Abstract
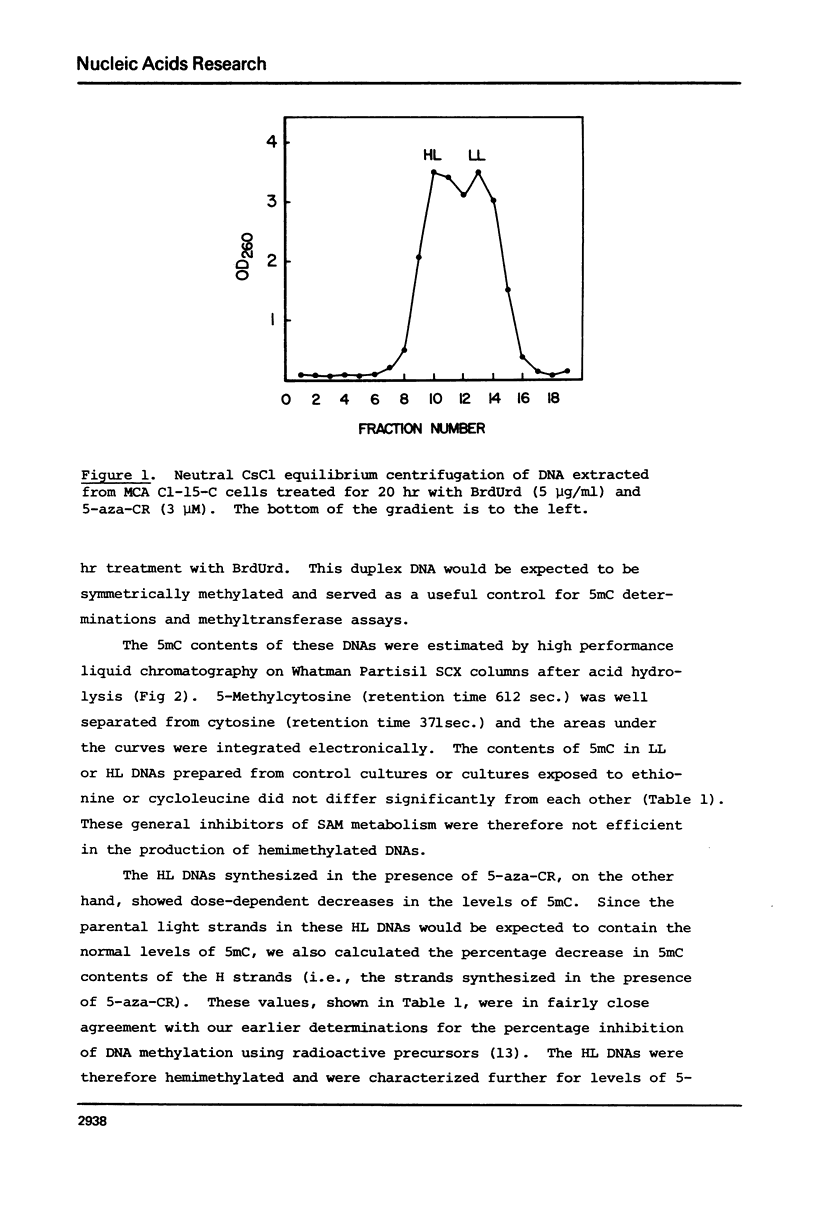
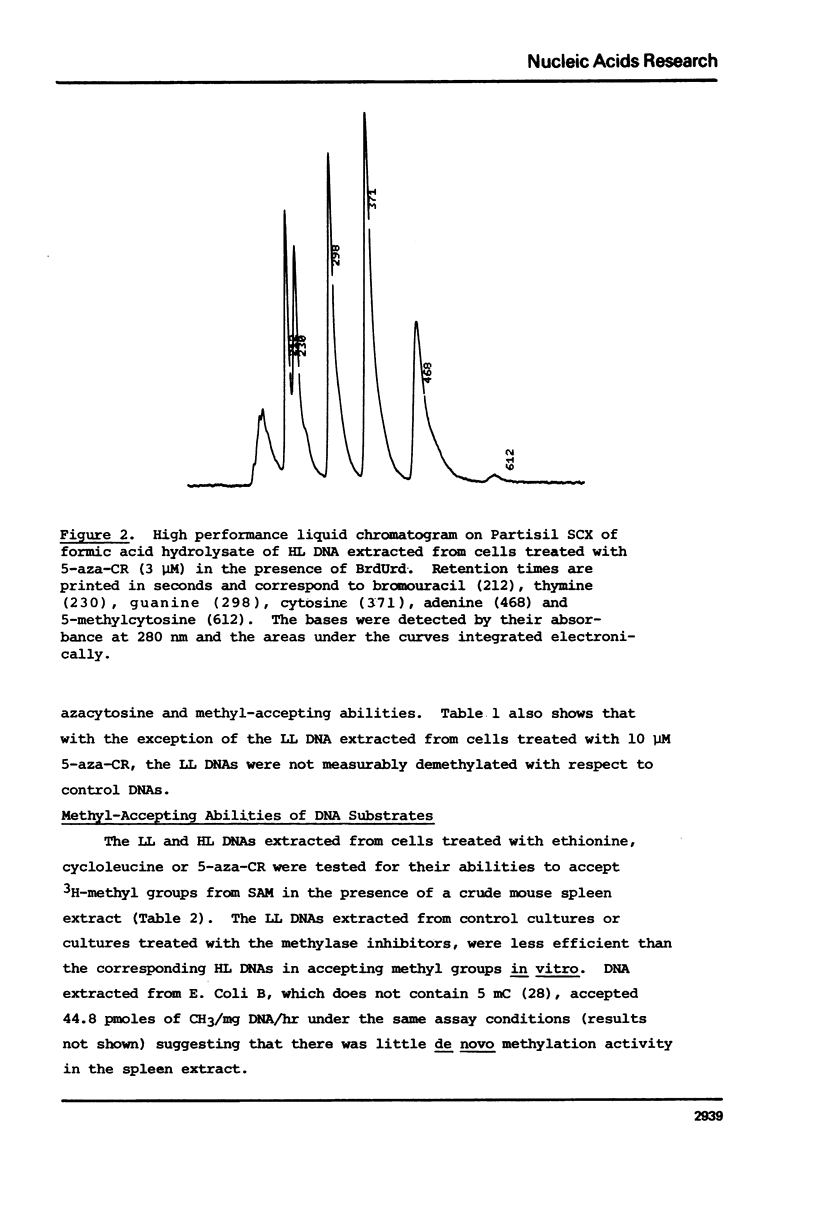
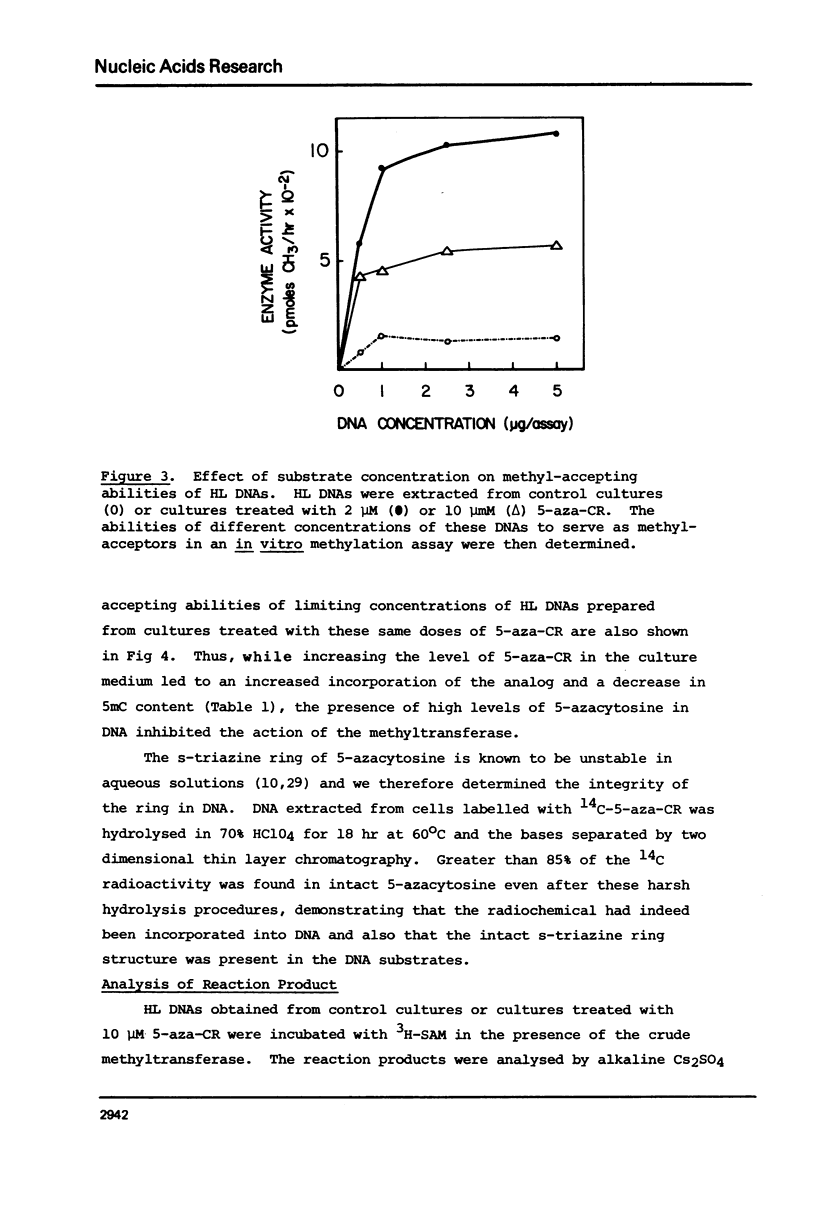
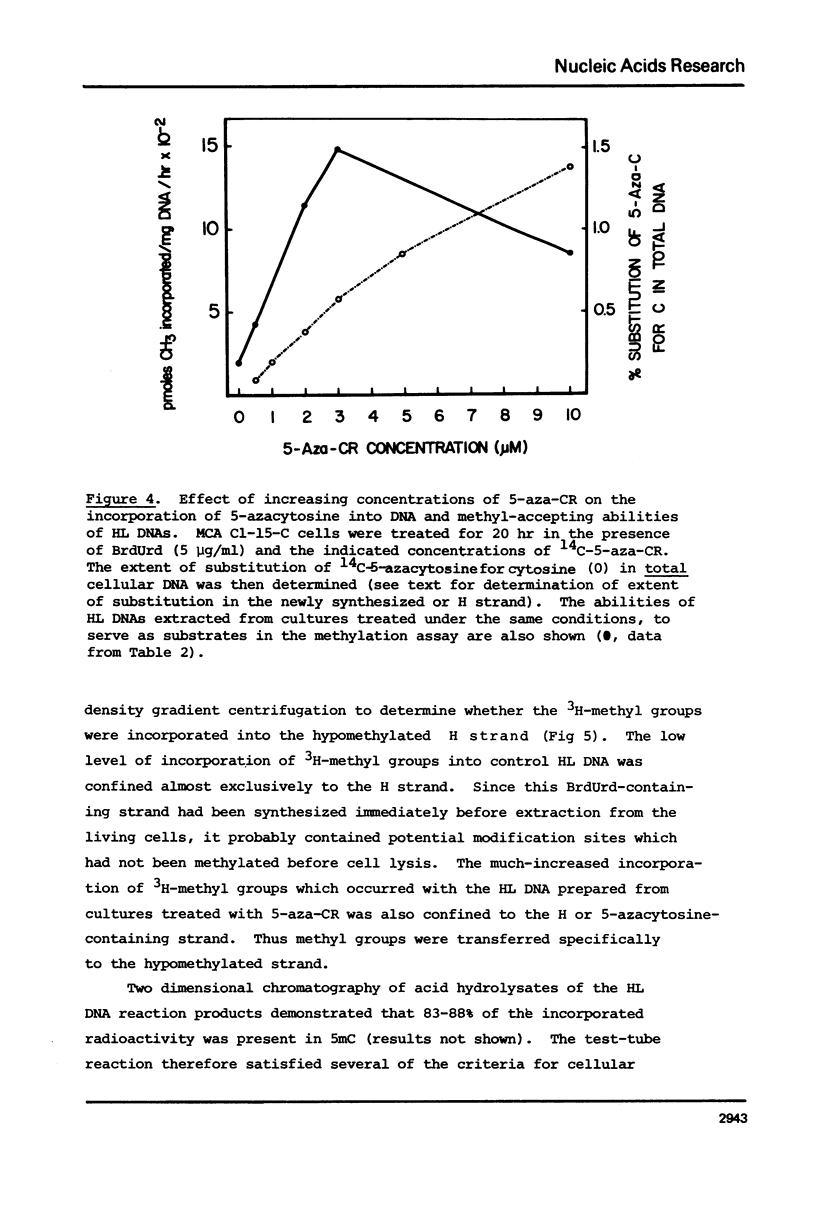
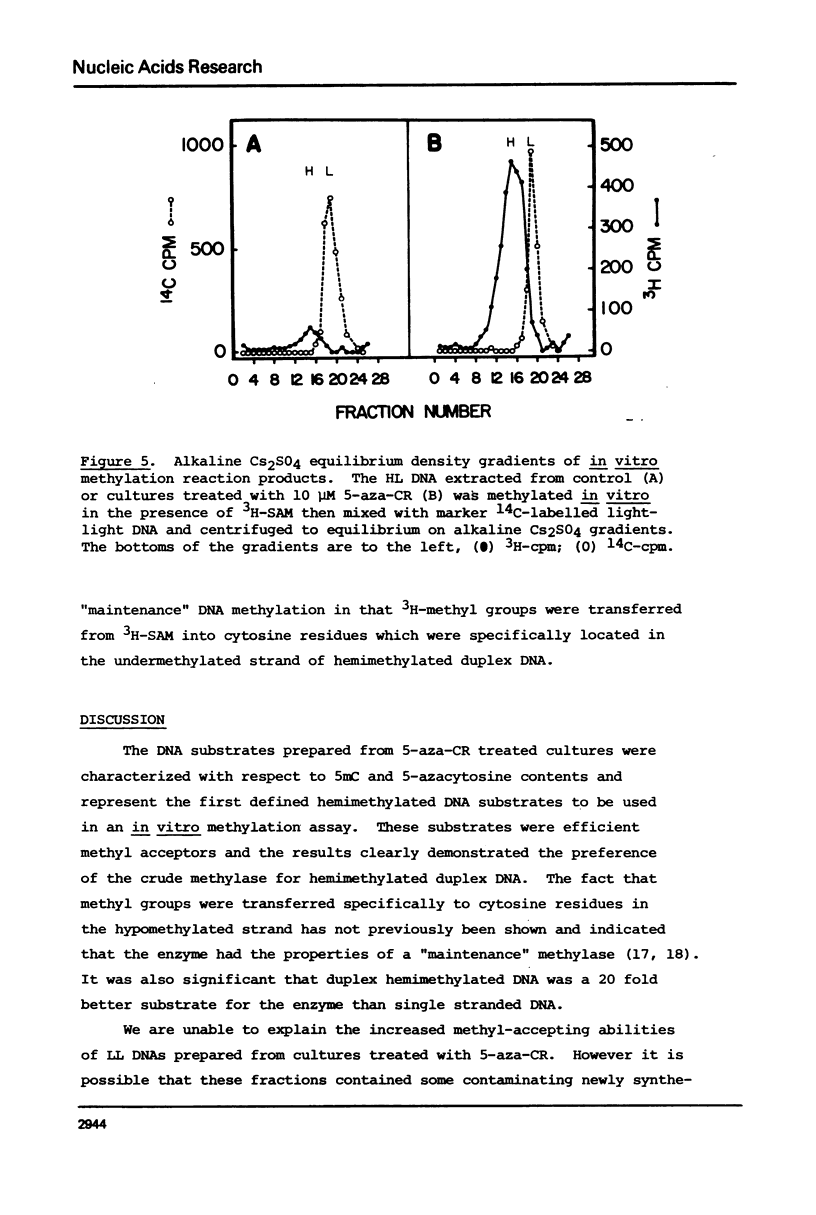
Duplex heavy-light (HL) DNAs synthesized in the presence of brdUrd and methylation inhibitors were separated from bulk cellular DNA by CsCl density gradient centrifugation and analysed for 5-methylcytosine (5mC) contents by HPLC. DNAs synthesized in the presence of 5 mM ethionine or 2 mg/ml cycloleucine were not detectably hypomethylated, was undermethylated with respect to control DNA. The heavy, or H-strand, in which up to 5% of the cytosine residues were replaced by intact 5-azacytosine, was undermethylated and the HL duplex DNA was therefore strand asymmetrically methylated. This duplex DNA served as an efficient substrate for a crude DNA methyltransferase preparation which transferred the methyl group from S-adenosylmethionine specifically into cytosine residues within the hypomethylated H strand. Increasing levels of incorporated 5-azacytosine inhibited the action of the methyltransferase suggesting that incorporation of 5-azacytosine into DNA may be responsible for the inhibitory effect of 5-azacytidine on DNA methylation.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham G. N., Scaletta C., Vaughan J. H. Modified diphenylamine reaction for increased sensitivity. Anal Biochem. 1972 Oct;49(2):547–549. doi: 10.1016/0003-2697(72)90460-5. [DOI] [PubMed] [Google Scholar]
- Adams R. L., McKay E. L., Craig L. M., Burdon R. H. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979 Feb 27;561(2):345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Price P., Pedrinan L., Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977 Nov 15;81(1):53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Weich N., Schoenbrun B., Schneiderman N., Acs G. Hypomethylation of DNA during differentiation of Friend erythroleukemia cells. J Cell Biol. 1980 Aug;86(2):366–370. doi: 10.1083/jcb.86.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides P. G., Jones P. A., Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977 May 26;267(5609):364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- Constantinides P. G., Taylor S. M., Jones P. A. Phenotypic conversion of cultured mouse embryo cells by aza pyrimidine nucleosides. Dev Biol. 1978 Sep;66(1):57–71. doi: 10.1016/0012-1606(78)90273-7. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Notari R. E., DeYoung J. L. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975 Jul;64(7):1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Reznikoff C. A., Bertram J. S., Brankow D. W., Heidelberger C. Quantitative and qualitative studies of chemical transformation of cloned C3H mouse embryo cells sensitive to postconfluence inhibition of cell division. Cancer Res. 1973 Dec;33(12):3239–3249. [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Stellwagen R. H., Roberts-Ems J., Riggs A. D. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J Biol Chem. 1977 Aug 10;252(15):5509–5513. [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Hibasami H., Nagai J., Ikeda T. Effect of 5-azacytidine on DNA methylation in Ehrlich's ascites tumor cells. Aust J Exp Biol Med Sci. 1980 Aug;58(4):391–396. doi: 10.1038/icb.1980.39. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979 Aug;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]


