNA strand obtained from the intermediate duplex is then loaded into the RNA-induced silencing complex (RISC), which also consists of the Argonaute protein and the target mRNA (see figure). miRNAs repress mRNA translation by base-pairing with partial complementarity to the 3′-UTR of target mRNAs (see figure). miRNAs can also induce mRNA cleavage through perfect base-pairing to the target mRNA; although direct mRNA degradation by miRNAs commonly occurs in plants only, a few studies have reported this phenomenon in animals (Bagga et al., 2005; Yekta et al., 2004). It is worth noting that each step of the miRNA biogenesis pathway, as well as miRNA turnover, can be potentially regulated by a variety of co-factors, which might result in differential expression patterns of mature miRNAs (reviewed by Krol et al., 2010).
The role of miRNAs in regulating aging processes has only recently been established, beginning with the report that the founding miRNA, lin-4, regulates lifespan in C. elegans (Boehm and Slack, 2005). To date, numerous miRNAs have been shown to be significantly up- or down-regulated with aging; many of these miRNAs have been identified as regulators of aging, on the tissue or organism level, or of cellular senescence, such as miR-71 in C. elegans and miR-17-92 in mammals (de Lencastre et al., 2010; Grillari et al., 2010). These developments have provided an understanding of specific factors that alter aging signaling pathways in various species from C. elegans to humans.
In this Commentary, we will first review the signaling pathways known to be involved in aging and then discuss the miRNAs that regulate longevity in C. elegans, together with recent advances uncovering roles of miRNAs in mammalian aging. We will address studies that have identified specific miRNAs that target components of signaling pathways that govern aging, as well as how these interactions might affect cellular senescence. Senescent cells can contribute to both aging and age-related diseases; although research on the mechanisms of aging-related diseases can provide important insight into the mechanisms of aging, this Commentary will only focus on aging per se. However, it should be noted that miRNAs have regulatory roles in many aging-related diseases, such as heart disease, diabetes, neurodegeneration and cancer, and many recent discussions on these topics are available (Eacker et al., 2009; Esquela-Kerscher and Slack, 2006; Jordan et al., 2011; Provost, 2010; Trajkovski et al., 2011). We will close by highlighting several fruitful areas for future investigations of miRNAs and aging. Because of the high degree of interest in this topic, we note other related recent reviews on the importance of miRNAs and other non-coding RNAs in aging (Bates et al., 2009; Chen et al., 2010; Grillari and Grillari-Voglauer, 2010; Ibanez-Ventoso and Driscoll, 2009).
Pathways involved in aging
Aging is a multifactorial process resulting from the accumulation of molecular and cellular damage over time, which leads to general physiological decline, increased mortality and eventual death. Although many environmental and stochastic factors contribute to aging processes in individuals, aging also inherently exhibits a strong genetic component (e.g. Kenyon et al., 1993). It has been proposed that gene expression patterns that modulate senescence and aging might be the result of secondary effects of mechanisms that are important during cellular growth and organism development (Campisi, 2005). Moreover, a number of these factors have been shown to regulate lifespan in C. elegans and have conserved functions across species, including signaling through the IIS pathway, heat-shock factors (HSFs), AMP-activated protein kinases (AMPKs), mitogen-activated protein kinases (MAPKs), sirtuins, target of rapamycin (TOR) and mitochondria, as discussed below.
Insulin signaling pathway and aging
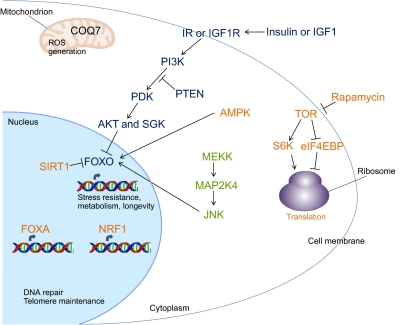
Perhaps being the most researched aging pathway, the IIS pathway was first shown to regulate longevity in C. elegans (Friedman and Johnson, 1988; Kenyon et al., 1993). Here, activation of the insulin or IGF-1 receptor, DAF-2 in C. elegans, triggers a downstream cascade, which, among other factors, is ultimately responsible for regulating organism lifespan. DAF-2 acts during development to control the decision of the animal to enter a quiescent diapause state known as dauer in crowded and resource-limited environments; during adulthood, however, DAF-2 functions to regulate lifespan (Dillin et al., 2002). On the cellular level, DAF-2 functions in response to signals from insulin-like peptides that are primarily released from neurons. DAF-2 inhibits the activity of the forkhead box FOXO transcription factor, DAF-16 in C. elegans, through a phosphorylation cascade that includes PI3K (AGE-1 in C. elegans), the phosphatidylinositol (PtdIns)-dependent kinase (PDK) (which is inhibited by DAF-18, the C. elegans homolog of the PtdInsP3 phosphatase PTEN), AKT and SGK (Fig. 1). When phosphorylated, DAF-16 remains in the cytoplasm; however, when this phosphorylation cascade is inactive, DAF-16 translocates to the nucleus. Here, it either activates or represses numerous genes that are important for controlling the cellular stress response (such as heat-shock proteins, superoxide dismutase, catalase and metallothionein), pathogen resistance and metabolism; the combined effect of these responses results in increased longevity (Murphy et al., 2003; Ogg et al., 1997).
Fig. 1.
Pathways involved in aging. There are numerous cellular mechanisms that regulate aging, many of which were first shown to regulate lifespan in C. elegans (see text for details and for names of the genetic homologs in C. elegans). Regarding hormonal signaling, the IIS pathway consists of a phosphorylation cascade triggered by the insulin receptor (IR) or the IGF1 receptor (IGF1R). This cascade prevents the translocation of the FOXO transcription factor to the nucleus. When the pathway is blocked, however, FOXO functions in the nucleus to activate or repress genes important for promoting longevity. FOXO can also be regulated through phosphorylation by AMPK and JNK, and through deacetylation by SIRT1. SIRT1 activation occurs during dietary restriction, as does inhibition of TOR signaling. TOR function can also be inhibited by rapamycin; in either case, this results in an increase in autophagy, as well as a decrease in translation through activation of eIF4EBP and suppression of S6K. The FOXA and NRF transcription factors are also required to extend longevity through dietary restriction. Additionally, factors that monitor components of the mitochondrial electron transport chain and ROS generation function during aging, such as COQ7, which is required for the production of ubiquinone (coenzyme Q).
In C. elegans, DAF-16 acts primarily in the intestine, but also in neurons, to coordinate aging throughout the entire animal (Libina et al., 2003). Studies of Drosophila FOXO have revealed that the IIS pathway functions cell non-autonomously to regulate aging in the adult Drosophila pericerebral and abdominal fat body (Hwangbo et al., 2004). In mice, the IGF1 receptor (IGF1R) has been shown to function in adult mice brain and adipose tissue to regulate lifespan (Holzenberger et al., 2003). Furthermore, analyses of numerous human cohorts have identified various polymorphisms in IIS factors that correlate with highly extended longevity, such as in IGF1 and in FOXO3 (FOXO3A genotype; FOXO function is blocked in response to IGF1 binding to IGF1R in the liver) (Suh et al., 2008; Willcox et al., 2008). Taken together, these studies illustrate a striking and conserved mechanistic role for IIS signaling in regulating aging.
Other factors involved in the aging process
Along with cues from IIS, DAF-16 can also be regulated through phosphorylation by AMPK and the MAPK family member Jun kinase (JNK), and through nicotinamide adenine dinucleotide (NAD)-dependent deacetylation by the sirtuin SIR-2 (the C. elegans homolog of the mammalian SIRT1) (Fig. 1) (Kenyon, 2010). Additionally, DAF-16, together with the transcription factor HSF-1, promotes gene expression in response to stress, which results in extended lifespan. The process of dietary restriction is also closely linked to stress resistance and results in increased lifespan across different species. Intermittent feeding in C. elegans has shown that DAF-16 is important for this response; however, various other dietary restriction models do not depend on DAF-16 (Kenyon, 2010). Inhibition of TOR signaling occurs during chronic dietary restriction or by administration of rapamycin, and this results in an increase in autophagy, as well as a decrease in translation through activation of the translational repressor eIF4EBP and downregulation of ribosomal S6 kinase (S6K) (Fig. 1). Proper function of the forkhead box transcription factor PHA-4 (a homolog of FOXA) and the basic-leucine-zipper (bZIP) transcription factor SKN-1 (a homolog of nuclear respiratory factor 1, NRF1) is also required to extend longevity upon dietary restriction (Fig. 1) (Bishop and Guarente, 2007; Panowski et al., 2007).
There are also other mechanisms independent of DAF-16 that regulate aging. As the mitochondrial electron transport chain produces reactive oxygen species (ROS) during the process of respiration, factors that monitor components of the electron transport chain have also been shown as important for longevity. For example, the clock gene CLK-1 (a homolog of COQ7) is required for the production of ubiquinone (coenzyme Q), and loss-of-function mutations in clk-1 increase lifespan independent of IIS (Fig. 1) (Lakowski and Hekimi, 1998). DNA damage checkpoint factors, such as CDC-25.1 and CHK-1, regulate cell division and proliferation but also control cell survival and maintenance in post-mitotic C. elegans adult tissues to increase longevity (Olsen et al., 2006). Finally, it has been known for many years that factors involved in telomere maintenance are required for a normal lifespan, but a highly-debated issue is whether the activation of individual genes involved in these pathways is sufficient to extend lifespan.
miRNA functions in C. elegans aging
miRNAs are a class of regulatory non-coding RNAs that have been shown to function during aging. As mentioned above, work in C. elegans has established an important role for miRNAs in regulating organism aging. It has been shown that an adult-specific knockdown of alg-1, a C. elegans Argonaute gene, results in a significantly shorter lifespan compared with that of wild-type animals, indicating that a large-scale perturbation of miRNA maturation and function affects longevity (Kato et al., 2011). Furthermore, a few miRNA mutants with altered longevity phenotypes have been reported, such as lin-4, mir-71, mir-238, mir-239 and mir-246, which are discussed in detail below.
lin-4 and lin-14 regulate lifespan
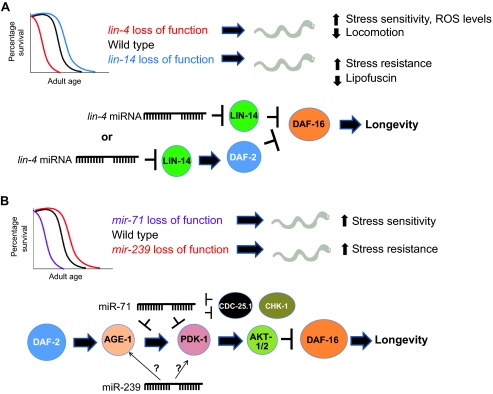
The miRNA lin-4 and its canonical target lin-14 have been shown to control lifespan in C. elegans. Specifically, a lin-4 loss-of-function mutation or a lin-14 gain-of-function mutation, which lacks the lin-4-binding sites in the lin-14 3′-untranslated region (3′-UTR), leads to decreased longevity; conversely, overexpression of lin-4 or knockdown of lin-14 [through RNA interference (RNAi) or loss-of-function alleles] results in increased longevity (Fig. 2A) (Boehm and Slack, 2005). Interestingly, knockdown of lin-14 only during adulthood is sufficient to increase longevity and suppress the lin-4 short-lived phenotype, indicating that these genes function in adulthood to modulate aging processes (Boehm and Slack, 2005). In support of these lifespan studies are analyses of stress resistance or sensitivity, as well as a demonstration of the accumulation of a potent aging biomarker known as autofluorescent lipofuscin, which is found in aging post-mitotic cells owing to the inability of lysosomes to properly degrade heavily oxidized macromolecules (Terman and Brunk, 2006). Using these types of analyses, it has been shown that lin-4 mutants display an increased rate of aging compared with that in wild-type animals, whereas lin-14 mutants display the opposite phenotypes (Fig. 2A) (Boehm and Slack, 2005). Further evidence for lin-4 function in aging comes from the observation that lin-4 mutants have increased ROS levels and decreased locomotion rates, which is indicative of an increased rate of aging (Fig. 2A) (Zhu et al., 2010).
Fig. 2.
Role of miRNAs in C. elegans aging. (A) lin-4 and lin-14 regulate lifespan in C. elegans. lin-4 loss-of-function mutants display shortened lifespans and various accelerated aging phenotypes, whereas lin-14 loss-of-function mutants exhibit extended longevity [not drawn to scale; see Boehm and Slack for lifespan curves (Boehm and Slack, 2005)]. lin-4 and LIN-14 are thought to function in the same pathway as DAF-2 and DAF-16, as illustrated in the lower part of this panel. Therefore, LIN-14 function during aging might result in direct repression of DAF-16 or indirect repression of DAF-16 through DAF-2 activation, which would in either case prevent DAF-16 from activating longevity genes. (B) miR-71, 238, 239, and 246 regulate lifespan in C. elegans. mir-71 mutants (as well as mir-238 and mir-246 mutants) have a shorter lifespan than wild-type animals, whereas mir-239 mutants have a longer lifespan [not drawn to scale; see de Lencastre et al for lifespan curves (de Lencastre et al., 2010)]. Shorter-lived C. elegans also display hallmarks of accelerated aging, such as increased stress sensitivity; the opposite holds true for longer-lived animals. The targets for some of these miRNAs have been established, as illustrated in the lower part of this panel: miR-71 targets AGE-1 and PDK-1 in the IIS pathway, as well as CDC-25.1 and CDK-1, which are involved in cell cycle checkpoints for DNA damage. Additionally, miR-239 activates AGE-1 and PDK-1 through unknown mechanisms (represented with question marks on the diagram).
lin-4 and LIN-14 function in the same pathway as DAF-2, DAF-16 and HSF-1 (Boehm and Slack, 2005). This suggests a model in which DAF-2 and LIN-14 negatively regulate DAF-16 function in parallel, or in which LIN-14 might act upstream of DAF-2 (Fig. 2A). Furthermore, DAF-16 has been shown to repress lin-4, indicating a possible negative feedback regulatory loop between DAF-16 and lin-4, although this mechanism has not yet been shown to specifically occur during aging (Baugh and Sternberg, 2006).
Other miRNAs that affect lifespan
In addition to lin-4, several other miRNAs that modulate longevity have recently been characterized, specifically miR-71, miR-238, miR-239 and miR-246 (de Lencastre et al., 2010). Interestingly, these miRNAs do not affect the developmental progression of C. elegans (Miska et al., 2007). mir-71, -238 and -246 mutants display a significantly shorter lifespan than those of wild-type animals, and overexpressing miR-71 or miR-246 increases lifespan, indicating that these miRNAs function to promote longevity (Fig. 2B) (de Lencastre et al., 2010). Conversely, mir-239 mutants exhibit an increase in lifespan compared with that of wild-type animals, and miR-239 overexpression produces the opposite effect, demonstrating that miR-239 antagonizes longevity (Fig. 2B) (de Lencastre et al., 2010). The individual miRNA deletion mutants are either sensitive (mir-71, mir-238 and mir-246) or resistant (mir-239) to heat or oxidative stress, as would be predicted by their respective lifespan phenotypes, suggesting that these mutations modulate aging per se and do not simply cause generic ‘sickness’ or other unrelated phenotypes (Fig. 2B) (de Lencastre et al., 2010). Moreover, at least two of these miRNAs, miR-71 and miR-239, target components of the IIS and DNA damage checkpoint pathways to regulate aging. miR-71 targets the PI3K AGE-1 and PDK-1 components of the IIS pathway, whereas miR-239 appears to upregulate the gene expression of these proteins through mechanisms that are yet to be determined (Fig. 2B) (de Lencastre et al., 2010). Additionally, miR-71 targets CDC-25.1 and CHK-1 in a negative-feedback loop (Fig. 2B) (de Lencastre et al., 2010). Interestingly, miR-71 is upregulated during the starvation-induced diapause state known as the dauer and during the post-dauer recovery stage, as well as during a starvation-induced early dormancy, whereas both miR-71 and miR-238 are upregulated during a starvation-induced early dormancy (Karp et al., 2011). Furthermore, miR-71 is required for both survival during, and recovery from, this early dormancy period (Zhang et al., 2011). This suggests that these miRNAs function in the cellular response to environmental stress, in addition to promoting longevity. In this context, the aging process might be viewed as a general stressor to the organism, and identifying miRNAs that function during environmental stress could help to identify miRNAs that are important for regulating aging.
There are very few examples of miRNA mutants in other model organisms that display lifespan phenotypes. Some work has been done in Drosophila; for example, miR-14, which is known to be a suppressor of cell death, reduces lifespan when mutated and has also been shown to affect metabolism and stress resistance, which are processes closely linked to aging-related mechanisms (Xu et al., 2003). Additionally, a study of Drosophila body size showed that miR-8 (homologous to miR-200 in humans) targets USH, known as ZFPM2 or FOG2 in humans, which subsequently inhibits PI3K in the IIS pathway; thus, miR-8 promotes cell growth to regulate body size cell non-autonomously (Hyun et al., 2009). mir-8-null flies have a small size and are defective in insulin signaling in the fat body, which is also the tissue in which the IIS pathway functions to modulate aging in Drosophila (Hwangbo et al., 2004; Hyun et al., 2009). As miR-8 and miR-200 regulate insulin signaling, these miRNAs might also be important for controlling Drosophila and human aging (Hyun et al., 2009).
miRNAs that are differentially regulated with aging
In addition to studies of miRNA mutants, the analysis of miRNAs that are differentially expressed with aging has led to the discovery of numerous miRNAs that might be important for controlling aging processes. For example, both microarray and deep-sequencing analyses in C. elegans have identified numerous miRNAs that are differentially expressed during aging (de Lencastre et al., 2010; Ibanez-Ventoso et al., 2006; Kato et al., 2011). In total, more than 50 of the ~200 miRNAs in C. elegans reported in miRBase are differentially expressed during aging, and more than half of these have conserved sequences in humans (Ibanez-Ventoso and Driscoll, 2009; Kozomara and Griffiths-Jones, 2011). One particular miRNA of note is miR-34, which is upregulated during aging, as well as during the dauer stage and early dormancy (de Lencastre et al., 2010; Ibanez-Ventoso et al., 2006; Karp et al., 2011; Kato et al., 2011). miR-34 has also been shown to target genes involved in aging pathways during cellular senescence, demonstrating a conserved role for this miRNA in regulating aging (e.g. Yamakuchi and Lowenstein, 2009; Zhao et al., 2010).
Although some key miRNAs, such as miR-34 and miR-71, are upregulated during aging, the vast majority of C. elegans miRNAs that are differentially expressed during aging are in fact downregulated (de Lencastre et al., 2010; Ibanez-Ventoso et al., 2006). This is in contrast to tissue-specific aging studies in mice, which have reported a general upregulation in miRNA expression during aging, indicating possibly important differences between global and tissue-specific miRNA expression, or between nematodes and mice (e.g. Li et al., 2011a; Maes et al., 2008). Nevertheless, it will be important to determine the factors that are responsible for actively up- or down-regulating miRNA expression during aging.
Roles of miRNAs in mammalian aging
Although work in C. elegans has demonstrated that miRNAs function in regulating pathways that control organism lifespan, studies focusing on mammalian aging also suggest that miRNAs have roles in regulating tissue- and cell-type-specific aging phenotypes.
Differential expression of miRNAs during tissue aging
Recent studies comparing expression levels of miRNAs in younger and older tissues in mice, humans and primates have reported that various miRNAs are differentially expressed during aging. These patterns of miRNA expression during aging appear to be generally tissue-specific, which is consistent with the tissue-specific functions of aging signaling pathways (e.g. Holzenberger et al., 2003). We discuss results from a few tissues below.
Liver
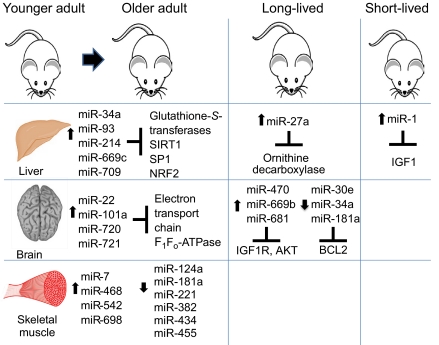
miR-669c and miR-709 levels are increased in mid-age (18-month to 33-month) murine liver tissue, whereas miR-93 and miR-214 are increased in extremely old (33-month) mice compared with 4- or 10-month-old mice (Fig. 3) (Maes et al., 2008). Furthermore, proteomic profiling indicates that miR-93, miR-214 and miR-669c all target glutathione-S-transferases, which are crucial for oxidative defense and decline in activity with age of the liver tissue (Fig. 3) (Maes et al., 2008). Additionally, proteins important for mitochondrial function or maintenance, such as a component of the cytochrome c complex (UQCRC1), have also been shown to decline in older liver tissue, and these are also targeted by miR-93, miR-214 and miR-709; this indicates a general trend of increasing miRNA expression accompanied by a decrease in those targets that normally function to maintain a healthy liver (Maes et al., 2008). Another study in rat liver has also shown an increase in miR-34a and miR-93 expression with age (Li et al., 2011b). These miRNAs target the glutathione-S-transferase MGST1 and the sirtuin SIRT1, as well as SP1 and NRF2, which are transcription factors that mediate the expression of MGST1 and SIRT1; all of these factors are important for oxidative stress defense and decrease with age (Fig. 3) (Li et al., 2011b).
Fig. 3.
Role of miRNAs in aging of mice. miR-34a, miR-93, miR-214, miR-669c and miR-709 are upregulated in aged murine liver tissue. Moreover, miR-27a is increased in the liver of the long-lived Ames dwarf mouse compared with levels in wild-type mice. miR-1 is upregulated in a progeroid mouse model and targets IGF1 in the liver. In aging brain tissue, miR-22, miR-101a, miR-720 and miR-721 are upregulated. Furthermore, miR-470, miR-669b and miR-681 have increased levels in the brains of long-lived Ames dwarf mice and growth hormone receptor knockout mice, whereas miR-30e, miR-34a and miR-181a are decreased in long-lived calorie-restricted mice. Finally, miR-7, miR-468, miR-542 and miR-698 are increased in aging murine skeletal muscle, whereas miR-124a, miR-181a, miR-221, miR-382, miR-434 and miR-455 are decreased. The targets for some of these miRNAs have been identified and shown to have numerous functions during aging, such as hormonal signaling (IIS pathway), apoptosis (BCL2) and mitochondrial activity (electron transport chain components); see text for details.
Although the above studies were carried out in the context of normal rodent aging, a study of liver aging in the Ames dwarf mouse, which lives up to 70% longer than wild-type mice owing to deficiencies in pituitary hormones, identified significantly increased levels of ten miRNAs, including miR-27a (Bates et al., 2010). These miRNAs appear to target genes that are important for glutathione metabolism and other components of intermediate metabolism, such as the urea cycle and polyamine biosynthesis. Moreover, miR-27a begins to repress expression of ornithine decarboxylase, an enzyme important for polyamine biosynthesis, earlier in the lifespan of the Ames dwarf mouse than in wild-type mice, which might be the underlying reason for its increased longevity (Fig. 3) (Bates et al., 2010). The polyamine spermidine is also important during aging, and spermidine synthase is also targeted by miR-27a (Bates et al., 2010). Spermidine naturally decreases during aging, but administration of this polyamine lengthens the lifespan of various organisms, and specifically prevents oxidative stress and promotes autophagy in mice (Eisenberg et al., 2009).
Mouse models of decreased longevity have also indicated that miRNAs have a role in tissue aging. A mouse model of Hutchinson–Gilford progeria syndrome (in which there is accelerated aging due to improper maturation of lamin A in the nuclear envelope) identified higher levels of miR-1 in the liver (as well as kidney and muscle) compared with its levels in wild-type animals (Marino et al., 2010). Interestingly, IGF1 is a validated target of miR-1, indicating that this miRNA functions through the IIS pathway by suppressing IGF1 levels in progeroid mice, and perhaps also during normal aging (Fig. 3) (Marino et al., 2010). IGF1 levels in the liver are also suppressed in the progeroid XPF mouse model for xeroderma pigmentosum, an autosomal recessive genetic disorder of DNA repair, suggesting a mechanism for how DNA damage can induce IIS to shift from monitoring growth to promoting somatic maintenance and longevity (Niedernhofer et al., 2006).
Brain
Another tissue in which miRNA expression during aging has been profiled is the mammalian brain. Similar to the trends observed during normal murine liver aging, about 70 miRNAs are upregulated during murine brain aging, starting from mid-adulthood, with this upregulation being observed from 18 months of age (Li et al., 2011a). Of these, some were identified both in liver and brain aging studies, whereas others were only upregulated during brain aging, such as miR-22, miR-101a, miR-720 and miR-721 (Fig. 3) (Li et al., 2011a). Additionally, 27 of the 70 miRNAs were predicted to target components of the mitochondrial electron transport chain and F1Fo-ATPase, which play crucial roles during oxidative phosphorylation and decrease in expression during aging, perhaps accounting for decreased respiration rates (Fig. 3) (Li et al., 2011a).
Various miRNAs have also been shown to be upregulated in the hippocampus of both long-lived Ames dwarf mice and growth-hormone-receptor-knockout strains (Liang et al., 2011). Specifically, miR-470, miR-669b and miR-681 suppress expression of IGF1R and AKT, as well as phosphorylation of AKT, which results in decreased phosphorylation of FOXO3, further highlighting the importance of the IIS aging pathway (Fig. 3) (Liang et al., 2011). However, another study has reported that under conditions of caloric restriction (known to have pro-longevity effects), instead of the expected age-dependent increase in miRNA expression, there is an age-dependent decrease in miR-30e, miR-34a and miR-181a in murine brain tissue (Fig. 3) (Khanna et al., 2011). These miRNAs have the common target BCL2, a known regulator of apoptosis, and the subsequent increase in BCL2 expression results in a decrease in apoptotic mechanisms, which might contribute to the increased neuronal survival that is apparent in these longer-lived mice (Fig. 3) (Khanna et al., 2011). Overexpression of miR-30e, miR-34a and miR-181a does indeed result in decreased BCL2 levels and increased apoptosis in transfected cell lines (Khanna et al., 2011). Furthermore, it will be important to determine whether suppression of these miRNAs can help to prevent neuronal apoptosis and promote longevity in wild-type mice not placed under caloric restriction.
Investigation of the aging human brain has further established important connections between miRNA expression and aging. For example, one study identified miRNAs that are upregulated during aging of the cortex and cerebellum of humans, chimpanzees and macaques (Persengiev et al., 2010). Most notably, the level of miR-144, which targets ataxin-1, the gene responsible for spinocerebellar ataxia type 1 (SCA1), has been shown to increase in all three species (Persengiev et al., 2010). Thus, this miRNA might be activated during aging to suppress the function of ataxin-1 in the brain, which could be important for the onset of SCA1 and other polyglutamine diseases (Persengiev et al., 2010).
Skeletal muscle
Aging of skeletal muscle has also been associated with changes in miRNA expression levels (Hamrick et al., 2010). A total of 57 miRNAs were differentially expressed in aging murine muscle tissue, including miR-221, which has been shown to regulate differentiation of myogenic precursors (Hamrick et al., 2010). Levels of miR-7, miR-468, miR-542 and miR-698 were increased the most substantially, whereas miR-124a, miR-181a, miR-221, miR-382, miR-434 and miR-455 levels were most decreased upon muscle aging (Fig. 3) (Hamrick et al., 2010). When miRNA expression in younger human skeletal muscle was compared with that in older muscle, 18 miRNAs were found to be differentially expressed, with let-7b and let-7e upregulated (Drummond et al., 2010). These let-7 family members repress the cell cycle regulators cyclin-dependent kinase 6 (CDK6), CDC25A and CDC34, as well as PAX7, which is important for satellite cell turnover; thus, miRNA-mediated regulation of these factors during aging could affect muscle cell proliferation (Drummond et al., 2010).
miRNA-mediated control of cellular senescence
In addition to measuring miRNA expression levels during tissue aging, it has been shown that miRNAs target various factors that are crucial for cellular senescence. The progressive accumulation of senescent cells contributes to tissue-specific aging and the regulation of organism lifespan; moreover, senescence, as a programmed state of growth arrest, is thought to have evolved as a protective mechanism against cancer (Campisi, 2005). miRNAs control the switch from a replicating to a senescent cell by targeting factors that respond to cellular stress, tumor suppressor pathways and signaling pathways previously shown to regulate longevity.
Stress-induced premature senescence
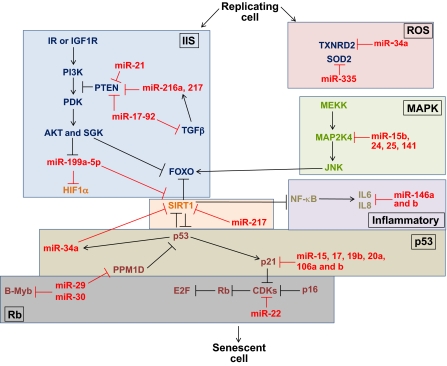
Components of the MAPK pathway, such as MAP2K4 (also known as MKK4), function during stress-induced senescence in response to mitogens. MAP2K4, which also activates JNK and p38 MAPK (MAPK14), is increased in senescent diploid fibroblasts; miR-15b, miR-24, miR-25 and miR-141 collectively target MAP2K4, but only the joint reduction of the expression levels of these miRNAs during senescence results in an upregulation of MAP2K4 (Fig. 4) (Marasa et al., 2009). The inflammatory mediators interleukin-6 and -8 (IL6 and IL8) are also secreted during stress-induced cellular senescence; miR-146a and b, which target and suppress these interleukins, have been shown to be upregulated in senescent primary fibroblasts, presumably in order to prevent excessive inflammatory response (Fig. 4) (Bhaumik et al., 2009). Oxidative stress is another factor that is important for mediating stress-induced premature senescence. When human diploid fibroblast and human trabecular meshwork cells are exposed to this type of stress, 25 miRNAs are differentially expressed (Li et al., 2009). For example, miR-182, which targets the retinoic acid receptor gamma, is upregulated (Li et al., 2009).
Fig. 4.
miRNAs regulate factors that are important for cellular senescence. Numerous miRNAs regulate gene expression of components of aging pathways (outlined in separate boxes) on the cellular level. Stress-induced cellular senescence can be regulated by the MAPK pathway and interleukins, among other factors. Regarding MAPK, miR-15b, 24, 25 and 141 collectively target MAP2K4. The inflammatory interleukins IL6 and IL8 are secreted during stress-induced cellular senescence; miR-146a and b target and suppress these interleukins. Various miRNAs also regulate the p53 and Rb pathways. p53 activates expression of miR-34a, which targets and suppresses SIRT1 (as does miR-217), and SIRT1 normally deacetylates p53 and FOXO. Moreover, miR-29 suppresses the phosphatase PPM1D, which normally dephosphorylates p53. Furthermore, transcription of the CDK inhibitor p21 is correlated with downregulation of miR-15, miR-17, miR-19b, miR-20a and miR-106a and 106b. miR-22 targets CDK6, which then targets Rb. miR-29 and miR-30 repress the MYBL2 (B-Myb) oncogene, and their upregulation during senescence depends on Rb. The miR-17-92 cluster, as well as miR-21, miR-216a and miR-217, target PTEN, which normally blocks the phosphorylation cascade of the IIS pathway. miR-17-92 additionally target TGFβ, which also induces expression of miR-216a and miR-217. Further downstream in the IIS phosphorylation cascade, AKT induces downregulation of miR-199a-5p, which normally targets HIF1α and SIRT1. Finally, a few miRNAs regulate mitochondrial senescence-associated factors: miR-34a targets the antioxidative enzyme TXNRD2, and miR-335 targets superoxide dismutase 2 (SOD2), both of which are crucial for monitoring ROS generation.
p53 and Rb pathways
In response to different stressors, cellular senescence can be induced and controlled by the tumor suppressor protein p53 and retinoblastoma (Rb)-associated protein (RB1) (Campisi, 2005). Numerous miRNAs post-transcriptionally regulate expression of the p53 pathway through regulation of its target p21 (also known as WAF1 or cyclin-dependent kinase inhibitor 1A, CDKN1A), as well as targeting the Rb pathway. Regarding the p53 pathway, there appears to be a positive-feedback loop linking the functions of miR-34a, SIRT1 and p53: p53 activates expression of miR-34a, and miR-34a targets and suppresses SIRT1, which prevents SIRT1-mediated deacetylation of p53; thus, this feedback loop promotes p53 activity (Fig. 4) (Yamakuchi and Lowenstein, 2009). Furthermore, miR-34a has been shown to induce senescence of endothelial progenitor cells by inhibiting SIRT1, which results in increased levels of acetylated FOXO1 (Zhao et al., 2010). Thus, the transcriptional activity of FOXO1 in the nucleus is no longer supressed when FOXO1 is acetylated. It has also been shown that miR-217 induces premature endothelial cell senescence through inhibition of both SIRT1 and its consequent deacetylation of FOXO1 (Fig. 4) (Menghini et al., 2009).
The general loss of mature miRNAs due to improper miRNA processing also results in an upregulation of p53 and p19 (also known as ARF, an upstream regulator of p53) and induction of premature senescence in embryonic fibroblasts; this premature senescence can be rescued by deleting the INK4A-ARF locus (encoding p16 and p19) or TP53 locus (encoding p53) (Mudhasani et al., 2008). Moreover, another study using the mouse model of Hutchinson–Gilford progeria syndrome has shown that miR-29 is upregulated during progeroid and normal aging, and that this increase in miR-29 transcription is both dependent on p53 and in response to DNA damage (Ugalde et al., 2011). Specifically, miR-29 suppresses the phosphatase PPM1D, which normally dephosphorylates p53, resulting overall in the stabilization of p53 activity (Fig. 4) (Ugalde et al., 2011). Regarding p21, it has also been shown that transcription of this CDK inhibitor is tightly correlated with downregulation of miR-15, 17, 19b, 20a and 106a and b (members of the miR-17-92 cluster, described below), in various models of replicative cell and organism aging (Fig. 4) (Hackl et al., 2010; Li et al., 2009).
In addition to regulating the p53 pathway, some of these miRNA families also affect the Rb tumor suppression pathway. miR-22 was recently shown to be upregulated in senescent human fibroblasts and epithelial cells, and this senescence-associated miRNA targets CDK6 (Fig. 4) (Xu et al., 2011). The miR-29 and miR-30 families are upregulated during cellular senescence, and their upregulation requires activation of the Rb pathway; conversely, interference with expression of miR-29 and miR-30 inhibits Rb-dependent senescence (Martinez et al., 2011). Furthermore, these miRNAs repress the MYBL2 gene (encoding the transcription factor Myb-related protein B), which has been shown to play a role in growth arrest and senescence, and is also regulated by other Rb proteins (Fig. 4) (Martinez et al., 2011).
IIS and miR-17-92
The miR-17-92 cluster, along with its paralogous clusters miR-106a-363 and miR-106b-25, regulate cellular senescence and are all downregulated in several aging models, whereas they are all upregulated in some cancers (Faraonio et al., 2011; Grillari et al., 2010). These miRNAs target PTEN, and it is thought that their downregulation during aging causes an increase in PTEN levels, which results in suppression of the IIS pathway (Fig. 4) (Grillari et al., 2010). Approximately 30 targets of miR-17-92 and its paralogous clusters have been identified, including BCL2, interferon regulatory factor (IRF), JNK2 (also known as MAPK9), transforming growth factor beta (TGFβ), hypoxia-inducible factor 1 alpha (HIF1α), p57 (also known as CDKN1C) and p27 (also known as CDKN1B), many of which are important for control of the cell cycle or apoptosis (Grillari et al., 2010). Additionally, miR-18a, miR-19a and miR-19b, from the miR-17-92 cluster, have been shown to target the extracellular matrix proteins connective tissue growth factor (CTGF) and thrombospondin 1 (TSP1); an increase in the levels of CTGF and TSP1 has been linked to decreasing levels of these miRNAs in cardiomyocytes during aging-associated heart failure (van Almen et al., 2011).
Other miRNAs beside miR-17-92 target components of the IIS pathway, namely PTEN and AKT. miRNAs that target PTEN include miR-216a and miR-217, which function in a feedback loop with TGFβ and AKT that increases cell survival through AKT activation (Kato et al., 2009). To activate AKT, TGFβ, along with miR-192, induces expression of miR-216a and 217, which both target PTEN, thus inhibiting subsequent deactivation of AKT through PTEN (Fig. 4) (Kato et al., 2009). Interestingly, miR-21 also targets PTEN and is upregulated upon AKT activation (Fig. 4) (Sayed and Abdellatif, 2010). Conversely, AKT induces downregulation of miR-199a-5p, resulting in an upregulation of its targets HIF1α and SIRT1 during hypoxia pre-conditioning, which is an oxidative stress state that affects induction of senescence (Fig. 4) (Sayed and Abdellatif, 2010).
ROS
A few miRNAs have been shown to regulate senescence-associated factors in the mitochondria. Along with miR-34a, miR-335 is upregulated in senescent kidney mesangial cells, and these miRNAs target the mitochondrial antioxidative enzymes thioredoxin reductase 2 (TXNRD2) and superoxide dismutase 2 (SOD2), respectively (Fig. 4) (Bai et al., 2011). There is a subsequent increase in ROS levels as these enzymes are downregulated, which might contribute to senescence of renal cells (Bai et al., 2011).
Taken together, the above studies indicate the importance of miRNA-mediated regulation of various responses to senescence-inducing stressors, as well as components of the p53, Rb and other pathways that mediate cellular senescence. It is evident that many different miRNAs can target the same senescence-associated factors, such as PTEN and SIRT1; in addition, some miRNAs, such as miR-34 and miR-17-92, can target multiple pathways. The role of miRNAs in targeting various factors that are crucial for the induction of cellular senescence introduces further mechanistic regulation of the transition from a replicating to a senescent cell.
Conclusions and future perspectives
The role of miRNAs in aging, although a relatively new area of research, has already shed much light on how aging and senescence-associated processes are controlled. miRNAs that regulate invertebrate aging at the organism level have been identified, and numerous miRNAs have been characterized that are differentially expressed during tissue-specific mammalian aging. Studies of cellular senescence have identified many miRNAs that not only target components of conserved aging signaling pathways but also target tumor suppressor pathways, and, as expected, exhibit completely opposite functions during senescence when compared with their roles during cancer. Analyses of miRNA functions have provided insights into how aging mechanisms are regulated at the cell, tissue and organism level, yielding a better understanding of the process of aging and its relationship to tumor suppression and the onset of aging-related diseases. With the frequent discovery of new functions for miRNAs, there is no doubt that many more miRNAs that regulate aging will be found.
We might also one day be able to measure miRNA levels to diagnose aging rates in individuals. For example, it has been shown that the expression patterns of a few miRNAs in C. elegans are predictive of the future longevity of individual animals (Pincus et al., 2011). Additionally, recent studies have reported that miRNA levels measured in peripheral blood mononuclear cells correlate with aging of both mice and humans, as well as a report of a single miRNA that can function as a marker for brain aging, neurodegeneration and memory impairment (Li, X. et al., 2011; Noren Hooten et al., 2010; Zovoilis et al., 2011). These studies suggest that miRNAs can be powerful biomarkers of aging.
Nevertheless, many important questions regarding the roles of miRNAs in aging remain. There appear to be discrepancies between reports of general upregulation, downregulation, and a mixture of both up- and down-regulation of miRNAs during aging at the cell, tissue and organism level. It will, therefore, be crucial to investigate whether different miRNAs are activated or repressed in particular contexts, such as whether there can be an overall activation of miRNA expression in a certain tissue but specific up- or down-regulation of those miRNAs at the cellular level. Interestingly, many miRNAs target genes whose functions promote longevity, whereas many other miRNAs target genes that antagonize longevity; thus, it does not appear that miRNAs, as a general class of molecules, have specific effects on aging. Instead, individual miRNAs contribute to accelerating or decelerating aging in specific contexts.
Furthermore, although much progress has been made in elucidating roles for miRNAs and their aging-associated targets, it will be crucial to understand the upstream factors that control the differential expression of miRNAs during aging. Thus far, we have only limited knowledge of how miRNA expression is controlled during aging, although insights have been made with studies of the miR-17-92 cluster and miRNA feedback loops, which have demonstrated that factors important for senescence both target and are targeted by miRNAs (Grillari et al., 2010; Kato et al., 2009; Yamakuchi and Lowenstein, 2009). Along with examining how the upstream regulation of miRNAs alters senescence, we can also investigate how experimentally modulating cellular levels of individual or multiple miRNAs can increase or decrease the rate of senescence towards potentially therapeutic levels.
Moreover, it is also important to note that not all miRNAs that are up- or down-regulated during aging necessarily play crucial roles during aging. Thus, functional studies of miRNA knockouts or overexpression are required to provide direct evidence for the role of specific miRNAs in regulating aging. Progress has been made in this regard in C. elegans by observing alterations in lifespan of lin-4, mir-71 and other mutants (Boehm and Slack, 2005; de Lencastre et al., 2010). Additionally, it has recently been shown that overexpression of several miRNAs that are upregulated during cellular senescence results in the formation of characteristic features of senescent cells (Faraonio et al., 2011). Nevertheless, studies in mammalian models that constitutively or conditionally lack or overexpress miRNAs will provide compelling evidence for any conserved aging mechanism mediated by miRNAs. These studies would elucidate how individual miRNAs contribute to tissue aging and possibly affect mammalian lifespan. In addition, in vivo analysis of knockouts or overexpression of miRNA processing machinery would illustrate how alteration of the function of essentially all mature miRNAs can affect the transition from replicating to senescent cells in various tissues, as well as potentially affecting organism lifespan, as was recently shown in one report (Mudhasani et al., 2008). One particular challenge of these in vivo models might be to separate an aging phenotype from an aging-related sickness or disease. Nevertheless, a complete understanding of how miRNAs both modulate cellular senescence and affect aging of the entire animal will allow us to determine additional roles of miRNAs in aging, as well as highlight the potential for therapeutic miRNA delivery that might modulate aging and aging-related diseases.
Acknowledgements
The authors would like to thank Zachary Pincus, Masaomi Kato and Alexandre de Lencastre for helpful discussions and comments.
Footnotes
Funding
T.S.-V. was supported by a National Institutes of Health Genomics and Proteomics Training Grant. This work was supported by the National Institutes of Health [grant number R01 AG0339201]. Deposited in PMC for release after 12 months.
References
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553-563 [DOI] [PubMed] [Google Scholar]
- Bai X. Y., Ma Y., Ding R., Fu B., Shi S., Chen X. M. (2011). miR-335 and miR-34a promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 22, 1252-1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. J., Wijshake T., Tchkonia T., LeBrasseur N. K., Childs B. G., van de Sluis B., Kirkland J. L., van Deursen J. M. (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281-297 [DOI] [PubMed] [Google Scholar]
- Bates D. J., Liang R., Li N., Wang E. (2009). The impact of noncoding RNA on the biochemical and molecular mechanisms of aging. Biochim. Biophys. Acta 1790, 970-979 [DOI] [PubMed] [Google Scholar]
- Bates D. J., Li N., Liang R., Sarojini H., An J., Masternak M. M., Bartke A., Wang E. (2010). MicroRNA regulation in ames dwarf mouse liver may contribute to delayed aging. Aging Cell 9, 1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Sternberg P. W. (2006). DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16, 780-785 [DOI] [PubMed] [Google Scholar]
- Bhaumik D., Scott G. K., Schokrpur S., Patil C. K., Orjalo A. V., Rodier F., Lithgow G. J., Campisi J. (2009). MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany N. Y.) 1, 402-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N. A., Guarente L. (2007). Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545-549 [DOI] [PubMed] [Google Scholar]
- Boehm M., Slack F. (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954-1957 [DOI] [PubMed] [Google Scholar]
- Campisi J. (2005). Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 120, 513-522 [DOI] [PubMed] [Google Scholar]
- Chen L. H., Chiou G. Y., Chen Y. W., Li H. Y., Chiou S. H. (2010). microRNA and aging: a novel modulator in regulating the aging network. Ageing Res. Rev. 9 Suppl. 1, S59-S66 [DOI] [PubMed] [Google Scholar]
- de Lencastre A., Pincus Z., Zhou K., Kato M., Lee S. S., Slack F. J. (2010). MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20, 2159-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Crawford D. K., Kenyon C. (2002). Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830-834 [DOI] [PubMed] [Google Scholar]
- Drummond M. J., McCarthy J. J., Sinha M., Spratt H. M., Volpi E., Esser K. A., Rasmussen B. B. (2010). Aging and MicroRNA expression in human skeletal muscle: A microarray and bioinformatics analysis. Physiol. Genomics 43, 595-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker S. M., Dawson T. M., Dawson V. L. (2009). Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 10, 837-841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Buttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305-1314 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F. J. (2006). Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259-269 [DOI] [PubMed] [Google Scholar]
- Faraonio R., Salerno P., Passaro F., Sedia C., Iaccio A., Bellelli R., Nappi T. C., Comegna M., Romano S., Salvatore G., et al. (2011). A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. doi: 10.1038/cdd.2011.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. B., Johnson T. E. (1988). A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J., Grillari-Voglauer R. (2010). Novel modulators of senescence, aging, and longevity: small non-coding RNAs enter the stage. Exp. Gerontol. 45, 302-311 [DOI] [PubMed] [Google Scholar]
- Grillari J., Hackl M., Grillari-Voglauer R. (2010). miR-17~92 cluster: ups and downs in cancer and aging. Biogerontology 11, 501-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M., Brunner S., Fortschegger K., Schreiner C., Micutkova L., Muck C., Laschober G. T., Lepperdinger G., Sampson N., Berger P., et al. (2010). miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 9, 291-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick M. W., Herberg S., Arounleut P., He H. Z., Shiver A., Qi R. Q., Zhou L., Isales C. M., Mi Q. S. (2010). The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem. Biophys. Res. Commun. 400, 379-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Geloen A., Even P. C., Cervera P., Le Bouc Y. (2003). IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182-187 [DOI] [PubMed] [Google Scholar]
- Hwangbo D. S., Gershman B., Tu M. P., Palmer M., Tatar M. (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562-566 [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee J. H., Jin H., Nam J., Namkoong B., Lee G., Chung J., Kim V. N. (2009). Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139, 1096-1108 [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C., Driscoll M. (2009). MicroRNAs in C. elegans aging: molecular insurance for robustness? Curr. Genomics 10, 144-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C., Yang M., Guo S., Robins H., Padgett R. W., Driscoll M. (2006). Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 5, 235-246 [DOI] [PubMed] [Google Scholar]
- Jordan S. D., Kruger M., Willmes D. M., Redemann N., Wunderlich F. T., Bronneke H. S., Merkwirth C., Kashkar H., Olkkonen V. M., Bottger T., et al. (2011). Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434-446 [DOI] [PubMed] [Google Scholar]
- Karp X., Hammell M., Ow M. C., Ambros V. (2011). Effect of life history on microRNA expression during C. elegans development. RNA 17, 639-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., et al. (2009). TGF-beta activates akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 11, 881-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Chen X., Inukai S., Zhao H., Slack F. J. (2011). Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17, 1804-1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461-464 [DOI] [PubMed] [Google Scholar]
- Kenyon C. J. (2010). The genetics of ageing. Nature 464, 504-512 [DOI] [PubMed] [Google Scholar]
- Khanna A., Muthusamy S., Liang R., Sarojini H., Wang E. (2011). Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany N. Y.) 3, 223-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152-D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597-610 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S. (1996). Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272, 1010-1013 [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858-862 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854 [DOI] [PubMed] [Google Scholar]
- Li G., Luna C., Qiu J., Epstein D. L., Gonzalez P. (2009). Alterations in microRNA expression in stress-induced cellular senescence. Mech. Ageing Dev. 130, 731-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Bates D. J., An J., Terry D. A., Wang E. (2011a). Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol. Aging 32, 944-955 [DOI] [PubMed] [Google Scholar]
- Li N., Muthusamy S., Liang R., Sarojini H., Wang E. (2011b). Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 132, 75-85 [DOI] [PubMed] [Google Scholar]
- Li X., Khanna A., Li N., Wang E. (2011). Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging (Albany, N. Y.) 3, 985-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R., Khanna A., Muthusamy S., Li N., Sarojini H., Kopchick J. J., Masternak M. M., Bartke A., Wang E. (2011). Post-transcriptional regulation of IGF1R by key microRNAs in long-lived mutant mice. Aging Cell 10, 1080-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J. R., Kenyon C. (2003). Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489-502 [DOI] [PubMed] [Google Scholar]
- Maes O. C., An J., Sarojini H., Wang E. (2008). Murine microRNAs implicated in liver functions and aging process. Mech. Ageing Dev. 129, 534-541 [DOI] [PubMed] [Google Scholar]
- Marasa B. S., Srikantan S., Masuda K., Abdelmohsen K., Kuwano Y., Yang X., Martindale J. L., Rinker-Schaeffer C. W., Gorospe M. (2009). Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci. Signal. 2, ra69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Ugalde A. P., Fernandez A. F., Osorio F. G., Fueyo A., Freije J. M., Lopez-Otin C. (2010). Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc. Natl. Acad. Sci. USA 107, 16268-16273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Cazalla D., Almstead L. L., Steitz J. A., DiMaio D. (2011). miR-29 and miR-30 regulate B-myb expression during cellular senescence. Proc. Natl. Acad. Sci. USA 108, 522-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menghini R., Casagrande V., Cardellini M., Martelli E., Terrinoni A., Amati F., Vasa-Nicotera M., Ippoliti A., Novelli G., Melino G., et al. (2009). MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120, 1524-1532 [DOI] [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., McGonagle S. M., Bartel D. P., Ambros V. R., Horvitz H. R. (2007). Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3, e215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R., Zhu Z., Hutvagner G., Eischen C. M., Lyle S., Hall L. L., Lawrence J. B., Imbalzano A. N., Jones S. N. (2008). Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 181, 1055-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283 [DOI] [PubMed] [Google Scholar]
- Niedernhofer L. J., Garinis G. A., Raams A., Lalai A. S., Robinson A. R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., et al. (2006). A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038-1043 [DOI] [PubMed] [Google Scholar]
- Noren Hooten N., Abdelmohsen K., Gorospe M., Ejiogu N., Zonderman A. B., Evans M. K. (2010). microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE 5, e10724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. (1997). The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994-999 [DOI] [PubMed] [Google Scholar]
- Olsen A., Vantipalli M. C., Lithgow G. J. (2006). Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science 312, 1381-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski S. H., Wolff S., Aguilaniu H., Durieux J., Dillin A. (2007). PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447, 550-555 [DOI] [PubMed] [Google Scholar]
- Persengiev S., Kondova I., Otting N., Koeppen A. H., Bontrop R. E. (2010). Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 32, 2316.e17-2316.e27 [DOI] [PubMed] [Google Scholar]
- Pincus Z., Smith-Vikos T., Slack F. J. (2011). MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 7, e1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P. (2010). Interpretation and applicability of microRNA data to the context of Alzheimer's and age-related diseases. Aging (Albany N. Y.) 2, 166-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901-906 [DOI] [PubMed] [Google Scholar]
- Sayed D., Abdellatif M. (2010). AKT-ing via microRNA. Cell Cycle 9, 3213-3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y., Atzmon G., Cho M. O., Hwang D., Liu B., Leahy D. J., Barzilai N., Cohen P. (2008). Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. USA 105, 3438-3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A., Brunk U. T. (2006). Oxidative stress, accumulation of biological ‘garbage’, and aging. Antioxid. Redox Signal. 8, 197-204 [DOI] [PubMed] [Google Scholar]
- Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M. H., Stoffel M. (2011). MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649-653 [DOI] [PubMed] [Google Scholar]
- Ugalde A. P., Ramsay A. J., de la Rosa J., Varela I., Marino G., Cadinanos J., Lu J., Freije J. M., Lopez-Otin C. (2011). Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 30, 2219-2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Almen G. C., Verhesen W., van Leeuwen R. E., van de Vrie M., Eurlings C., Schellings M. W., Swinnen M., Cleutjens J. P., van Zandvoort M. A., Heymans S., et al. (2011). MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 10, 769-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox B. J., Donlon T. A., He Q., Chen R., Grove J. S., Yano K., Masaki K. H., Willcox D. C., Rodriguez B., Curb J. D. (2008). FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA 105, 13987-13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Takeshita F., Hino Y., Fukunaga S., Kudo Y., Tamaki A., Matsunaga J., Takahashi R. U., Takata T., Shimamoto A., et al. (2011). MiR-22 represses cancer progression by inducing cellular senescence. J. Cell Biol. 193, 409-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A. (2003). The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13, 790-795 [DOI] [PubMed] [Google Scholar]
- Yamakuchi M., Lowenstein C. J. (2009). MiR-34, SIRT1 and p53: the feedback loop. Cell. Cycle 8, 712-715 [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I. H., Bartel D. P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594-596 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zabinsky R., Teng Y., Cui M., Han M. (2011). microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA 108, 17997-18002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Li J., Chen A. F. (2010). MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am. J. Physiol. Endocrinol. Metab. 299, E110-E116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Ji C. B., Zhang C. M., Gao C. L., Zhu J. G., Qin D. N., Kou C. Z., Zhu G. Z., Shi C. M., Guo X. R. (2010). The lin-4 gene controls fat accumulation and longevity in Caenorhabditis elegans. Int. J. Mol. Sci. 11, 4814-4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A., Agbemenyah H. Y., Agis-Balboa R. C., Stilling R. M., Edbauer D., Rao P., Farinelli L., Delalle I., Schmitt A., Falkai P., et al. (2011). MicroRNA-34c is a novel target to treat dementias. EMBO J. 30, 4299-4308 [DOI] [PMC free article] [PubMed] [Google Scholar]