SUMMARY
Small ubiquitin-like modifier (SUMO) modification has emerged as an important regulatory mechanism during embryonic development. However, it is not known whether SUMOylation plays a role in the development of the immune system. Here, we show that SUMO-specific protease 1 (SENP1) is essential for the development of early T and B cells. STAT5, a key regulator of lymphoid development, is modified by SUMO-2 and is specifically regulated by SENP1. In the absence of SENP1, SUMO-2 modified STAT5 accumulates in early lymphoid precursors, resulting in a block in its acetylation and subsequent signaling. These results demonstrate a crucial role of SENP1 in the regulation of STAT5 activation during early lymphoid development.
INTRODUCTION
SUMOylation has been shown to regulate a number of cellular processes, including transcription, DNA repair, cell-cycle progression and signal transduction from yeast to human (Mukhopadhyay and Dasso, 2007; Yeh, 2009). Many transcription factors are SUMOylated, resulting in alteration of their function (Gill, 2004). SUMOylation is catalyzed by SUMO-specific E1, E2, and E3 ligases and can be reversed by SUMO-specific proteases (SENPs). In mammalian cells, six SENPs have been identified. They can be divided into three subfamilies on the basis of their sequence homology, cellular localization and substrate specificity (Yeh, 2009). SENP1 belongs to the first subfamily, which deSUMOlates SUMO-1 or SUMO-2/3-conjugated proteins. Previously, we reported that inactivation of the murine SENP1 gene results in embryonic lethality prior to embryonic day 16.5 (E16.5), due to a severe defect in definitive erythropoiesis stemming from deficient erythropoietin (Epo) production (Cheng et al., 2007). SENP1 knockout (KO) mouse study revealed that SENP1 controls EPO production by regulating the stability of hypoxia-inducible factor 1α (HIF1α) (Cheng et al., 2007). Despite extensive biochemical studies on SENPs in vitro, the physiological significance and in vivo functions of the SENPs remain poorly understood.
Lymphoid development is tightly regulated by transcription factors and cytokines (Busslinger, 2004; Rothenberg and Taghon, 2005). Interleukin-7 (IL-7) is a non-redundant and essential cytokine for the development of both T and B cells (von Freeden-Jeffry et al., 1995). Its receptor consists of two chains, IL-7Rα and common cytokine receptor γ-chain (γC), which bind the Janus kinase 3 (Jak3) (Goodwin et al., 1990; Noguchi et al., 1993). STAT5 is a key signal molecule downstream of IL-7R (Mazzucchelli and Durum, 2007). It contains two highly related isoforms, STAT5A and STAT5B, which are encoded by separate genes and play critical roles in the development and function of immune cells (Hennighausen and Robinson, 2008). Complete inactivation of the murine STAT5A and STAT5B genes results in severe defects in early T and B cell development (Hoelbl et al., 2006; Malin et al., 2010; Yao et al., 2006).
In unstimulated cells, quiescent STAT5 exists in the cytoplasm as a monomer. A ligand such as Epo, prolactin, or IL-7, binds to its cognate receptor to activate the receptor associated Jak tyrosine kinase, resulting in STAT5 activation by tyrosine phosphorylation. Tyrosine phosphorylated STAT5 dimerizes and translocates to the nucleus, where it drives transcription of target genes. Nuclear STAT5 can be dephosphorylated and return to the cytoplasm to complete the activation-inactivation cycle (Leonard and O’Shea, 1998; Levy and Darnell, 2002). Recent findings indicate that STAT5 activity is also under synchronous acetylation regulation, which plays an important role in regulating STAT5 dimerization (Ma et al., 2010). However, little is known about the mechanisms of STAT5 inactivation regarding simultaneous dephosphorylation and deacetylation.
In the present study, we investigated the role of SENP1 in lymphoid development using SENP1 KO mice that we described earlier (Cheng et al., 2007). Our data demonstrated that SENP1 KO mice exhibit severe defects in early T and B cell development. In the absence of SENP1, accumulation of SUMOylated STAT5 results in inhibition of STAT5 activity and lymphoid development. We further showed that SENP1 regulates the SUMOylation status of STAT5. These results reveal a specific role of SENP1 in the regulation of STAT5 transcriptional activity during lymphoid development.
RESULTS
SENP1 deficiency impairs T cell development
To investigate the role of SENP1 in lymphopoiesis, we first analyzed its expression pattern in lymphoid cells. Interestingly, SENP1 was highly expressed at the early stages of T and B cell development, including DN2, DN3 and DN4 T cells, and proB and Pre-B cells, respectively (Figure S1), raising the possibility that SENP1 might participate in T and B lymphopoiesis.
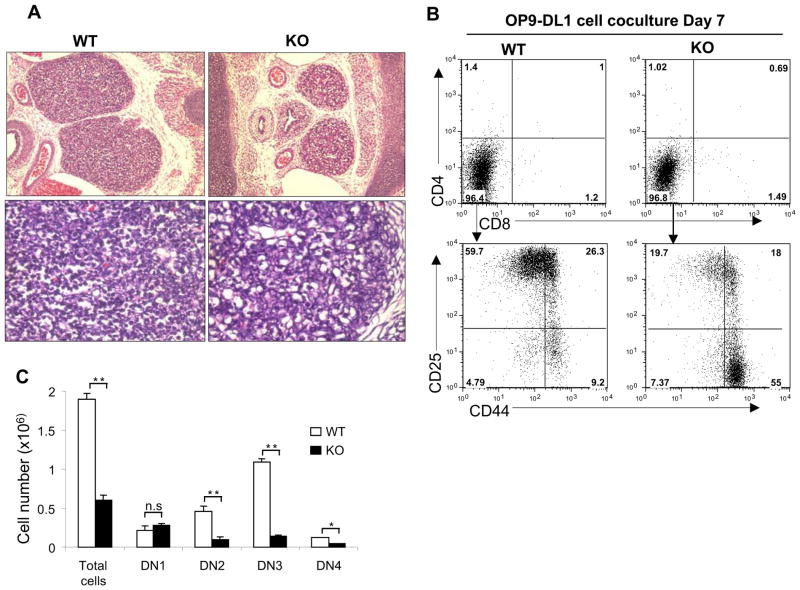
We next examined the phenotype of thymus in SENP1−/− embryos. Histological examination of thymic sections revealed that both the size and cellularity of E15.5 SENP1−/− thymi were markedly decreased compared with those of wild-type littermates (Figure 1A). To check whether a partial or complete block in T cell development could explain thymic hypocellularity and smaller thymus size in SENP1−/− embryos, we cocultured hematopoietic stem cells (HSCs) isolated from fetal livers (FL) in E14.5 wild-type and SENP1−/− embryos with OP9 stromal cells expressing Delta-like 1 (OP9-DL1 cells) in the presence of cytokines Flt3L (FMS-like tyrosine kinase 3 ligand) and IL-7 to induce T cell differentiation in vitro (Schmitt and Zuniga-Pflucker, 2002). After 7 days of coculture, flow cytometric analysis revealed that most of the cells derived from both wild-type and SENP1−/− HSCs were immature CD4−CD8− (DN) thymocytes (Figure 1B, top panel). However, further segregation of DN thymocytes into distinct subsets (DN1-DN4) showed an increased frequency of DN1 subset, and decreased frequencies of DN2 and DN3 subsets generated from SENP1−/− HSCs (Figure 1B, lower panel). Furthermore, the absolute numbers of DN2, DN3 and DN4 T cells derived from SENP1−/− HSCs were significantly decreased compared with those of wild-type controls (Figure 1C). These results indicate that SENP1 is required for early T cell development at or before the DN2 stage.
Figure 1. SENP1 deficiency results in a severe defect in early T cell development.
(A) Hematoxylin and eosin staining of thymic sections from E15.5 wild-type (WT) and SENP1−/− (KO) embryos. Original magnifications 100x (top) and 400x (bottom). The results shown are a representative of at least four independent experiments.
(B) FL-HSCs (Lin−c-Kit+Sca1+) from wild-type and SENP1−/− embryos at E14.5 were cocultured on OP9-DL1 stromal cells. After 7 days of coculture, cells were isolated and analyzed by flow cytometry for T cell markers CD4 and CD8 (top panel). Gated CD4−CD8− population was further analyzed based on CD44 and CD25 (lower panel). Data shown are representative of three independent experiments.
(C) Absolute numbers of the indicated subpopulations of thymocytes were calculated based on flow cytometry (shown in B) and total thymocytes counts, and represented as mean ± SD; n= 5 each. *, P<0.05; **, P<0.01; n.s. is not significant.
Under OP9-DL1 cell coculture conditions, wild-type and SENP1−/− HSCs were unable to generate B and myeloid cells (Figure S2A). After 14 days of coculture on OP9-DL1 cells, despite a substantial decrease in the absolute cell numbers, CD4/CD8 double positive (DP), CD4 single positive (SP) and CD8 SP cells could still be generated from SENP1−/− HSCs (Figure S2B). These results strongly suggest that SENP1 deficiency leads to an incomplete block in early T cell development.
SENP1 deficiency impairs B cell development
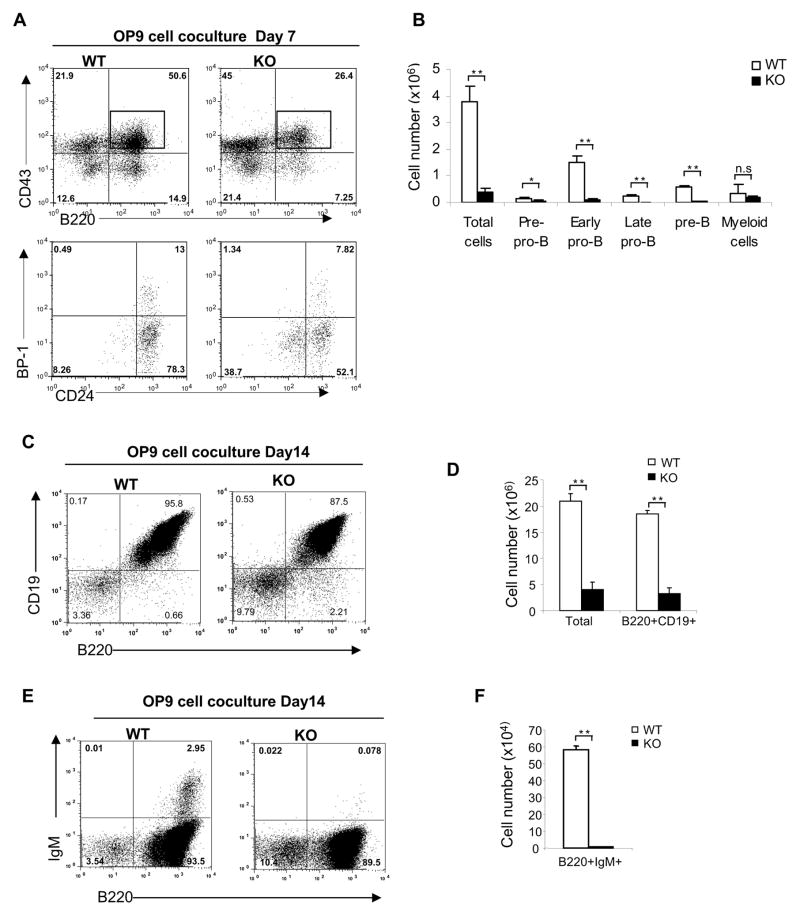
Next, we investigated whether B cell development is also affected by the absence of SENP1. We first examined B lymphopoiesis in fetal livers from E14.5 SENP1−/− embryos and found that SENP1−/− embryos displayed a severe reduction in the absolute number of B220+CD19+ B cells compared with wild-type littermate controls (Figure S2C). To further assess the effects of SENP1 deficiency on B cell development, we cocultured FL-HSCs with OP9 stromal cells in the presence of Flt3L and IL-7 to promote B cell differentiation in vitro (Vieira and Cumano, 2004). B cell development occurs through various stages, which can be characterized by their expression of cell-surface markers according to the Hardy nomenclature (Hardy and Hayakawa, 2001). After 7 days of coculture, flow cytometric analysis revealed an increased frequency of early pro-B (B220+CD43+BP-1−CD24+) cells generated from wild-type HSCs. In contrast, most B cells generated from SENP1−/− HSCs displayed markers of pre-pro-B (B220+CD43+BP-1−CD24−) and early pro-B (B220+CD43+BP-1−CD24+) cells (Figure 2A). Furthermore, the absolute numbers of pre-pro-B, early pro-B, late pro-B and pre-B cells derived from SENP1−/− HSCs were significantly reduced compared with wild-type controls (Figure 2B), suggesting a critical role of SENP1 at the earliest stages of B cell development.
Figure 2. SENP1 deficiency results in a severe defect in early B cell development.
(A) Sorted FL-HSCs from wild-type and SENP1−/− embryos at E14.5 were cocultured on OP9 stromal cells. After 7 days of coculture, cells were isolated and analyzed by flow cytometry for B cell markers B220 and CD43 (top panel), CD24 and BP-1 (Gated CD43+B220+ population, lower panel). Data shown are representative of three independent experiments.
(B) Absolute number of B cells in each developmental stage was calculated based on flow cytometry (shown in D) and total cell number, and represented as mean ± SD; n= 5 each. *, P<0.05; **, P<0.01; n.s. is not significant.
(C-F) Wild-type and SENP1−/− FL-HSCs were cocultured on OP9 stromal cells for 14 days. B cells were analyzed by flow cytometry using B220 and CD19 antibodies (C), and B220 and IgM antibodies (E), and their absolute cell numbers were calculated and presented as means ± SD (D end F, respectively); n=5 each. **, P<0.01. Data shown are representative of three independent experiments.
Under OP9 cell coculture conditions, myeloid development was not affected in the absence of SENP1 since the absolute number of myeloid cells derived from both wild-type and SENP1−/− HSCs remained similar (Figures 2B and S3A). After 14 days of coculture, we also found significant reductions in the frequencies and absolute numbers of pro-B and pre-B cells (B220+CD19+) (Figures 2C and 2D), and immature B cells (B200+IgM+) derived from SENP1−/− HSCs (Figures 2E and 2F). These results clearly show that SENP1 is required for B cell development at the early stages.
SENP1 deficiency does not affect myeloid cell development
To demonstrate that the hematopoietic defects caused by SENP1 deficiency are restricted to the erythroid (Cheng et al., 2007; Yamaguchi et al., 2005; Yu et al., 2010) and lymphoid lineages, but not the myeloid lineage, we cultured wild-type and SENP1−/− HSCs on OP9 stromal cells in the presence of IL3, IL6, SCF, and Flt3L, and analyzed myeloid cells expressing Gr-1 and CD11b markers after 7 days of culture. Consistent with our previous report (Cheng et al., 2007), no defect in myeloid-lineage development was observed in the absence of SENP1 since the frequency and absolute number of myeloid cells (Gr-1+CD11b+) generated from both wild-type and SENP1−/− HSCs were comparable (Fig. S3B).
SENP1 deficiency results in intrinsic defects in T and B development
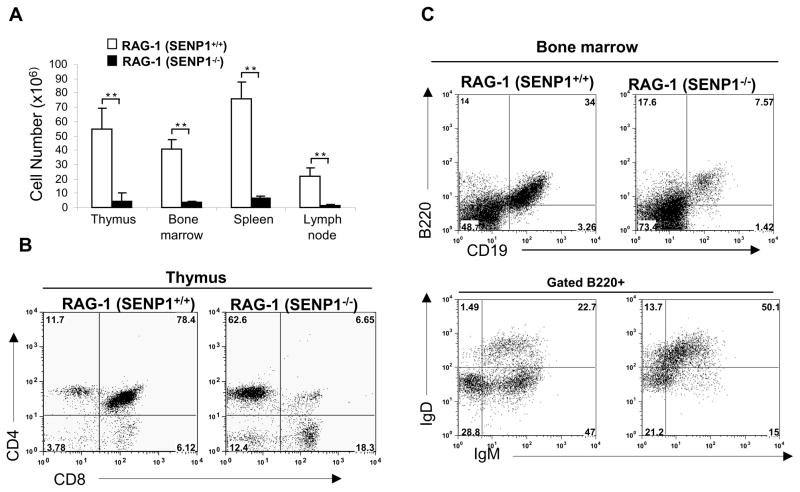
To further confirm that SENP1 deficiency resulted in defects that are intrinsic to T and B cell development, we performed FL transplantation experiments using RAG-1−/− mice, which lack mature B and T cells (Mombaerts et al., 1992). FL cells from E14.5 wild-type and SENP1−/− embryos were transplanted into lethally irradiated RAG-1−/− mice. Six weeks after transplantation, the lymphoid compartments of the recipients were harvested, counted and analyzed by flow cytometry. Compared with RAG-1−/− mice reconstituted with wild-type FL cells (wild-type recipients), the total cell numbers in lymphoid organs of RAG-1−/− mice reconstituted with SENP1−/− FL cells (SENP1−/− recipients), exhibited a 10- to 20-fold reduction (Figure 3A). Analysis of T cell development in the thymi of recipients showed that the frequency of DP T cells in the thymi of SENP1−/− recipients was greatly reduced compared with wild-type recipients (Figure 3B). Similarly, analysis of B cell development in the bone marrow of recipients revealed that the frequencies of CD19+B220+ and B220+IgM+IgD− B cells were significantly reduced in SENP1−/− recipients (Figure 3C). Taken together, our data demonstrate that SENP1 deficiency results in intrinsic defects in T and B development.
Figure 3. SENP1 deficiency impairs T and B cell development in fetal liver transplantation experiments.
(A) Cell counts in lymphoid compartments including thymus, bone marrow, spleen and lymph node of RAG1−/− mice reconstituted with wild-type and SENP1−/− fetal liver cells. Total cell number was calculated and represented as mean ± SD; n= 6 each. **, P<0.01.
(B) Flow cytometric analysis for T cell markers including CD4 and CD8 in the thymi of RAG1−/− mice reconstituted with wild-type and SENP1−/− fetal liver cells. Data shown are representative of two independent experiments.
(C) Flow cytometric analysis of B cells in bone marrow from RAG1−/− mice reconstituted with wild-type and SENP1−/− fetal liver cells using B cell markers B220 and CD19 (upper panel). Gated B220+ cells were further analyzed based on IgM and IgD (lower panel). Data shown are representative of two independent experiments.
SENP1 regulates SUMOylation of STAT5
To check whether SENP1 deficiency may affect transcription of key transcription factors crucial for early lymphoid development, we analyzed their expression in fetal livers of E14.5 embryos by quantitative real-time PCR. Expression levels of PU.1, STAT5, GATA3 and E2A from SENP1−/− fetal livers remained unchanged compared with those from wild-type littermate controls (Figure S4A). To determine whether the impaired development of B and T cells in SENP1 deficiency is due to reduced expression of IL-7R components, real-time PCR analysis was performed. The mRNA levels of IL-7R components including IL-7Rα and γc chains, Jak1 and Jak3 in fetal livers of E14.4 SENP1−/− embryos were undisturbed, suggesting that the expression of genes involved in the IL-7R signaling pathway is intact (Figure S4B). Indeed, the lymphoid defects in SENP1 deficiency described here are quite similar to the one observed in STAT5 deficiency (Hoelbl et al., 2006; Malin et al., 2010; Yao et al., 2006).
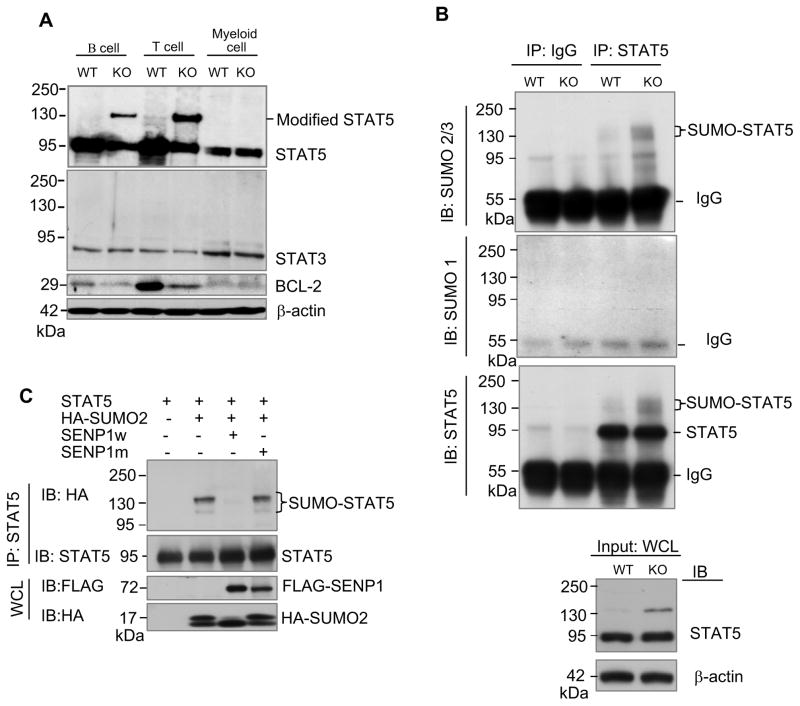
Given that SENP1 regulated activity of transcription factors (Cheng et al., 2007; Yu et al., 2010), we hypothesized that SENP1 may control early development of T and B cells through altering the SUMOylation status of STAT5. To explore this possibility, we first examined SUMOylation of STAT5 in wild-type and SENP1−/− B, T and myeloid cells derived from HSC differentiation in vitro after 7 days of coculture by Western blot analysis. As shown in Figure 4A, the ~95-kDa full-length STAT5 was detected in all wild-type and SENP1−/− B, T and myeloid cells, whereas a modified form of STAT5 migrating at 135 kDa (~ 40 kDa shift) was observed only in SENP1−/− B and T cells. On the other hand, the protein level of another STAT family member, STAT3 in SENP1−/− B and T cells was unchanged compared with wild-type B and T cells, respectively. To verify that the ~ 135-kDa band was a SUMOylated form of STAT5, lysates of wild-type and SENP1−/− B cells were subjected to denaturing immunoprecipitation with anti-STAT5 antibody, followed by immunoblotting with anti-SUMO-1 and anti-SUMO-2/3 antibodies. This ~ 135-kDa band displayed in SENP1−/− B cells was readily detected with anti-SUMO-2/3, but not by anti-SUMO-1 or control IgG (Figure 4B). These results clearly demonstrate that the modified form of STAT5 accumulated in SENP1−/− B and T cells is indeed SUMOylated STAT5. Consistent with the accumulation of SUMOylated STAT5, the expression of BCL-2, a well-studied downstream target of STAT5 (Akashi et al., 1997; Maraskovsky et al., 1997) was markedly decreased in SENP1−/− B and T cells compared with wild-type B and T cells, respectively (Figure 4A, third panel). We also looked at the mRNA expression of STAT5 target genes, including bcl-2 and pim-1, and found that mRNA levels of bcl-2 and pim-1 in SENP1−/− T cells were significantly reduced (Figure S4C). These data suggest that deletion of SENP1 results in the accumulation of SUMOylated STAT5, which alters its transcriptional activity in early B and T cells.
Figure 4. SENP1 regulates SUMOylation of STAT5.
(A) Modified STAT5 accumulated in SENP1−/− B and T cells. Sorted FL-HSCs from wild-type and SENP1−/− embryos at E14.5 were cocultured on OP9 and OP9-DL1 cells to differentiate into B and T cells, respectively. After 7 days of coculture, B and T cells were isolated and lysed in RIPA buffer. The whole cell lysates were analyzed by immunobloting with anti-STAT5 (top panel), anti-STAT3 (second panel), anti-BCL-2 (third panel), and anti-β-actin (bottom) antibodies. Wild-type and SENP1−/− myeloid cells derived from HSC differentiation on OP9 stromal cells for 7 days were used as controls. The results shown are a representative of at least three independent experiments.
(B) Endogenous STAT5 is SUMOylated by endogenous SUMO2/3 in vivo. B cells isolated from differentiation of wild-type and SENP1−/− FL-HSCs after 7 days of coculture on OP9 cells were lysed and boiled in denaturing lysis buffer. Cell lysates were aliquoted equally for performing immunoprecipitation (IP) by control IgG or a mixture of anti-STAT5A and B antibodies. Bound proteins were detected by immunoblotting (IB) with anti-SUMO2/3 (top panel), anti-SUMO1 (middle panel) or anti-STAT5 (bottom panel) antibodies. Loading samples were immunoblotted with anti-STAT5 or anti-β actin antibodies. The results shown are a representative of at least three independent experiments.
(C) SENP1 de-SUMOylates SUMOylated STAT5 in vivo. COS-1 cells were transfected with the indicated plasmids. Protein extracts were precipitated with anti-STAT5B antibody and analyzed with HA (top) and STAT5 (bottom) antibodies. The results shown are a representative of at least three independent experiments.
To further confirm that SUMOylated STAT5 can be regulated by SENP1, we SUMOylated STAT5 by cotransfecting FLAG-tagged STAT5A or STAT5B with HA-tagged SUMO-2 constructs in COS-1 cells. In the presence of HA-tagged SUMO-2 construct, two major bands migrating at 115 kDa and 135 kDa were detected (Figures S5A and S5B). In COS-1 cells, PIAS3 greatly enhanced STAT5 SUMOylation in a RING-domain-dependent manner (Figure S5C). Notably, overexpression of SENP1 completely abrogated SUMOylated STAT5, while a SENP1 catalytic mutant (C603A mutation) could not deconjugate SUMOylated STAT5 (Figure 4C). Together, SENP1 plays a critical role in regulating the SUMOylation state of STAT5 in vivo.
SUMOylation inhibits tyrosine phosphorylation of STAT5
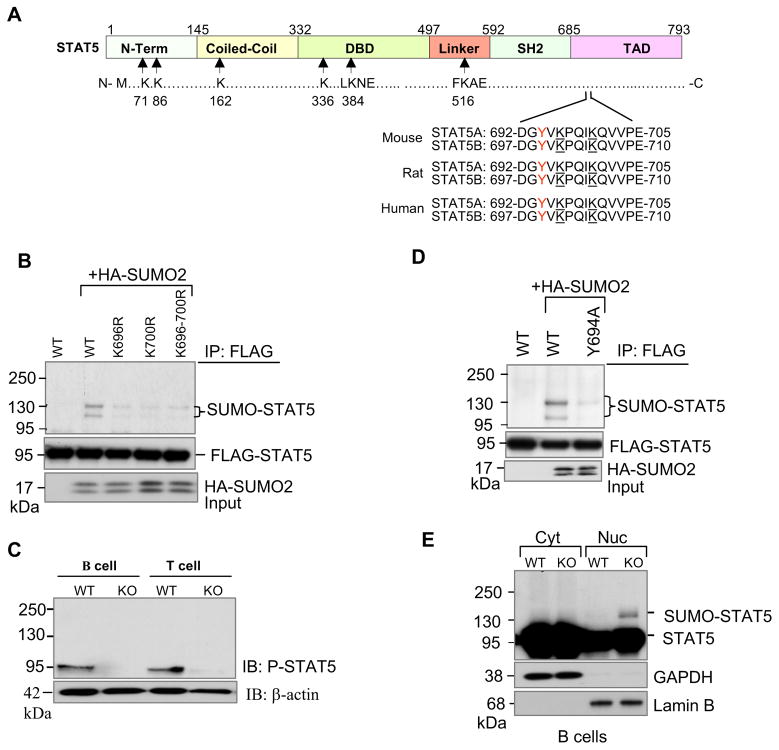
SUMOylation often, but not always, occurs on lysine residues found in the consensus motif ΨKXE (where Ψ is any large hydrophobic residue, and X is any residue). STAT5 has two SUMO consensus motifs at lysines 384 and 516, and four high-scoring non-consensus motifs at lysines 71, 86, 163 and 336 (Figure 5A). We examined the ability of these sites to be SUMOylated in vivo by introducing single or combined mutations (six lysine-to-arginine mutations). Compared with SUMOylation of wild-type STAT5, SUMOylation of single mutants or combined mutants remained unchanged, suggesting that these lysine residues are not involved in SUMOylation of STAT5 (Figure S6A). Previous studies indicate that STAT1 is SUMOylated at lysine 703, close to tyrosine 701 (Rogers et al., 2003; Song et al., 2006; Ungureanu et al., 2003). Indeed, STAT5A contains two lysines at 696 and 700, close to tyrosine 694 (Figure 5A). These lysine residues of STAT5A/B are well conserved across species including mouse, rat and human. SUMOylation of single mutants (K696R and K700R) or double mutant (K696/700R) of STAT5A was drastically reduced (Figure 5B). These results indicate that lysine residues 696 and 700 are the major sites for SUMOylation of STAT5A in vivo. Consistently, the major SUMOylation sites in STAT5B were lysine residues 701 and 705 (Figure S6B).
Figure 5. SUMOylation inhibits tyrosine phosphorylation of STAT5.
(A) Schematic diagram of full-length murine STAT5 protein structure. STAT5 contains amino terminal, coiled-coil, DNA binding, linker, SH2, and transcriptional activation domains. Arrows indicate approximate locations of two putative SUMO consensus motifs at lysines 384 and 516, and four other non-consensus motifs at lysines 71, 86, 163 and 336. Two lysine residues 696 and 700 located close to Y694 and Y699 of STAT5A and STAT5B respectively are underlined.
(B) STAT5A is modified by SUMO at lysine residues 696 and 700. COS-1 cells were transfected with FLAG-tagged STAT5A and its mutants along with HA-SUMO2 plasmids. Protein extracts were immunoprecipitated with anti-FLAG and then revealed by Western blot analysis with antibodies to anti-HA (top) and anti-FLAG (bottom). The results shown are representative of at least three independent experiments.
(C) Tyrosine phosphorylation of STAT5 is diminished in SENP1−/− B and T cells. Sorted FL-HSCs from wild-type and SENP1−/− embryos at E14.5 were cocultured on OP9 and OP9-DL1 cells in the presence of IL-7 and Flt3L to differentiate into B and T cells, respectively. After 7 days of coculture, B and T cells were isolated and lysed in RIPA buffer. The whole cell lysates (prepared and analyzed in Fig. 4A) were further analyzed by immunobloting with anti-P-STAT5 (top panel) and anti-β-actin (bottom) antibodies. The results shown are a representative of at least three independent experiments.
(D) Phosphorylation is required for STAT5 SUMOylation. COS-1 cells were transfected with FLAG-tagged wild-type STAT5A, or STAT5A phosphorylation mutant Y694A (mutation of tyrosine 694 to alanine), and HA-tagged SUMO-2 plasmids. Twenty four hours after transfection, cellular protein extracts were immunoprecipitated with anti-FLAG antibody followed by Western blot analysis with anti-HA (top panel) or anti-FLAG antibodies (second panel). Cell lysates were analyzed by immunoblotting with anti-HA antibody (bottom panel). The results are a representative of at least three independent experiments.
(E) SUMOylated forms of STAT5 were localized in the nucleus. Cytoplasmic (Cyt) and nuclear (Nuc) fractions from wild-type and SENP1−/− B cells harvested from OP9 coculture at day 7 were prepared, followed by Western blot analysis with anti- STAT5 antibody (top panel), and fraction purity was determined using antibodies against GAPDH (cytoplasmic, second panel) and Lamin B (nuclear, bottom panel).
Since the SUMOylation sites are in close proximity to tyrosine, whose phosphorylation is a prerequisite for STAT5 activation, we therefore assessed the interplay between STAT5 phosphorylation and SUMOylation. We checked the level of STAT5 phosphorylation in SENP1−/− B and T cells derived from HSC differentiation in vitro after 7 days of coculture on OP9 and OP9-DL1 cells in the presence of IL-7 and Flt3L, respectively. Western blot analysis showed a marked reduction of STAT5 phosphorylation in SENP1−/− B and T cells (Figure 5C), correlating with an increased SUMOylation (Figure 4A). Notably, the phosphorylation of STAT5 SUMOylated form could not be detected in SENP1−/− B and T cells. These results suggest that in the absence of SENP1, SUMOylation inhibits STAT5 phosphorylation and that the accumulation of SUMOylated STAT5 impairs the activation–inactivation cycle of STAT5 in lymphocytes.
To further address the interplay between STAT5 phosphorylation and SUMOylation, we introduced a tyrosine to alanine mutation at residue 694 in STAT5A (Y694A mutant) and examined its SUMOylation. As shown in Figure 5D, SUMOylation of Y694A mutant was completely abolished as compared to the wild-type STAT5A. In contrast, the constitutively active form of STAT5A (N642H mutant) is still a good substrate (Figure S6C). These data suggest that tyrosine phosphorylation is required for STAT5 SUMOylation, most likely through regulation of nuclear translocation. To determine the subcellular localization of SUMOylated STAT5, we first coexpressed FLAG-tagged STAT5A alone or with HA-tagged SUMO2 and MYC-tagged PIAS3 plasmids in COS-1 cells. Twenty-four hours after transfection, we prepared nuclear and cytoplasmic fractions of these cells. Equal amounts of each sample were immunoprecipitated with anti-FLAG antibody followed by Western blot analysis with indicated antibodies. SUMOylated forms of STAT5 were found predominantly in the nuclear fraction (Figure S6D). We next examined the subcellular localization of SUMOylated STAT5 in SENP1-deficient B cells, and also found that SUMOylated STAT5 accumulated in the nucleus (Figure 5E).
SENP1 controls acetylation/SUMOylation status of STAT5
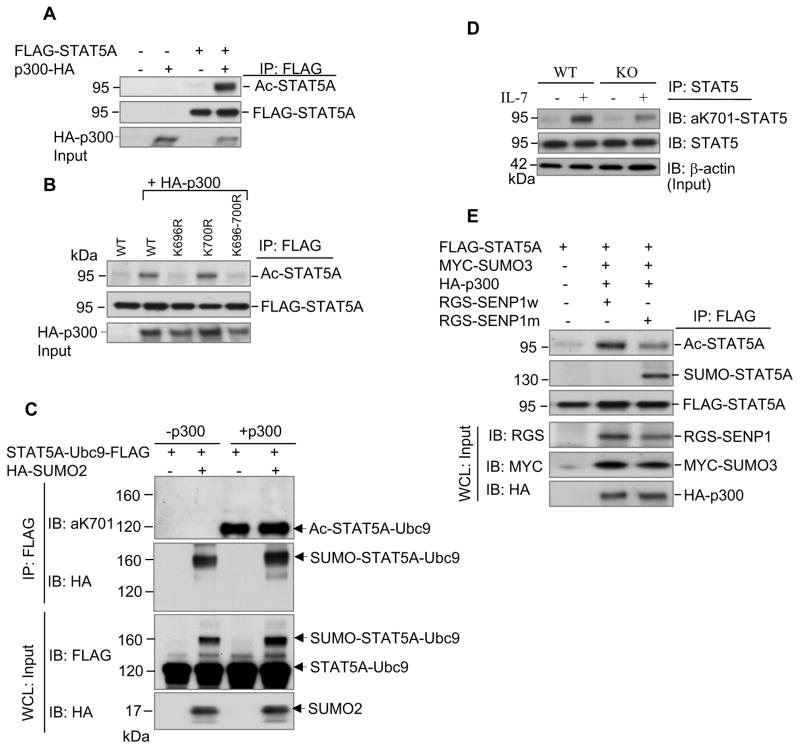
To further understand the effect of SUMOylation on STAT5-mediated transcriptional activity, we compared wild-type and SUMOylation-deficient STAT5A for their ability to activate transcription in reporter assays. STAT5-deficient MEF cells stably expressing wild-type STAT5A and its mutants were transiently transfected with a luciferase reporter pGL4-β-Casein. Compared with wild-type STAT5A, STAT5A(K700R) exhibited a similar transactivation activity, suggesting that SUMOylation of STAT5A at lysine 700 did not play a role in transcription regulation (Figure S7). However, STAT5A(K696R) or STAT5A(K696-700R) showed a markedly decreased in transactivation activity, raising an intriguing possibility that lysine 696 may also be subjected to another form of post-translational modification.
Lysine residues can be targeted by multiple modifications, such as ubiquitination, SUMOylation, methylation, and acetylation. The carboxyl-terminal transactivation domain of STAT5 is known to interact with the histone acetyltransferase p300/CBP, resulting in enhanced STAT5-dependent transcription (Pfitzner et al., 1998). We hypothesized that lysine 696 is also a target for acetylation. To test this possibility, we first checked whether STAT5 can be acetylated by p300. Co-expression of p300 in HEK-293 cells significantly enhances STAT5A acetylation (Figure 6A). Wild-type STAT5A and STAT5A(K700R) mutant, but not STAT5A(K696R) and STAT5A(K696-700R) mutants, were acetylated, suggesting that lysine 696 is also a major acetylation site (Figure 6B). Consistent with our data, a recent study indicates that STAT5B acetylation on lysine residues including lysine 701 (corresponding to lysine 696 on STAT5A) is essential for STAT5B dimerization since mutation of lysine 701 to arginine impaired STAT5B dimerization and transcriptional activity (Ma et al., 2010). To further determine the interplay between SUMOylation and acetylation on a lysine 696/701 of STAT5A/B, we constructed a STAT5A-Ubc9 fusion protein that is efficiently SUMOylated in an Ubc9-dependent manner (Jakobs et al., 2007). COS-1 cells were transfected with FLAG-tagged STAT5A-Ubc9 alone or with HA-tagged SUMO-2 construct. In the presence of HA-tagged SUMO-2, SUMOylation of STAT5A-Ubc9 was greatly increased (Figure S8). Then, purified STAT5A-Ubc9 and SUMOylated STAT5A-Ubc9 (SUMO-STAT5A-Ubc9) were incubated with p300 (HAT domain, Millipore) and acetyl-CoA in vitro. Western blot analysis using an antibody specific for K701-acetylated STAT5B (corresponding to K696-acetylated STAT5A) revealed that only the non-SUMOylated form of STAT5A-Ubc9 was acetylated (Figure 6C), demonstrating that SUMOylation inhibits acetylation through competing the lysine 696 of STAT5A. It is also possible that SUMOylation of K700 could block the acetylation of K696.
Figure 6. SENP1 regulates acetylation/SUMOylation status of STAT5.
(A) STAT5A is acetylated by p300. HEK-293 cells (in 6-well plate) were transfected with indicated FLAG-tagged STAT5A and HA-tagged p300 plasmids. Immunoprecipitation was performed with anti-FLAG and analyzed with anti-acetyl lysine (top panel) and anti-FLAG (second panel) antibodies. The results are a representative of at least three independent experiments.
(B) Lysine 696 is also a target for acetylation. Lysates from HEK-293 cells transfected with plasmids expressing FLAG-tagged STAT5A or SUMOylation-deficient mutants of STAT5A and HA-p300 were immunoprecipitated with anti-FLAG and analyzed with the indicated antibodies. The results are representatives of at least three independent experiments.
(C) SUMOylation of STAT5 inhibits its acetylation. COS-1 cells were transfected with the plasmids expressing FLAG-tagged STAT5A-Ubc9 alone or with HA-tagged SUMO-2. Twenty four to thirty six hours after transfection, cellular protein extracts were immunoprecipitated with anti-FLAG antibody. In vitro acetylation assay using purified STAT5A-Ubc9 and SUMO-STAT5A-Ubc9 was done in the absence or the presence of p300, followed by Western blot analysis with antibodies against K701-acetylated STAT5B (corresponding to K696-acetylated STAT5A) (top panel) and HA (second panel). Cellular extracts were analyzed by immunoblotting with anti-FLAG (third panel) and anti-HA (bottom panel) antibodies. The results are representatives of at least three independent experiments.
(D) Endogenous STAT5 is acetylated in FL cells treated with IL-7. FL cells isolated from three to five E14.5 wild-type and SENP1−/− embryos were treated with IL-7 for 10 min and lysed in RIPA buffer. Cellular extracts were immunoprecipitated with anti-STAT5 antibody and analyzed with anti-aK701-STAT5B (top panel) and anti-STAT5 (second panel) antibodies. Loading samples were immunoblotted with anti-β actin antibody. The results shown are a representative of three independent experiments.
(E) SENP1 regulates the SUMOylation/acetylation status of STAT5. Lysates from HEK-293 Cells (in 6-well plate) transfected with plasmids expressing FLAG-tagged STAT5A, MYC-tagged SUMO3, HA-tagged p300 and RGS-tagged SENP1w (wild-type) or RGS-tagged SENP1m (mutant) were immunoprecipitated with anti-FLAG and analyzed with anti-acetyl lysine (top panel), anti-SUMO2/3 (second panel) and anti-FLAG (third panel) antibodies. The input was immunoblotted with anti-RGS, anti-Myc and anti-HA antibodies. The results shown are a representative of at least three independent experiments.
Next, we assessed endogenous STAT5 acetylation in wild-type and SENP1−/− FL cells. Protein extracts were immunoprecipitated with anti-STAT5 antibody and analyzed by immunoblotting with the specific K701-acetylated STAT5B antibody. As shown in Figure 6D, endogenous STAT5 was acetylated in both wild-type and SENP1−/− FL cells treated with IL-7. Notably, STAT5 acetylation in IL7-treated SENP1−/− FL cells was significantly decreased compared with that of IL7-treated wild-type FL cells, suggesting that SENP1 regulates the acetylation status of STAT5. To confirm the role of SENP1 in the regulation of STAT5 acetylation and SUMOylation, HEK-293 cells were transfected with FLAG-tagged STAT5A, MYC-tagged SUMO3, HA-tagged p300 in the presence of RGS-tagged SENP1 or RGS-tagged SENP1 mutant plasmids. Overexpression of SENP1 enhanced acetylation and inhibited SUMOylation of STAT5A (Figure 6E). In contrast, overexpression of SENP1 mutant decreased acetylation and enhanced SUMOylation of STAT5A (Figure 6E). Taken together, these results demonstrate that SENP1 plays a critical role in regulating the SUMOylation status of STAT5 and that SUMOylation of STAT5 inhibits its phosphorylation, acetylation and subsequent signaling.
DISCUSSION
In this study, we discovered that SENP1 controls lymphoid development through regulation of SUMOylation status of STAT5. Our data revealed that inactivation of SENP1 results in severe defects in early T and B cell development. We further showed that SENP1 deficiency causes accumulation of SUMOylated STAT5, resulting in inhibition of STAT5 activation and subsequent signaling. Moreover, biochemical studies indicated that both acetylation and SUMOylation occur on the same lysine residue in STAT5. SUMOylation of this lysine in STAT5 blocks its acetylation. These results suggest a critical role for SENP1 in regulating STAT5 transcriptional activity. SUMOylation of STAT5 observed in the absence of SENP1 appears to be lymphocyte specific, since no SUMOylated STAT5 could be detected in SENP1−/− myeloid cells.
Protein function is tightly regulated by reversible posttranslational modifications to create an on and off state that is crucial for many biological processes. Many proteins are dynamically modified at multiple sites by different modifications. The interplay between phosphorylation and SUMOylation of neighboring sites has been shown to play an important role in regulating the transcriptional activity of several transcription factors. For example, heat-shock factors (HSFs), GATA-1 and myocyte enhancer factor 2 (MEF2), containing a SUMO consensus site and an adjacent proline-directed phosphorylation site (ΨKxExxSP), are regulated by phosphorylation-dependent SUMOylation (Gregoire et al., 2006; Hietakangas et al., 2003; Hietakangas et al., 2006). The motif ΨKxExxSP couples sequential phosphorylation and SUMOylation and has been referred to as a “Phospho-SUMOyl switch” (Yang and Gregoire, 2006). We have identified a previously undescribed motif in which two SUMOylation sites of STAT5A at lysines 696 and 700 are located in close proximity to tyrosine 694, whose phosphorylation is a prerequisite for STAT5 activation. This suggests a possible interplay between SUMOylation and phosphorylation in regulating STAT5 activity. Indeed, SUMOylation of STAT5 is phosphorylation-dependent, since STAT5 phosphorylation mutant Y694A abolished SUMOylation of STAT5. We also found that SENP1 deficiency causes increased STAT5 SUMOylation, correlating with diminished STAT5 phosphorylation and activity in lymphocytes, indicating that SUMOylation of STAT5 inhibits its phosphorylation and subsequent signaling.
Since lysine can be a target of different posttranslational modifications, SUMOylation can block alternative lysine-targeted modifications, such as ubiquitination, methylation or acetylation. It has been reported that transcriptional activity of several transcription factors, such as SP3, HIC1 and MEF2A can be regulated by interplay between SUMOylation and acetylation on the same lysine residue (Sapetschnig et al., 2002; Shalizi et al., 2006; Stankovic-Valentin et al., 2007). In addition to tyrosine phosphorylation, acetylation of different STATs has been shown to play a critical role in regulating their activity (Kramer et al., 2009; Shankaranarayanan et al., 2001; Tang et al., 2007; Yuan et al., 2005). For example, STAT3 acetylation at lysine 685 is essential for its dimerization and transcriptional activity (Yuan et al., 2005). Here, our data clearly show that STAT5A is acetylated at lysine 696, which is also a target for SUMOylation. Acetylation of STAT5A at lysine 696 is essential for STAT5A activation, since mutation of this lysine diminished the transcriptional activity of STAT5A. Consistent with our finding, a recent study has reported that STAT5B acetylation on lysine 701 (corresponding to lysine 696 on STAT5A) is essential for STAT5B dimerization and transcription (Ma et al., 2010). Notably, our data provide direct evidence that SENP1 regulates the activation of STAT5.
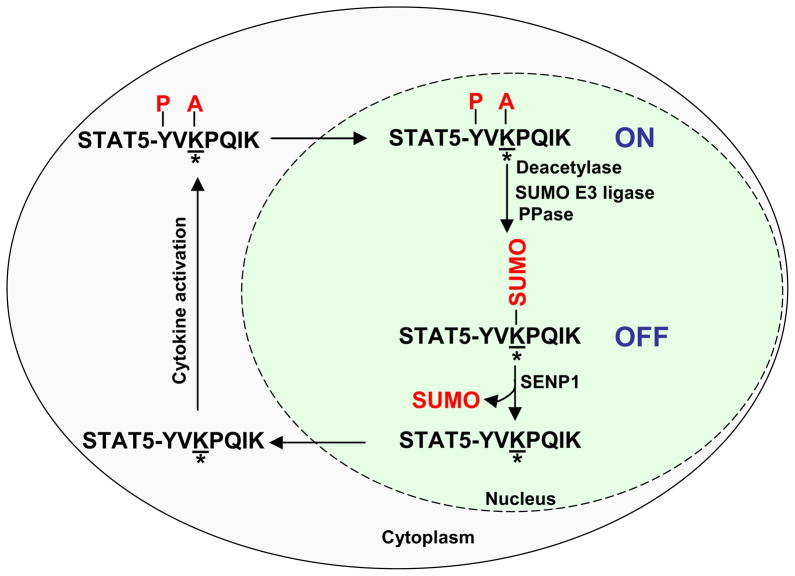
Based on the findings reported here, we propose a model for the role of SENP1 in the regulation of STAT5 activation (Figure 7). Upon activation, tyrosine-phosphorylated and acetylated STAT5 dimerizes, translocates to the nucleus, and activates transcription. We currently do not know which signal induces SUMOylation of STAT5. SENP1 protein, which is predominantly present in the nucleus (Gong et al., 2000), is required for de-conjugating SUMOylated STAT5 before it returns to the cytoplasm to complete an activation-inactivation cycle. In the absence of SENP1, STAT5 is accumulated in the SUMOylation state, leading to inhibition of STAT5 phosphorylation and acetylation, and subsequent signaling.
Figure 7. A model for the role of SENP1 in the regulation of STAT5 activation.
Upon activation, phosphorylated and acetylated STAT5 dimerizes, translocates to the nucleus, and activates transcription. SUMOylation switches STAT5 from an active state to an inactive state. SENP1 is required to remove SUMO from conjugated STAT5, allowing it to re-enter the activation-inactivation cycle. Where * is lysine 696/701 in STAT5A/B; A is acetylation; P is phosphorylation.
It is not clear why SENP1 deficiency selectively affected the lymphoid, but not the myeloid lineage. There are several possibilities. First, lymphoid and myeloid precursors utilize different cytokines during development. For example, IL-7R signaling is critical for early lymphoid, but not myeloid development (Mazzucchelli and Durum, 2007). Second, lymphoid cells may have a SUMO-specific E3 ligase that catalyzes SUMOylation of STAT5. This E3 ligase may not be present in myeloid precursors so STAT5 is not SUMOylated, which obviates the requirement for SENP1 to remove SUMO.
PIAS (Protein Inhibitor of Activated STAT) proteins were initially identified as negative regulators of STAT signaling that inhibit the activity of STAT-transcription factors (Chung et al., 1997; Liu et al., 1998). It has been shown that PIAS proteins function as SUMO-specific E3 ligases, raising the possibility that STAT activity might be regulated by the SUMOylation pathway (Schmidt and Muller, 2003). PIAS3 is known to bind to STAT5 and suppress STAT5-mediated transcription (Rycyzyn and Clevenger, 2002), but the precise molecular mechanism how PIAS3 negatively regulates STAT5 transcriptional activity is unknown. Although we also found that SUMOylation of STAT5 is greatly enhanced by PIAS3 in an overexpression system, further studies will be required to define whether this SUMO-specific E3 ligase is involved in regulation of STAT5 activity in lymphocytes.
In conclusion, SENP1 is essential for early T and B lymphopoiesis. Our data clearly demonstrate that SENP1 controls STAT5 activity by regulating the SUMOylation status of STAT5. Our findings establish a specific role of SENP1 in the regulation of STAT5 activation at the early stages of T and B cells.
EXPERIMENTAL PROCEDURES
Mice
The generation and screening of SENP1−/− fetuses has been described previously (Cheng et al., 2007). SENP1+/− mice were intercrossed, and E14.5 fetuses (date of plug = E0.5) were obtained for FL isolation. RAG1−/− mice were obtained from The Jackson Laboratory. All animal protocols used in this study were approved by the Institutional Animal Care and Use Committee at The University Texas M. D. Anderson Cancer Center.
Purification of FL-HSCs
E14.5 fetal liver cells were harvested and treated with red blood cell lysis buffer (Sigma). Then, cells were stained with anti-c-Kit-APC and anti-Sca-1-PE antibodies, and a mixture of antibodies to lineage markers: Gr-1, CD11b. TER-119, CD19, CD4 and CD8. FL-HSCs (lin- c-KithighSca-1high) were sorted using FACSAria cell sorter (BD Biosciences).
B and T cell differentiation in vitro
OP9 and OP9-DL1 stromal cell lines (generous gifts from Dr. Juan Carlos Zúñiga-Pflücker, University of Toronto, Toronto, Canada) were co-cultured as previously described (Schmitt and Zuniga-Pflucker, 2002). Briefly, stromal cells were maintained in alpha-MEM (Gibco BRL) containing 20% FBS (Gibco BRL) and penicillin–streptomycin (Sigma-Aldrich). Wild-type and SENP1−/− FL-HSCs (1000–3000 cells/well in 24-well plates) were placed on OP9 and OP9-DL1 monolayers for B and T lineage differentiation, respectively. Co-culture media contained: alpha-MEM, 10% FBS, 10 mM HEPES (Gibco BRL), 1 mM sodium pyruvate (Gibco BRL), 2 mM GlutaMax, penicillin–streptomycin, 50 mM 2-mercaptoethanol, 5 ng/ml IL-7 and 5 ng/ml Flt3L. After 4 days of culture, the cells were passed onto newly prepared OP9 and OP9-DL1 cells. For myeloid differentiation, wild-type and SENP1−/− FL-HSCs were placed on OP9 monolayers in the presence of IL3 (10ng/ml), IL6 (10ng/ml), SCF (50 ng/ml) and Flt3L (50 ng/ml).
Transplantation experiments
Two × 106 fetal liver cells isolated from E14.5 wild-type and SENP1−/− embryos were injected intravenously into lethally (950 rad) irradiated RAG-1-deficient mice. Mice were sacrificed 5–6 weeks after transplantation. The lymphoid compartments of the recipients were analyzed by flow cytometry. The experiments shown in this study represent results from two independent experiments.
Supplementary Material
HIGHLIGHTS.
SENP1 is essential for early T and B lymphopoiesis.
SENP1 regulates the SUMOylation status of STAT5.
SUMOylation of STAT5 blocks its acetylation and subsequent signaling.
SENP1 is required for STAT5 signaling during early T and B cell development.
Acknowledgments
We would like to thank Drs. L.-Y. Yu-Lee, Fang Liu, Koichi Ikuta, Yong-Jun Liu & Stephanie S. Watowich for plasmids; Dr. J. C. Zúñiga-Pflücker for OP9-GFP and OP9-DL1 cells, and Dr J. N. Ihle for STAT5-deficient MEF cells. We would also thank Drs. Shao-Cong Sun, Phillip Carpenter, Sue-Hwa Lin and Jingxiong Wang for reading of the manuscript, and the flow cytometry core facility (Karen Martinez, David He, and Amy Cortez) of MD Anderson Cancer Center for technical assistance. This work was supported in part by National Institute of Health Grant to E.T.H.Y (CA239520). T.V.N receives a fellowship from Vietnam Education Foundation. E.T.H.Y. is the McNair Scholar of the Texas Heart Institute/St. Luke’s Episcopal Hospital.
Footnotes
The authors declare no conflicting financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Goodwin RG, Friend D, Ziegler SF, Jerzy R, Falk BA, Gimpel S, Cosman D, Dower SK, March CJ, Namen AE, et al. Cloning of the human and murine interleukin-7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell. 1990;60:941–951. doi: 10.1016/0092-8674(90)90342-c. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, Beug H, Hennighausen L, Moriggl R, Sexl V. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–250. doi: 10.1038/nmeth1006. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, Stauber RH, Bohmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, Zhang Z, Xu L, Hyun YM, Kim M, et al. Acetylation modulates prolactin receptor dimerization. Proc Natl Acad Sci U S A. 2010;107:19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, Leonard WJ. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci U S A. 2002;99:6790–6795. doi: 10.1073/pnas.092160699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S. Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J Biol Chem. 2001;276:42753–42760. doi: 10.1074/jbc.M102626200. [DOI] [PubMed] [Google Scholar]
- Song L, Bhattacharya S, Yunus AA, Lima CD, Schindler C. Stat1 and SUMO modification. Blood. 2006;108:3237–3244. doi: 10.1182/blood-2006-04-020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3313. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- Vieira P, Cumano A. Differentiation of B lymphocytes from hematopoietic stem cells. Methods Mol Biol. 2004;271:67–76. doi: 10.1385/1-59259-796-3:067. [DOI] [PubMed] [Google Scholar]
- von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng EC, Chen H, Krause DS, Min W. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J Exp Med. 2010;207:1183–1195. doi: 10.1084/jem.20092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.