Summary
Recent advances of biological drugs have broadened the scope of therapeutic targets for a variety of human diseases. This holds true for dozens of RNA-based therapeutics currently under clinical investigation for diseases ranging from genetic disorders to HIV infection to various cancers. These emerging drugs, which include therapeutic ribozymes, aptamers, and small interfering RNAs (siRNAs), demonstrate the unprecedented versatility of RNA. However, RNA is inherently unstable, potentially immunogenic, and typically requires a delivery vehicle for efficient transport to the targeted cells. These issues have hindered the clinical progress of some RNA-based drugs and have contributed to mixed results in clinical testing. Nevertheless, promising results from recent clinical trials suggest that these barriers may be overcome with improved synthetic delivery carriers and chemical modifications of the RNA therapeutics. This review focuses on the clinical results of siRNA, RNA aptamer, and ribozyme therapeutics and the prospects for future successes.
Keywords: RNAi, siRNA, shRNA, miRNA, aptamer, ribozyme, clinical trial, delivery
Introduction
Since the milestone discoveries of catalytic RNAs in the early 1980s and RNA interference in the late 1990s, the biological understanding of RNA has evolved from simply an intermediate between DNA and protein to a dynamic and versatile molecule that regulates the functions of genes and cells in all living organisms [1–3]. These and similar breakthroughs have led to the emergence of numerous types of RNA-based therapeutics that broaden the range of “drug-able” targets beyond the scope of existing pharmacological drugs [4]. RNA-based therapeutics can be classified by the mechanism of activity, and include inhibitors of mRNA translation (antisense), the agents of RNA interference (RNAi), catalytically active RNA molecules (ribozymes), and RNAs that bind proteins and other molecular ligands (aptamers).
Despite a number of hurdles encountered along the way, more than 50 RNA or RNA-derived therapeutics have reached clinical testing. Challenges with the delivery, specificity, stability, and immune activation of RNA therapeutics have spawned improvements in synthetic and natural nucleic acid carriers and the development of chemically modified oligonucleotides [5]. In this review, we will discuss many of these refinements and highlight several promising therapeutics currently in the clinic.
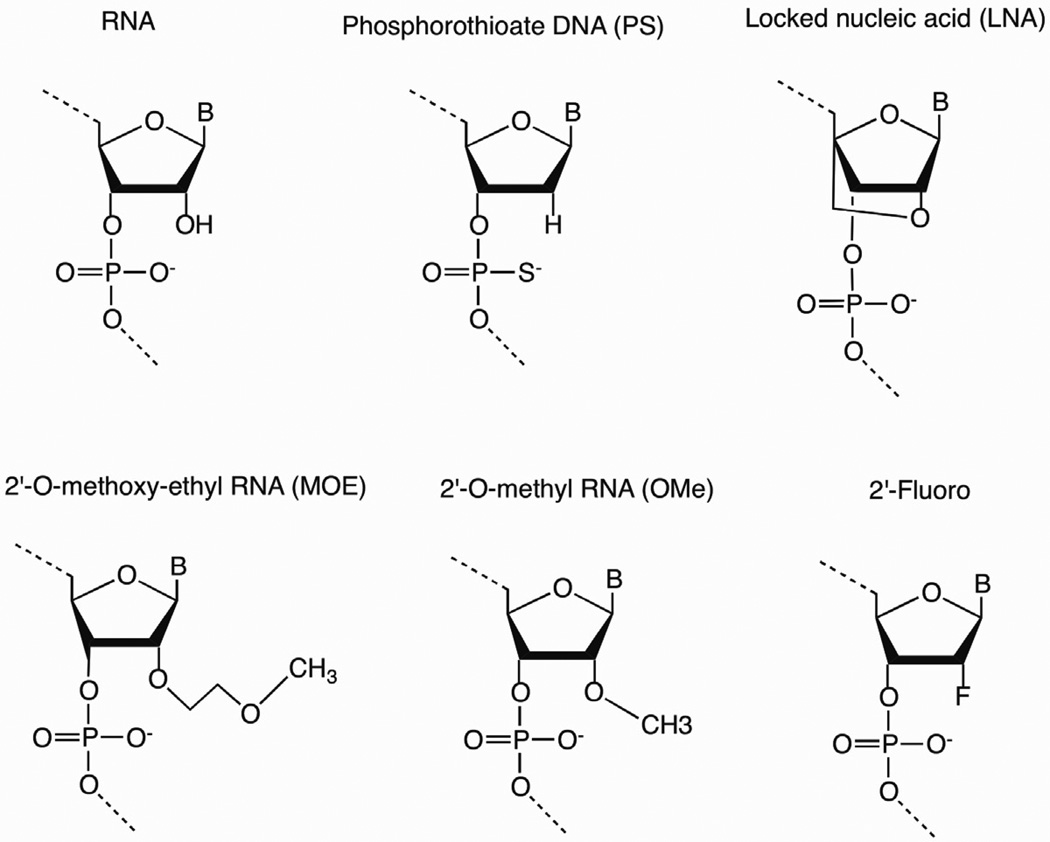
RNA is unstable in vivo due to the plethora of ribonucleases in serum and in cells, and chemical modifications can enhance desired properties without reducing activity. Chemical modifications to small interfering RNAs (siRNAs), aptamers, ribozymes, antisense (AS) oligonucleotides (ONs), and miRNAs may improve the pharmacokinetic (PK), pharmacodynamic (PD) properties and reduce immunogenicity. Such modifications in short synthetic ONs include changes in the sugar, base, or backbone and may increase target affinity and specificity, decrease susceptibility to nuclease degradation, improve PK, and improve RNAi silencing efficiency. Recent advances in the process of synthesizing modified RNA and DNA molecules have increased the efficiency and reliability of manufacturing while also reducing production costs [6]. While dozens of different sugar, base, and backbone modifications are available by ON synthesis, the variety of chemical modifications for RNA-derived ONs destined for the clinic include phosphorothioate (PS) backbone modification; 2’-O-methyl (2’-OMe), 2’-fluoro (2’-F), 2’-O-methoxyethyl (2’-MOE) sugar substitutions; 2’-O, 4’-C-methylene linked bicyclic ribonucleotides known as a locked nucleic acid (LNA); and L-RNA (enantiomer of natural RNA) ONs known as spiegelmers (Figure 1) [7].
Figure 1.
Common chemical modifications of therapeutic nucleic acid analogs. The unmodified RNA structure is shown next to backbone (5’-phosphorothioate), LNA, and 2’-substitutions (2’-O-methoxy-ethyl, 2’-O-methyl, and 2’-fluoro).
The PS backbone modification, in which the nonbridging phosphate oxygen atom is replaced by a sulfur atom, was one of the earliest ON modifications and remains widely used in DNA antisense therapeutics, and to a lesser degree, in aptamers and siRNAs [7]. This simple and inexpensive chemical modification improves resistance to nucleolytic degradation, elicits RNase H-mediated cleavage of the target mRNA for antisense applications, and increases affinity for plasma proteins to hinder renal clearance of the ON [6, 8]. Chemical substitutions at the 2’-hydroxyl group with 2’-OMe, 2’-F, or 2’-MOE groups often improve the ON potency, stability, and overall PK and PD properties. Even greater potency and stability improvements are observed with the LNA modification [6]. Collectively, modifications at the 2’ position of the sugar ring – including 2’-OMe, 2’-F, 2’-MOE, and LNA – confer the ON to adopt an RNA-like C3’-endo (N-type) sugar pucker, which is the most energy-favorable conformation of RNA. Thus, such modifications increase Watson-Crick binding affinity and, due to the proximity of the 2’-substituent and the 3’-phosphate, improve nuclease resistance [8]. In contrast to the aforementioned modifications, spiegelmers do not contain any chemical substitutions, but rather are enantiomers of natural RNAs and thus are utilized as nuclease-resistant aptamers.
Inhibitors of mRNA translation: Antisense ONs
Sequence-specific antisense ONs inhibit gene expression by altering mRNA splicing, arresting mRNA translation, and inducing degradation of targeted mRNA by RNase H. Like other RNA- and DNA-derived therapeutics, antisense ONs often include chemical modifications to the backbone, base, or sugar to enhance the properties of the drug, such as PS backbones, 2’-O-Me, 2’-F, 2’-MOE, and LNA substitutions. A detailed survey of these technologies is beyond the scope of this review due to the wide variety of RNA and DNA-based antisense therapeutics. However, recent reviews have addressed the current status of clinical antisense drugs [6, 8].
Though most current antisense therapeutics target mRNA, one clinical drug employs antisense technology to inhibit an endogenous microRNA (miRNA). The dysregulation of endogenous miRNAs has been linked to numerous diseases, including many types of cancers [9]. Emerging therapeutic strategies to regulate miRNA activity include antisense-miRNA ONs (antagomirs) and RNA competitive inhibitors or decoys (miRNA sponges) [10]. Santaris Pharma A/S developed miravirsen (SPC3649), a locked nucleic acid (LNA)-modified ON that specifically inhibits the endogenous microRNA-122 (miR-122), a liver specific miRNA required for the infection of Hepatitis C virus (HCV). Given the high mutation rate of HCV, miravirsen targets a critical host factor and thus may provide an elevated barrier for emergence of viral resistance [11]. Two completed Phase I trials (NCT00688012 and NCT00979927) have indicated that the drug is well tolerated and safe. Miravirsen has recently advanced into a Phase IIa clinical study (NCT01200420) to test safety, tolerability and efficacy for treatment-naïve patients with chronic HCV genotype 1 infection.
Agents of RNA Interference
The cellular process of RNA interference (RNAi) uses small RNAs to silence gene expression through post-transcriptional gene silencing (PTGS) or transcriptional gene silencing (TGS), though TGS pathway is not currently explored for clinical purposes [12]. PTGS is regulated by two distinct mechanisms: translational repression and degradation of mRNAs with imperfect complementarity, and sequence-specific cleavage of perfectly complementary mRNAs. Endogenous microRNAs (miRNAs) induce translational repression and mRNA degradation when the guide (antisense) strand has limited complementary to the target mRNA. The sequence-specific cleavage mechanism is exploited by exogenous small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) having perfect or near-perfect Watson-Crick base pairing with the intended mRNA target. The production and processing of miRNAs requires an ensemble of host machinery that is ultimately guided by one of the two miRNA strands to the target mRNA (Figure 2). Likewise, siRNAs and shRNAs utilize many of the same endogenous factors, and siRNA/shRNA therapeutics may compete with the production/function of natural miRNAs.
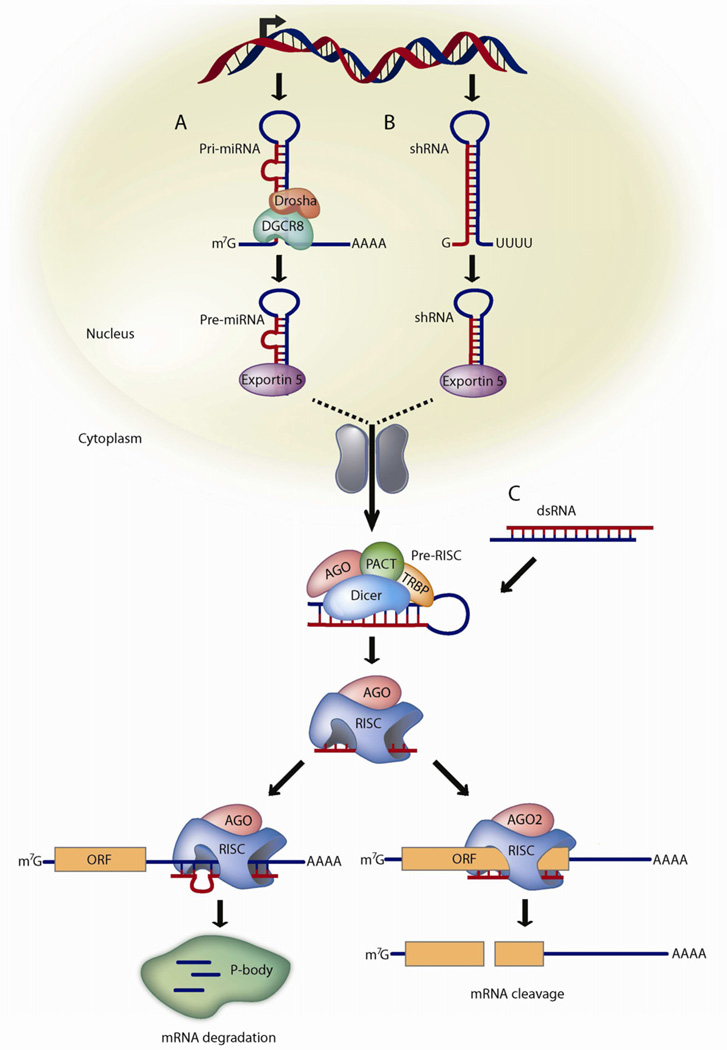
Figure 2.
Mammalian PTGS pathway for miRNAs, shRNAs, and siRNAs
(A) miRNAs are transcribed from DNA as primary miRNAs (pri-miRNAs) and processed into 70-nt stem-loop precursor miRNAs (pre-miRNAs) by Drosha and DGCR8. The pre-miRNAs are transported to the cytoplasm by dsRNA-binding protein exportin 5, where they are processed into ~22 nt miRNA duplexes by the Dicer/TRBP complex. The imperfectly complementary miRNA duplexes associate with an AGO protein and are loaded into RISC, where the passenger strand is removed and the guide strand remains to target mRNA for silencing. The resulting mature RISC complex may silence gene expression either by inhibiting the initiation of translation or by transporting the complex to cytoplasmic processing bodies (p-bodies) where the mRNA is deadenylated and destroyed.
(B) Like miRNAs, shRNAs are transcribed from DNA and undergo similar processing. However, the perfect Watson-Crick base-pairing between the guide strand and the target mRNA triggers AGO2-mediated cleavage of the mRNA target.
(C) In contrast to shRNAs, siRNAs are artificially introduced into the cytoplasm. All steps of siRNA and shRNA are the same after processing by Dicer/TRBP.
In general, primary miRNAs (pri-miRNAs) are processed by a complex containing Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) and incorporated into the pre-RISC complex with Dicer and TAR RNA-binding protein (TRBP) (Figure 2A) [13]. Similarly, the Dicer/TRBP complex directs the processing of shRNA and double-stranded RNA (dsRNA) molecules into ~21–23-nt siRNAs (Figure 2B–C) [14]. One strand (guide or passenger) of the siRNA is loaded into RNA-Induced Silencing Complex (RISC) and may direct sequence-specific cleavage of mRNA by perfect or near-perfect Watson-Crick base pairing [15]. The Argonaute 2 (AGO2) component of RISC contains an endonuclease activity that cleaves the target mRNA and the resulting mRNA fragments are destroyed by cellular exonucleases [16]. While protected inside RISC, the guide siRNA strand can be repeatedly used to target other complementary mRNAs. These remarkable properties have prompted the widespread usage of synthetic siRNA molecules for the therapeutic knockdown of endogenous and viral mRNA [17].
PTGS may be induced by delivering siRNA molecules to cells in dsRNA form or by shRNAs that are transcribed within the cell and processed into siRNAs (Figure 2). To mimic the Dicer cleavage products that are loaded into RISC, many RNAi applications in mammals commonly deliver synthetic siRNAs that are ~19–23 base pairs (bp) with 2-nt overhangs at both 3’ ends. However, this symmetric design often allows either strand (guide or passenger) to be selected into RISC. As an alternative strategy to bias the strand selection of RISC, the dsRNA may be designed asymmetrically with a 2-nt 3’ overhang at one end and a blunt end at the other [18]. The resulting Dicer substrate (D-siRNA), with a 25-nt passenger strand and a 27-nt guide strand, directs the preferential biogenesis of the guide strand, thereby increasing the potency of the siRNA and decreasing off-target effects [19].
In contrast to siRNAs, which transiently knock down gene expression after each drug treatment, shRNAs which are constitutively expressed from promoters can induce long-term gene silencing for the duration of their transcription and biogenesis. Although elevated levels of shRNAs may be desired to achieve maximum knockdown of the mRNA target, expression from strong RNA Pol III promoters can saturate the natural miRNA machinery resulting in severe toxicity [20, 21]. To avoid saturation of the natural RNAi machinery, multiple shRNAs can be expressed as a multicistronic transcript from a RNA Pol II promoter or combined with other non-RNAi therapies like ribozymes and RNA decoys [22, 23].
Over the past decade, interest in siRNA/shRNA technologies has surpassed many of the antisense strategies due to variety of reasons, such as ability of siRNAs to elicit potent target-specific knockdown of any mRNA, ease of siRNA design and screening against the mRNA target, and achievement of long-lasting silencing as the siRNA can retain its catalytic activity in RISC for long periods [12]. Additionally, reduction of off-target toxicity and increased potency can be accomplished by designing siRNA as asymmetric Dicer substrates to bias the loading of the guide strand [18]. Finally, unlike some antisense therapies, which act stoichiometrically on the mRNA target, siRNAs are constantly recycled after inducing mRNA cleavage. Moreover, with the expectation that siRNA-directed cleavage of the mRNA target occurs between nucleotides 10 and 11 relative to the 5’ end of the guide strand, 5’-rapid amplification of cDNA ends (5’-RACE) PCR is used to precisely confirm sequence-specific cleavage by AGO2 [24], an important consideration in validating that the siRNA is operating through the RNAi pathway [25–27].
Approximately 22 different siRNA/shRNA therapeutics have reached clinical testing for the treatment of at least 16 diseases (Table 1) [28]. Similar to other RNA-based therapeutics, the efficacy of siRNA/shRNA drugs relies on maximizing targeted delivery while minimizing off-target toxicity and degradation. Delivery methods can be categorized as ex vivo, local or systemic, and systemic methods can be further classified as active (targeted) or passive [5]. The current state of clinical trials using different siRNA/shRNA delivery methods are discussed in more detail below.
Table 1.
Anti-miRNA and siRNA/shRNA Therapeutics in Clinical Trials
| Company | Drug | Delivery route |
Target | Vehicle | Disease | Phase | Status |
|---|---|---|---|---|---|---|---|
| Santaris | SPC3649 (LNA) | SC | miR-122 | Naked LNA | HCV | IIa | Ongoing |
| Opko Health | Bevasiranib | IVT | VEGF | Naked siRNA | AMD/DME | III | Terminated |
| Allergan/Sirna | AGN-745 | IVT | VEGF-R1 | Naked siRNA | AMD | II | Terminated |
| Quark/Pfizer | PF-655 | IVT | RTP801 | Naked siRNA | AMD/DME | II | Completed |
| Quark Pharma | QPI- 1007 | IVT | Caspase 2 | Naked siRNA | NAION | I | Ongoing |
| TransDerm/IPCC | TD101 | Intralesional injection | KRT6A(N171K) | Naked siRNA | Pachyonychia Congenita | Ib | Completed |
| Sylentis | SYL040012 | Ophthalmic drops | ADRB2 | Naked siRNA | Intraocular Pressure | II | Ongoing |
| Sylentis | SYL1001 | Ophthalmic drops | TRPV1 | Naked siRNA | Dry eye syndrome | I | Ongoing |
| ZaBeCor | ExcellairTM | Inhalation | Syk kinase | unknown | Asthma | II | Ongoing |
| Alnylam/Cubist | ALN-RSV01 | Nebulization or intransal | RSV Nucleocapsid | Naked siRNA | RSV | IIb | Ongoing |
| Marina Biotech | CEQ508 | Oral | Beta catenin | tkRNAi in E. Coli | FAP/ colon cancer | I | Ongoing |
| Silenseed Ltd | siG12D LODER | EUS biopsy needle | KRASG12D | LODER polymer | PDAC | I | Ongoing |
| Tekmira | TKM-ApoB | IV | Apo B | SNALP | Hypercholesterolemia | I | Terminated |
| Tekmira | TKM-PLK1 | IV | PLK1 | SNALP | Solid tumors | I | Ongoing |
| Alnylam/Tekmira | ALN-VSP02 | IV | KSP and VEGF | SNALP | Solid tumors | I | Completed |
| Alnylam | ALN-TTR01 | IV | TTR | SNALP | TTR-mediated amyloidosis (ATTR) | I | Ongoing |
| University Duisburg | Bcr-Abl siRNA | IV | Bcr-Abl | Anionic liposome | CML | I | Completed |
| Silence Therapeutics | Atu027 | IV | PKN3 | siRNA-lipoplex | Advanced solid cancer | I | Ongoing |
| Quark Pharma | I5NP | IV | P53 | Naked siRNA | AKI and DGF | II | Ongoing |
| Calando Pharma | CALAA-01 | IV | RRM2 | Cyclodextrin nanoparticle, TF, and PEG | Solid tumors | I | Ongoing |
| Gradalis Inc. | FANG vaccine | Ex vivo IV | Furin and GM-CSF | Electroporation | Solid tumors | II | Ongoing |
| Duke University | iPsiRNA | Ex vivo intradermal injection | LMP2, LMP7, MECL1 | Transfection | Metastatic melanoma | I | Ongoing |
| City of Hope/Benitec | Tat/Rev shRNA | Ex vivo transplant | HIV Tat and Rev | Lentivirus | HIV | 0 | Ongoing |
Local delivery of siRNAs
Local delivery of siRNA is advantageous for tissues that are external and/or locally restricted including ocular, epidermal, pulmonary, colonic, and pancreatic tissue. Additionally, local delivery may be suitable for non-invasive therapies that require patient administration, such as eye drops and nasal sprays.
The treatment of vision loss in age-related macular degeneration (AMD) and diabetic macular edema (DME) using intravitreal (IVT) injections were some of the first clinical applications for siRNAs as these drugs can be delivered directly to ocular tissue to target well-characterized gene targets for these diseases [29]. However, despite some initial successes, many of these approaches ended with disappointing results. These siRNAs were designed against the VEGF pathway to inhibit neovascularization leading to retinal edema and damage. Opko Health Inc. developed Bevasiranib, a 21-mer siRNA containing two deoxythymidine (dT) residues on both 3’ ends, that was designed to knockdown the mRNA of VEGF A. Although the therapy demonstrated some biological activity in Phase I and II trials (NCT00722384 and NCT00259753, respectively), the Phase III trial (NCT00499590) of Bevasiranib for AMD was terminated as a result of poor efficacy in reducing vision loss [30]. Despite the completion of a Phase II trial (NCT00306904) using Bevasiranib in DME, no Phase III trial has been announced. Similarly, after the completion of a Phase I/II trial (NCT00363714), the Allergan siRNA AGN-745 against the VEGF receptor was discontinued in the Phase II trial (NCT00395057) due to an off-target effect [31, 32]. Quark Pharmaceuticals and Pfizer have tested the PF-655 siRNA therapeutic, which targets pro-angiogenic factor RTP801, in Phase I and II trials for AMD (NCT00725686 and NCT00713518, respectively) and a now-terminated Phase II trial for DME (NCT00701181). These siRNAs inadvertently activated Toll-like receptors (TLRs), leading to the suggestion that modifications to the backbone chemistry, the terminal nucleotides, and the particular siRNA sequences may reduce immunostimulatory responses [33].
Quark Pharmaceuticals has developed the siRNA QPI-1007 with proprietary modifications to the siRNA structure and chemistry that maintains drug efficacy while reducing off-target effects. QPI-1007 is specifically designed to down modulate caspase-2, a key activator in the apoptotic cascade, as a treatment for optic nerve-related visual loss (NAION) [34]. Pre-clinical studies suggest that QPI-1007 exhibits neuroprotective effects in animal models of NAION and glaucoma. A dose-escalated Phase I trial is currently in development (NCT01064505).
TransDerm, along with the International Pachyonychia Congenita Consortium (IPCC), has designed the first mutation-specific siRNA to be used for human therapy. The TD101 siRNA is directed at the mRNA sequence encompassing the dominant mutation (N171K) in the keratin 6a gene (KRT6A). This mutation causes pachyonychia congenita, a rare skin disorder characterized by painful calluses on weight-bearing areas and hypertrophic nails among other epidermal defects. The siRNA therapy was administered by intralesional injection in a single patient using a split body control. Since the Phase Ib therapy (NCT00716014) was well tolerated and efficacious in reducing the callus, TransDerm is developing less painful alternatives for delivering the drug [35], such as an ointment with lipid-based carriers (GeneCreme) and a dissolvable microneedle array (Protrusion Array Device).
Sylentis developed an siRNA drug (SYL040012) against the beta-2 adrenergic receptor (ADRB2) to inhibit the production of aqueous humor and thereby relieve intraocular hypertension [36]. The eye drop mode of delivery helps to direct SYL040012 to the affected ciliary epithelium. An initial Phase I trial (NCT00990743) evaluated the safety of SYL040012 in patients with ocular hypertension and glaucoma, while a scheduled Phase I/II study (NCT01227291) will continue to evaluate the tolerance and efficacy of this drug. In September 2011, Sylentis announced a Phase I trial (NCT01438281) for their second siRNA drug (SYL1001) for the treatment of ocular pain associated with “dry eye” syndrome. SYL1001 triggers the knockdown of transient receptor potential cation channel subfamily V member 1 (TRPV1), also known as the capsaicin receptor, on the ocular surface, which alleviates ocular surface irritation, inflammation, and pain in animal models [37].
ZaBeCor Pharmaceuticals reported improvements in asthmatic symptoms for patients treated with Excellair™ in a Phase I study [28]. This siRNA drug inhibits spleen tyrosine kinase (Syk) which is involved in activating pro-inflammatory transcription factors. These benefits have helped usher Excellair™ into a Phase II trial.
Alnylam has developed a siRNA therapeutic (ALN-RSV01) against the respiratory syncytial virus (RSV) nucleocapsid (N) protein for prophylaxis against RSV infections in healthy patients (NCT00496821) and for treating RSV infections in lung transplant patients (NCT00658086 and NCT01065935). The drug is composed of a double-stranded RNA duplex with 19 base pairs of complementarity and 2-nt dT overhangs at both 3’ ends [27]. Nasal spray and electronic nebulizer aid the delivery of the therapeutics to healthy or lung transplant patients, respectively. The anti-viral effect of ALN-RSV01 was demonstrated by a reduced infection rate in healthy patients and by alleviating the daily symptoms in transplant patients [38, 39]. Although the clinical trials have not yet shown direct evidence for a human anti-viral RNAi mechanism, such mechanism is supported by animal studies [27].
Marina Biotech has launched a Phase I trial of the first orally administered shRNA drug for treating familial adenomatous polyposis (FAP), a rare hereditary disease that often leads to colon cancer. The CEQ508 shRNA therapy down-regulates β-catenin to slow the polyp growth in intestinal cells [40]. The drug is encapsulated by the company’s TransKingdom RNA™ interference (tkRNAi) technology, which utilizes non-pathogenic Escherichia coli to produce and deliver the shRNAs to target cells [41]. The bacterial vector is coated with Invasin protein from Yersinia pseudotuberculosis to facilitate the entry of the bacterial carrier into intestinal cells expressing the β-1 integrin receptor. Finally, the vector encodes lysteriolysin O pore forming protein (from Listeria monocytogenes) to permit the shRNA to escape the bacterial vehicle and enter the cytoplasm.
Silenseed Ltd is launching a Phase 0/I trial (NCT01188785) to evaluate an siRNA drug (siG12D) that targets somatic mutations in the KRAS oncogene (KRASG12D) for pancreatic ductal adenocarcinoma (PDAC). An endoscopic ultrasound (EUS) needle is used to inject the siRNA drug directly into the tumor. The siRNA is encapsulated in the Local Drug EluteR (LODER) biodegradable polymer, which enables slow release of the siRNA therapeutic over an eight-week period [28].
Systemic delivery of siRNAs
Upon intravenous (IV) injection, unmodified siRNAs tend to accumulate in the kidneys, while siRNAs encapsulated in liposomes and nanoparticles often become trapped in the liver. Thus, siRNA therapeutics designed for these tissues can be delivered by non-targeted (passive) systemic delivery, in which uptake of the therapeutics relies on the filtering organs of the reticuloendothelial system [5]. For applications that require targeted systemic delivery, synthetic carriers may be decorated with cell-specific ligands or aptamers that allow receptor-mediated uptake [42], and biodegradable nanoparticle carriers allow for slow drug release once inside the cell [43].
Tekmira Pharmaceuticals has developed two distinct siRNA drugs that are encapsulated in the stable nucleic acid lipid particle (SNALP). SNALP is a first generation lipid nanoparticle developed by Tekmira that is designed to deliver the siRNA to the targeted tissue by IV injection. The first drug (TKM-ApoB or PRO-040201) is an siRNA that targets the mRNA of ApoB and is designed to indirectly reduce the uptake of cholesterol in cells. A total of 17 patients received TKM-ApoB and one of the two that received the highest dosage of the drug exhibited flu-like symptoms that were consistent with siRNA induced immune stimulation [28]. Although the drug did not show evidence of toxicity in the liver, the Phase I clinical study (NCT00927459) was terminated because patients exhibited only transient reductions in cholesterol levels [44]. Nonetheless, the company is working on improvements to the nanoparticle carriers and siRNA design. The second drug known as TKM-PLK1, which targets polo-like kinase 1 (PLK1), will be tested in two Phase I trials (NCT01262235 and NCT01437007) for patients with advanced solid tumors that are resistant to current therapies since down-modulation of PLK1 levels prevents cell cycle progression into mitosis and induces apoptosis in tumor cells [45].
Alnylam Pharmaceuticals has partnered with Tekmira for the use of SNALP carriers to package siRNA therapies for two diseases, liver cancer and transthyretin (TTR)-mediated amyloidosis (ATTR). To treat hepatocellular carcinoma, Alnylam generated a therapeutic (ALN-VSP02) with two distinct siRNAs against vascular endothelial growth factor (VEGF) and kinesin spindle protein (KSP). The therapy was generally well tolerated in a Phase I trial (NCT00882180), and an anti-VEGF effect was observed in most patients and confirmed [46]. Long-term follow up of patients treated with ALN-VSP02 continues in a second Phase I trial (NCT01158079). Additionally, Alnylam has initiated Phase I study (NCT01148953) to determine the safety and tolerability of siRNA treatment for ATTR, ALN-TTR01. The ALN-TTR01 siRNA targets the TTR mRNA to reduce the accumulation of amyloid deposits in surrounding tissues [36]. Since TTR is mainly expressed in the liver, Alnylam has teamed with Tekmira to design SNALP carriers with high affinity for hepatocytes. In November 2011, Alnylam announced that ATTR patients receiving ALN-TTR01 exhibited a statistically significant reduction in serum TTR protein levels that was dose-dependent and durable after a single dose.
Taking a similar approach to the Tekmira-designed cationic SNALP carriers, multiple clinical trials utilize other design strategies for encapsulating the siRNA therapeutics. A Phase I trial sponsored by the University of Duisburg-Essen (Germany) incorporated an siRNA drug against bcr-abl with anionic liposomes. BCR-ABL is a fusion oncogene uniquely expressed in chronic myeloid leukemia (CML) resulting from a chromosomal defect. Despite transient knockdown of the BCR-ABL fusion, the inhibition of the oncogene mRNA was not maintained in patients [47]. Silence Therapeutics is conducting a Phase I trial (NCT00938574) for the treatment of advanced solid cancers. The Atu027 siRNA is designed to inhibit protein kinase N3 (PKN3), a down-stream target of the phosphoinositide 3-kinase (PI3K) signaling pathway that mediates metastasis of cancer cells [48]. The Atu027 therapeutic is formulated as an siRNA-lipoplex, a complex with negatively charged nucleic acids and cationic lipids, known as AtuPLEX™.
Quark Pharmaceuticals has opted to deliver uncoated siRNA therapeutics to the kidney for preventing acute kidney injury (AKI) and delayed graft function (DGF). The natural pathway of excretion by the kidneys allows internalization of the siRNAs making this tissue a tractable target for siRNA therapies. A Phase I trial indicated that the siRNA I5NP (or QPI-1002) temporarily suppresses the pro-apoptotic p53 protein as prophylaxis for AKI post cardiovascular surgeries (NCT00554359). This strategy is currently in a Phase I/II trial for DGF after kidney transplants (NCT00802347).
Calando Pharmaceuticals tested the first receptor-mediated delivery of siRNA nanoparticles as treatment for relapsed/refractory cancers. In this Phase I trial (NCT00689065), the siRNA is complexed in cyclodextrin nanoparticles that are coated with polyethylene glycol (PEG) for stability and the human transferrin (TF) protein for receptor mediated uptake via the transferrin receptor which is often highly expressed in tumor cells. The siRNA CALAA-01 is directed against the M2 subunit of ribonucleotide reductase (RRM2), which is essential in providing the ribonucleotide pool for DNA synthesis and repair. Interestingly, this landmark study also demonstrated first evidence for RNAi mechanism in humans as the investigators verified the occurrence of siRNA-induced cleavage of the target RRM2 mRNA [49].
Ex vivo delivery of siRNA/shRNA
Delivery of siRNA/shRNA via bacterial or viral carriers is often performed ex vivo, as the targeted cells are collected, modified, and re-infused back into the patient. This delivery method is often preferred when simultaneous delivery and expression of multiple therapeutic genes (mRNAs, ribozymes, aptamers, etc.) are required and/or when a specific cell type (generally leukocytic lineages) is targeted for therapeutic gene applications.
Two autologous immune cell therapies for cancer combine an siRNA/shRNA therapeutic with the expression of a recombinant gene or co-delivery of therapeutic mRNA. Gradalis Inc. is treating advanced solid cancers, including stage IIIc ovarian cancer, in Phase I (NCT01061840) and Phase II trials (NCT01309230) by expressing recombinant granulocyte-macrophage colony stimulating factor (GM-CSF) and a bifunctional shRNA against Furin (bi-shRNAfurin) in the FANG™ Vaccine [50]. Both stem loops in the bi-shRNAfurin target the Furin mRNA, but one contains a perfectly complementary guide strand which induces mRNA cleavage while the other has a guide strand which is mismatched to the target 3’ UTR and therefore functions as a miRNA. Down-regulation of Furin indirectly reduces the TGFβ1 and TGFβ2 isoforms that contribute to diminished T cell responsiveness in tumor cells. GM-CSF overexpression induces differentiation of dendritic cells (DCs) antigen presentation. Therefore, the combined down-regulation of TGFβ isoforms and overexpression of GM-CSF is designed to mobilize the patient’s immune cells to eradicate malignant cells.
A similar Phase I study (NCT00672542) conducted by Duke University is administering the siRNA/mRNA therapy for metastatic melanoma. Autologous DCs harvested from patients are transfected with siRNAs against immunoproteasome subunits LMP2, LMP7, and MECL1 (iPsiRNA) and subsequently transfected with melanoma antigens MART, MAGE-3, gp100, and tyrosinase [51]. The down regulation of immunoproteasomes is believed to enhance presentation of melanoma antigens by DCs. By boosting proteasome-mediated antigen presentation in autologous DCs, this strategy enhances the immune response towards melanoma cells.
The City of Hope, in partnership with Benitec, Inc. has conducted an all RNA based gene therapy human pilot feasibility (Phase 0) study for patients with AIDS-related non-Hodgkin’s lymphoma (NHL) [52]. The 4-patient cohort consisted of patients requiring transplantation of autologous CD34+ hematopoietic progenitor cells (HPCs) for the treatment of NHL. However, to protect the transplanted progenitor cells, and particularly the eventual subset of differentiated CD4+ T-cells, from HIV infection, a fraction of the cells were transduced ex vivo with a replication incompetent lentiviral vector that encoded three anti-HIV small RNAs (pHIV7-shI-TAR-CCR5RZ). The three small RNAs are each expressed from separate RNA polymerase III promoters and are designed to inhibit infection and/or replication of HIV-1 by a distinct mechanism: (1) an anti-CCR5 ribozyme intended to block viral entry, (2) an shRNA designed to destroy viral mRNA, and (3) an RNA hairpin known as the transactivating region (TAR) decoy that antagonizes viral transactivation [23]. The shRNA component is designed to induce RISC-mediated cleavage of viral mRNA at a site with overlapping, frame-shifted reading frames of Tat and Rev, which mediate transactivation of viral gene expression and nuclear export of viral mRNA transcripts, respectively. The therapy was well tolerated and genetic marking of the siRNA was detected in primary blood mononuclear cell (PBMCs) and/or primary blood granulocytic cell (PBGCs) of all patients up to 6 months after treatment, including at least 24 months in one patient [52]. A second clinical study with the same AIDS-related NHL cohort is scheduled to begin in early 2012 [28].
Ligand RNAs: Aptamers and decoys
Aptamers are single-stranded nucleic acids that bind to molecular targets with high affinity and specificity due to their stable-three dimensional shapes [53]. Many aptamers exist as hairpin-like monomers that bind targets via unpaired nucleotides, but some aptamers function as duplexes [54], triplexes [55], or quadruplexes [56, 57]. RNA and DNA aptamers are typically identified for a particular function through multiple rounds of in vitro or cell-based selection in a process known as systematic evolution of ligands by exponential enrichment (SELEX) [58, 59]. Using SELEX, aptamers of 20–100 nucleotides (nt) can be selected from libraries (up to 10~15 unique sequences) to bind with high affinity to a wide array of protein families to modulate the protein function similar to antibodies [60]. Like other RNA therapeutics, RNA aptamers are often modified during chemical synthesis to increase their resistance to nucleases and improve pharmacological properties. These modifications include 2’-F, 2’-OMe, and LNA sugar substitutions or the spiegelmer form of the aptamer. Additionally, aptamers may be conjugated with cholesterol or polyethylene glycol (PEG) to reduce renal filtration [60].
While the affinity and specificity of RNA aptamers for their target ligands rival the properties of antibodies, aptamers offer several advantages over their protein counterparts. Aptamers are evolved and identified in vitro using SELEX, and can be reproducibly and economically synthesized in large scale for clinical applications. Using chemical substitutions and other modifications (including L-RNA), aptamers elicit minimal immunogenicity relative to antibodies. The small size of aptamers allows for improved transport and tissue penetration compared to antibodies. Finally, aptamers are amenable to applications that require engineering, such as the conjugation of aptamers to ribozymes (riboswitches) and aptamer-siRNA chimeras.
There are at least 6 RNA-based aptamers or decoys that have been clinically tested (Table 2), including a VEGF-specific modified RNA aptamer (Macugen by Pfizer/Eyetech) that is now an FDA approved drug for the treatment of AMD [6, 60, 61]. In addition to their antibody-like abilities to inhibit or activate the functions of protein targets, aptamers also offer novel functions as therapeutic and diagnostic agents. By utilizing Watson-Crick pairing of nucleic acids, RNA aptamers can be engineered to undergo conformational changes in the presence and/or absence of other effector RNAs both in vitro and in vivo. Such a strategy has been tested in two Phase II trials (NCT00932100 and NCT00113997) with the REG1 (by Regado Biosciences) dual-aptamer therapy for acute coronary syndrome (ACS). Using this therapy, the first aptamer RNA aptamer (RB006 or pegnivacogin) is administered to bind coagulation factor IXa, but after the subsequent injection of the second “antidote” aptamer (RB007 or anivamersen), RB006 binds to RB007 thereby releasing it from its factor IXa target [62]. Also, aptamers can be combined with other types of therapeutic agents to serve as a delivery vehicle for siRNAs [26, 63, 64], enzymes [65], and anti-cancer drugs [66, 67]. Finally, although no clinical applications have yet been demonstrated, RNA aptamers can be engineered to form self-organizing RNA scaffolds [68], combined with ribozymes to create “riboswitches” [69], or conjugated to miRNAs to create ligand-responsive miRNAs [70]. While many of these therapeutic aptamers consist of RNA-based molecules with 2’-OMe and/or 2’-F substitutions and PEG conjugation, numerous examples of DNA and spiegelmer aptamers are also in the clinic [60].
Table 2.
Aptamers and Decoys in Clinical Trials
| Company | Drug | Route | Target | Modification(s) | Disease | Phase | Status |
|---|---|---|---|---|---|---|---|
| Eyetech / Pfizer | pegaptanib sodium (Macugen) | IVT | VEGF | 2'-OMe purine/2'-F pyrimidine with two 2'-ribo purines conjugated to 40 kDa PEG, 3' inverted dT | AMD | FDA approved | Approved |
| Archemix Corp. | ARC19499 (BAX499) | IV and SC | tissue factor pathway inhibitor (TFPI) | Unknown | Hemophilia | II | Not yet recruiting |
| Regado Biosciences | REG1 (RB006 & RB007) | IV | Factor IXa | 2'-ribo purine/2'-F pyrimidine (RB006); PEG and 2'-Ome antidote (RB007) | ACS | II | Completed |
| Ophthotech | ARC1905 | IVT | complement component 5 (C5) | 2'-ribo purine/2'-F pyrimidine conjugated to 40 kDa PEG, 3' inverted dT | AMD | I | Ongoing |
| City of Hope / Benitec | TAR decoy | Ex vivo transplant | HIV Tat protein | U6 snoRNA domain | HIV | 0 | Ongoing |
| Childrens Hospital Los Angeles | RRE decoy | Ex vivo transplant | HIV Rev protein | expressed by HIV promoter | HIV | 0 | Ongoing |
While the SELEX technique enables the discovery of novel RNA and DNA aptamers, mimics of protein-binding RNAs can be used as therapeutic decoys. Like RNA aptamers, RNA decoys bind target proteins due to their three-dimensional structure. Two examples of clinically tested RNA decoys have both been used to inhibit HIV-1 replication [71]. The Rev response element (RRE) decoy is composed of the 41-nt RRE portion of HIV-1 transcripts, which is a hairpin-like structure that binds to the viral Rev protein [72]. This therapeutic RNA is expressed from a retroviral vector following ex vivo transduction and re-infusion of CD34+ HPCs. A similar anti-HIV decoy is composed of the transactivating region (TAR) hairpin at the 5’-end of viral mRNA transcripts that recruits and binds the viral Tat protein [52]. While the natural TAR transcript is located in the nucleus at the site of viral transcription, the TAR decoy is designed to translocate to the nucleolus, where it binds and sequesters the Tat protein from its natural target [23]. As described earlier, this therapy also includes an shRNA and a ribozyme and is expressed from a lentiviral vector (pHIV7-shI-TAR-CCR5RZ) in autologous CD34+ HPCs.
Catalytic RNAs: Ribozymes
Ribozymes are catalytic RNAs that function as enzymes and do not require proteins for catalysis. Most known natural ribozymes are self-processing RNAs that catalyze RNA cleavage and ligation reactions. However, the substrate recognition domain of ribozymes can be artificially engineered to stimulate site-specific cleavage in cis (the same nucleic acid strand) or trans (a non-covalently linked nucleic acid) [73]. Moreover, ribozymes are amenable to in vitro selection and directed evolution to generate improved properties and new functions for therapeutic and diagnostic reagents. Ribozymes can be engineered to be allosterically activated by effector molecules, which has led to the development of artificial “riboswitches” as biosensors and synthetic biological tools [74, 75]. There are numerous types of ribozymes in biology, but the most common ribozyme therapeutics are derived from either “hammerhead” or “hairpin/paperclip” motifs.
Like siRNA/shRNA therapeutics, ribozymes can either be delivered to the target cells in RNA form or can be transcribed from therapeutic genes (Table 3). Due to poor stability of fully-RNA ribozymes, therapies that rely on direct delivery often require chemically stabilized ribozymes, including the following modifications: 5’-PS backbone linkage, 2’-O-Me, 2’-deoxy-2’-C-allyl uridine, and terminal inverted 3’-3’ deoxyabasic nucleotides. All of these modifications were incorporated for Angiozyme (RPI.4610), the first synthetic ribozyme tested in clinical trials [76]. Currently licensed by Merck-Sirna, Angiozyme is a ribozyme that targets the mRNA of the vascular endothelial growth factor receptor-1 (VEGFR-1) to block angiogenesis and tumor growth. A Phase I trial successfully demonstrated no pharmacokinetic interactions between Angiozyme and chemotherapeutic agents carboplatin and paclitaxel for 12 patients with advanced solid tumors [76]. Another Phase I study evaluated the maximum tolerated dose, pharmacokinetic variables, pharmacodynamic markers, clinical response, and safety of daily subcutaneous (SC) injection of Angiozyme for 28 patients with refractory solid tumors [77]. A Phase II trial (NCT00021021) for patients with metastatic renal cancer was completed in 2009, though details of this trial have not been published. The same company also developed Heptazyme, a synthetic ribozyme against hepatitis C virus (HCV). However, despite encouraging results in Phase I and II trials [78, 79], this drug was discontinued after the observation of vision loss in one animal during simultaneous testing in non-human primates [80].
Table 3.
Ribozymes in Clinical Trials
| Company | Drug | Delivery route | Target | Modification(s) | Disease | Phase | Status |
|---|---|---|---|---|---|---|---|
| Merck-Sirna | Angiozyme | SC | VEGFR-1 | 5'-PS, 2'-O-Me, 2'-deoxy-2'-C-allyl uridine, inveted 3'-3' dT | renal cancer | II | Completed |
| Merck-Sirna | Heptazyme | SC | HCV IRES | 5'-PS, 2'-O-Me, 2'-deoxy-2'-C-allyl uridine, inveted 3'-3' dT | HCV | II | Terminated |
| UCSD | MY-2 | Ex vivo, autologous CD4+ T-cells | HIV U5 and pol | Expressed in MMLV vector | HIV | I | Completed |
| Johnson & Johnson, St. Vincent's Hospital | RRz1 | Ex vivo, syngeneic CD4+ T-cells | HIV Tat and Vpr | Expressed in MMLV vector | HIV | I | Completed |
| Janssen-Cilag Pty Ltd, UCLA | OZ1 (RRz1) | Ex vivo, autologous HPCs | HIV Tat and Vpr | Expressed in MMLV vector | HIV | II | Ongoing |
| City of Hope, Benitec | CCR5 ribozyme | Ex vivo, autologous HPCs | CCR5 | Expressed in lentiviral vector | HIV | 0 | Ongoing |
| Ribozyme, City of Hope | L-TR/Tatneo | Ex vivo, autologous HPCs | HIV Tat and Rev | Expressed in MMLV vector | HIV | II | Completed |
Several ribozymes against HIV have been clinically tested using a gene therapy-based approach in CD4+ T-cells or CD34+ hematopoietic stem cells (HSCs), which differentiate into various hematopoietic lineages including CD4+ T-cells [81–84]. Since HIV-1 preferentially infects CD4+ T cells, the therapeutically modified CD4+ cells would be protected from producing functional HIV-1 virus. Each of these trials used either autologous (patient’s own cells) or syngeneic (cells from identical twin) cell therapy, in which the cells are harvested from the patient or healthy twin, treated with the ribozyme-embedded gene therapy, and re-infused back into the patient. Retroviral or lentiviral gene vectors were utilized for these trials, which facilitates integration of the therapeutic genes into the host genome and long-term gene expression after integration. While all of these trials have demonstrated the safety and feasibility of gene-delivered ribozyme therapy, none has proven a clear survival advantage for the protected cells vs. the empty vector (control) transduced cells. This might be due to the poor engraftment of transduced cells, limited efficacy of the therapeutic ribozyme, chromatin silencing of the integrated ribozyme, suboptimal ribozyme kinetics, or other possible factors [85].
As mentioned earlier in the siRNA/shRNA section, a Phase 0 clinical study at the City of Hope uses gene-modified autologous CD34+ HPCs to deliver three RNA-based for the treatment of HIV-1 [52]. In addition to the shRNA and the TAR decoy encoded in the lentiviral gene vector, this therapeutic expresses a hammerhead ribozyme that cleaves the mRNA of the chemokine receptor 5 (CCR5) protein [23]. The CCR5 receptor is expressed on a subset of CD4+ T-cells and serves as a co-receptor for HIV-1 infection. However, CCR5 is not essential for normal T-cell function and offers an attractive target for anti-HIV therapeutics since, unlike many viral targets, it is not prone to mutational escape.
Design and Delivery
In the development of RNA-based therapeutics, in vitro and animal studies have specifically and efficiently treated infectious diseases, gene disorders, and cancers by using siRNA/shRNAs to induce PTGS, ribozymes to cleave mRNA transcripts, and aptamers to bind and block targeted proteins. However, these therapies have encountered obstacles in clinical testing, including the efficiency and specificity of delivery, the stability of the RNA drug, the minimization of immune stimulation, and the prolonged duration of the drug. These issues have raised serious concerns for several RNA-based drugs and turmoil within the RNAi industry. In particular, Bevasiranib and AGN-745, two intravitreally injected naked siRNAs for the treatment of AMD and DME, were terminated due to the lack of patient improvement and TLR-mediated inflammation [30, 31]. Other RNA-based drugs, including the Bcr-Abl and TKM-ApoB siRNAs and several anti-HIV ribozymes, have been discontinued due to insufficient in vivo drug efficacy [47, 85, 86]. Therefore, the future progress of RNA therapeutics relies heavily on improvements in the design of RNA drugs and delivery technologies that can improve drug efficacy and minimize off-target effects.
As discussed, local delivery of naked siRNAs or aptamers, typically preferred for lung, eye, and skin applications, may generate a proinflammatory response due to activation of TLRs [31, 32] and suffer from poor cellular uptake and nuclease sensitivity of naked siRNAs. In some cases, these problems can be alleviated encapsulating the nucleic acid with a synthetic carrier, or introducing chemical modifications to ONs, including clinically promising LNA and spiegelmer conversions, which are expected to improve the specificity, stability, and immunoresistance of RNA-based drugs.
A major benefit of synthetic RNA drug carriers is the ability to engineer tissue specificity in local or systemic delivery applications, thereby preventing nonspecific delivery and degradation of the drug during transport. Biodegradable polymers, such as the LODER delivery system and the Calando cyclodextrin carrier, can be engineered to release the RNA drug over a localized tissue area for a controllable duration. Carriers designed from liposomes tend to accumulate in the liver, which is the intended target for the SNALP-encapsulated siRNAs ALN-TTR01 and ALN-VSP02.
Ex vivo delivery is ideal for bacterial or lentiviral vectors that express shRNAs, ribozymes, and/or aptamer-like RNA decoys. This delivery method is the most direct method of introducing therapeutic genes, though it is limited to certain cell types and patient cohorts. However, as observed with the cell-based gene therapy strategies of anti-HIV ribozymes, the efficacy of the therapy is dependent upon the success of the transplant. As an alternative, synthetic carriers can be modified to specifically deliver the RNA drug to desired cells or tissues. In particular, ligand-decorated nanoparticles [49] and aptamer-mediated siRNAs [26, 63] may increase drug efficiency while avoiding the effects of off-target toxicity.
In addition to improvements in drug delivery for RNA therapeutics, advances in drug design may also improve drug efficacy and reduce off-target toxicity. An example mentioned earlier, an asymmetric 25/27-R Dicer substrates that bias incorporation of the guide ssRNA stand into RISC, increases the drug potency and mitigate the off-target effects from loading of the passenger RNA strand [18]. Additionally, dual-targeting siRNAs are designed so that both strands will be incorporated into RISC and separately target different mRNA transcripts with complete complementarity [87]. In contrast to siRNAs, which tend to saturate the natural RNAi machinery and become toxic at high concentrations, ribozymes can be combined with each other or with another siRNA therapy to provide a multipronged approach. Other combinations of multiple RNA-based drugs, as with siRNA-aptamer chimeras, riboswitches, and gene therapy vectors, may offer the advantages of improving drug specificity, reducing the required drug dosage, and preventing disease resistance.
Future Prospects
While RNA-based therapeutics must overcome barriers in clinical testing for future success, results from previous trials have revealed important lessons. In general, siRNAs will require some chemical modifications to minimize nonspecific inflammation while natural or synthetic carriers should be employed for efficient and tissue-specific delivery. Many of these considerations have contributed to encouraging clinical results for several siRNA drugs including CALAA-01, TD101, ALN-VSP02, and ALN-RSV01. While these examples hint at the potential of siRNA therapeutics, they also affirm the need for tailored carriers that specifically target the intended cells.
Like siRNAs, ribozymes and aptamers face similar challenges of delivery and off-target toxicity. These RNA drugs are amenable to chemical modification and, for some applications, delivery via gene therapy. The chemically modified and highly specific aptamer Macugen has gained FDA approval for the treatment of AMD, signifies the most notable success in RNA-based therapeutics to date. The REG1 dual-aptamer therapeutic highlights the versatility of RNA therapeutics, as one aptamer serves as a controlling mechanism for the therapeutic aptamer.
Collectively, these and other examples of RNA-based therapeutics have demonstrated early promise in the treatment of cancers, viruses, and genetic disorders. However, advanced delivery strategies are critical to fully harness the power of RNAi and the flexibility of RNA-based therapeutics. Engineered designs, such as aptamer-siRNA chimeras and transferrin-decorated nanoparticles, will continue to dramatically improve the precision of delivery for RNA drugs. Therefore, the future prospects of RNA-based drugs will require biochemical refinements to maximize drug potency while minimizing off target toxicity and immunogenicity.
Acknowledgements
The authors would like to thank Anh Pham for artistic assistance. JJR is a co-founder of Dicerna Pharmaceuticals and Calando Pharmaceuticals, both are RNAi companies. JJR is supported by NIH grants AI29329, AI42552 and HL07470.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Melnikova I. RNA-based therapies. Nature Reviews Drug Discovery. 2007;6:863–864. doi: 10.1038/nrd2314. [DOI] [PubMed] [Google Scholar]
- 5.Peer D, Lieberman J. Special delivery: targeted therapy with small RNAs. Gene Ther. 2011 doi: 10.1038/gt.2011.56. [DOI] [PubMed] [Google Scholar]
- 6.Sanghvi YS. A status update of modified oligonucleotides for chemotherapeutics applications. Curr Protoc Nucleic Acid Chem. 2011;Chapter 4(Unit4 1) doi: 10.1002/0471142700.nc0401s46. [DOI] [PubMed] [Google Scholar]
- 7.Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. Rna. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 16.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nature Reviews Genetics. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 19.Amarzguioui M, Rossi JJ. Principles of Dicer substrate (D-siRNA) design and function. Methods Mol Biol. 2008;442:3–10. doi: 10.1007/978-1-59745-191-8_1. [DOI] [PubMed] [Google Scholar]
- 20.Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K, Storm TA, Kay MA. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Rossi JJ. Strategies in designing multigene expression units to downregulate HIV-1. Methods Mol Biol. 2010;623:123–136. doi: 10.1007/978-1-60761-588-0_8. [DOI] [PubMed] [Google Scholar]
- 23.Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 24.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 25.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, Smith DD, Swiderski P, Rossi JJ, Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3001581. 66ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez R, Elbashir S, Borland T, Toudjarska I, Hadwiger P, John M, Roehl I, Morskaya SS, Martinello R, Kahn J, Van Ranst M, Tripp RA, DeVincenzo JP, Pandey R, Maier M, Nechev L, Manoharan M, Kotelianski V, Meyers R. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob Agents Chemother. 2009;53:3952–3962. doi: 10.1128/AAC.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dejneka NS, Wan S, Bond OS, Kornbrust DJ, Reich SJ. Ocular biodistribution of bevasiranib following a single intravitreal injection to rabbit eyes. Mol Vis. 2008;14:997–1005. [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho WG, Albuquerque RJ, Kleinman ME, Tarallo V, Greco A, Nozaki M, Green MG, Baffi JZ, Ambati BK, De Falco M, Alexander JS, Brunetti A, De Falco S, Ambati J. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proc Natl Acad Sci U S A. 2009;106:7137–7142. doi: 10.1073/pnas.0812317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel-Abraham S, Leonard JN. Staying on message: design principles for controlling nonspecific responses to siRNA. FEBS J. 2010;277:4828–4836. doi: 10.1111/j.1742-4658.2010.07905.x. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem. 2002;277:13430–13437. doi: 10.1074/jbc.M108029200. [DOI] [PubMed] [Google Scholar]
- 35.Leachman SA, Hickerson RP, Schwartz ME, Bullough EE, Hutcherson SL, Boucher KM, Hansen CD, Eliason MJ, Srivatsa GS, Kornbrust DJ, Smith FJ, McLean WI, Milstone LM, Kaspar RL. First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez V, Jímenez A, Martínez T. SYL1001 Targeting TRPV1 Receptor for the Treatment of Ocular Pain associated to Dry Eye Syndrome. Presentation Abstract at the Association for Research Vision and Opthamology. 2011 Program # 3844/D3977. [Google Scholar]
- 38.DeVincenzo J, Lambkin-Williams R, Wilkinson T, Cehelsky J, Nochur S, Walsh E, Meyers R, Gollob J, Vaishnaw A. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2010;107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zamora MR, Budev M, Rolfe M, Gottlieb J, Humar A, Devincenzo J, Vaishnaw A, Cehelsky J, Albert G, Nochur S, Gollob JA, Glanville AR. RNA Interference Therapy in Lung Transplant Patients Infected with Respiratory Syncytial Virus. Am J Respir Crit Care Med. 2011;183:531–538. doi: 10.1164/rccm.201003-0422OC. [DOI] [PubMed] [Google Scholar]
- 40.Xiang S, Keates AC, Fruehauf J, Yang Y, Guo H, Nguyen T, Li CJ. In vitro and in vivo gene silencing by TransKingdom RNAi (tkRNAi) Methods Mol Biol. 2009;487:147–160. doi: 10.1007/978-1-60327-547-7_7. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen TA, Fruehauf JH. Transkingdom RNA interference (tkRNAi): a novel method to induce therapeutic gene silencing. Methods Mol Biol. 2009;514:27–34. doi: 10.1007/978-1-60327-527-9_3. [DOI] [PubMed] [Google Scholar]
- 42.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Chen YC, Tseng YC, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J Control Release. 2010;142:416–421. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watts JK, Corey DR. Clinical status of duplex RNA. Bioorg Med Chem Lett. 2010;20:3203–3207. doi: 10.1016/j.bmcl.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reagan-Shaw S, Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. FASEB J. 2005;19:611–613. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- 46.Vaishnaw AK, Cervantes A, Alsina M, Tabernero J, Infante JR, LoRusso P, Shapiro GI, Paz-Ares L, Schwartz G, Weiss G, Falzone R, Hill J, Cehelsky J, White A, Toudjarska I, Bumcrot D, Meyers R, Hinkle G, Svrzikapa N, Sah DW, Burris HA, Gollob JA. RNAi in Humans: Phase I Dose-escalation Study of ALN-VSP02, a Novel RNAi Therapeutic for Solid Tumors with Liver Involvement. Nucleic Acid Therapeutics. 2011;21:A44–A44. [Google Scholar]
- 47.Koldehoff M, Steckel NK, Beelen DW, Elmaagacli AH. Therapeutic application of small interfering RNA directed against bcr-abl transcripts to a patient with imatinib-resistant chronic myeloid leukaemia. Clin Exp Med. 2007;7:47–55. doi: 10.1007/s10238-007-0125-z. [DOI] [PubMed] [Google Scholar]
- 48.Santel A, Aleku M, Roder N, Mopert K, Durieux B, Janke O, Keil O, Endruschat J, Dames S, Lange C, Eisermann M, Loffler K, Fechtner M, Fisch G, Vank C, Schaeper U, Giese K, Kaufmann J. Atu027 prevents pulmonary metastasis in experimental and spontaneous mouse metastasis models. Clin Cancer Res. 2010;16:5469–5480. doi: 10.1158/1078-0432.CCR-10-1994. [DOI] [PubMed] [Google Scholar]
- 49.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maples PB, Kumar P, Yu YU, Wang Z, Jay C, Pappen BO, Rao DD, Kuhn J, Nemunaitis J, Senzer N. FANG Vaccine: Autologous Tumor Cell Vaccine Genitically Modified to Express GM-CSF and Block Production of Furin. BioProcessing Journal. 2010;8:4–14. [Google Scholar]
- 51.Dannull J, Lesher DT, Holzknecht R, Qi W, Hanna G, Seigler H, Tyler DS, Pruitt SK. Immunoproteasome down-modulation enhances the ability of dendritic cells to stimulate antitumor immunity. Blood. 2007;110:4341–4350. doi: 10.1182/blood-2007-04-083188. [DOI] [PubMed] [Google Scholar]
- 52.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, Mi S, Yam P, Stinson S, Kalos M, Alvarnas J, Lacey SF, Yee JK, Li M, Couture L, Hsu D, Forman SJ, Rossi JJ, Zaia JA. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3000931. 36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and Development of Therapeutic Aptamers. Annual Review of Pharmacology and Toxicology. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 54.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd, Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci U S A. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sussman D, Wilson C. A water channel in the core of the vitamin B(12) RNA aptamer. Structure. 2000;8:719–727. doi: 10.1016/s0969-2126(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 56.Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 59.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 60.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen MG, Purdy DA, Rossi JS, Grinfeld LR, Myles SK, Aberle LH, Greenbaum AB, Fry E, Chan MY, Tonkens RM, Zelenkofske S, Alexander JH, Harrington RA, Rusconi CP, Becker RC. First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation. 2010;122:614–622. doi: 10.1161/CIRCULATIONAHA.109.927756. [DOI] [PubMed] [Google Scholar]
- 63.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 65.Chen CH, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, Gonzales M, Baltasar FE, Fan AL, Myerowitz R, Neufeld EF. Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci U S A. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hwang do W, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, Lee DS, Kim S. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 67.Hicke BJ, Stephens AW, Gould T, Chang YF, Lynott CK, Heil J, Borkowski S, Hilger CS, Cook G, Warren S, Schmidt PG. Tumor targeting by an aptamer. J Nucl Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 68.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 69.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–2994. doi: 10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haasnoot J, Berkhout B. Nucleic acids-based therapeutics in the battle against pathogenic viruses. Handb Exp Pharmacol. 2009:243–263. doi: 10.1007/978-3-540-79086-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, Hao Q, Zhou C, Bahner I, Kearns K, Brody K, Fox S, Haden E, Wilson K, Salata C, Dolan C, Wetter C, Aguilar-Cordova E, Church J. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- 73.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 74.Wieland M, Berschneider B, Erlacher MD, Hartig JS. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem Biol. 2010;17:236–242. doi: 10.1016/j.chembiol.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 75.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell. 2011;43:915–926. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kobayashi H, Eckhardt SG, Lockridge JA, Rothenberg ML, Sandler AB, O'Bryant CL, Cooper W, Holden SN, Aitchison RD, Usman N, Wolin M, Basche ML. Safety and pharmacokinetic study of RPI.4610 (ANGIOZYME), an anti-VEGFR-1 ribozyme, in combination with carboplatin and paclitaxel in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2005;56:329–336. doi: 10.1007/s00280-004-0968-x. [DOI] [PubMed] [Google Scholar]
- 77.Weng DE, Masci PA, Radka SF, Jackson TE, Weiss PA, Ganapathi R, Elson PJ, Capra WB, Parker VP, Lockridge JA, Cowens JW, Usman N, Borden EC. A phase I clinical trial of a ribozyme-based angiogenesis inhibitor targeting vascular endothelial growth factor receptor-1 for patients with refractory solid tumors. Mol Cancer Ther. 2005;4:948–955. doi: 10.1158/1535-7163.MCT-04-0210. [DOI] [PubMed] [Google Scholar]
- 78.Sandberg JA, Rossi SJ, Gordon GS, Turik MA, Braun DK, Aitchison R, Babcock SA, Hansen B, Wright T, Blatt LM. Safety analysis of a phase I study of HEPTAZYME (TM), a nuclease resistant ribozyme targeting hepatitis C (HCV) RNA. Hepatology. 2001;34:333a–333a. [Google Scholar]
- 79.Tong M, Schiff E, Jensen DM, Jacobsen I, Eversen G, McHutchison JG, Aitchison R, Gordon GS, Babcock SA, Enright JH, Maloney L, Sandberg JA, Blatt L. Preliminary analysis of a phase II study of Heptazyme (TM), a nuclease resistant ribozyme targeting hepatitis C virus (HCV) RNA. Hepatology. 2002;36:360a–360a. [Google Scholar]
- 80.Berk PD. An editor's look-back. Hepatology. 2006;43:S13–S30. doi: 10.1002/hep.21056. [DOI] [PubMed] [Google Scholar]
- 81.Wong-Staal F, Poeschla EM, Looney DJ. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum Gene Ther. 1998;9:2407–2425. doi: 10.1089/hum.1998.9.16-2407. [DOI] [PubMed] [Google Scholar]
- 82.Macpherson JL, Boyd MP, Arndt AJ, Todd AV, Fanning GC, Ely JA, Elliott F, Knop A, Raponi M, Murray J, Gerlach W, Sun LQ, Penny R, Symonds GP, Carr A, Cooper DA. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J Gene Med. 2005;7:552–564. doi: 10.1002/jgm.705. [DOI] [PubMed] [Google Scholar]
- 83.Amado RG, Mitsuyasu RT, Rosenblatt JD, Ngok FK, Bakker A, Cole S, Chorn N, Lin LS, Bristol G, Boyd MP, MacPherson JL, Fanning GC, Todd AV, Ely JA, Zack JA, Symonds GP. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum Gene Ther. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- 84.Michienzi A, Castanotto D, Lee N, Li S, Zaia JA, Rossi JJ. RNA-mediated inhibition of HIV in a gene therapy setting. Ann N Y Acad Sci. 2003;1002:63–71. doi: 10.1196/annals.1281.008. [DOI] [PubMed] [Google Scholar]
- 85.Burnett JC, Rossi JJ. Stem cells, ribozymes and HIV. Gene Ther. 2009;16:1178–1179. doi: 10.1038/gt.2009.86. [DOI] [PubMed] [Google Scholar]
- 86.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiemann K, Hohn B, Ehsani A, Forman SJ, Rossi JJ, Saetrom P. Dual-targeting siRNAs. Rna. 2010;16:1275–1284. doi: 10.1261/rna.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]