ABSTRACT
Previously we described a method to estimate the average number of virus genomes expressed in an infected cell. By analyzing the color spectrum of cells infected with a mixture of isogenic pseudorabies virus (PRV) recombinants expressing three fluorophores, we estimated that fewer than seven incoming genomes are expressed, replicated, and packaged into progeny per cell. In this report, we expand this work and describe experiments demonstrating the generality of the method, as well as providing more insight into herpesvirus replication. We used three isogenic PRV recombinants, each expressing a fluorescently tagged VP26 fusion protein (VP26 is a capsid protein) under the viral VP26 late promoter. We calculated a similar finite limit on the number of expressed viral genomes, indicating that this method is independent of the promoter used to transcribe the fluorophore genes, the time of expression of the fluorophore (early versus late), and the insertion site of the fluorophore gene in the PRV genome (UL versus US). Importantly, these VP26 fusion proteins are distributed equally in punctate virion assembly structures in each nucleus, which improves the signal-to-noise ratio when determining the color spectrum of each cell. To understand how the small number of genomes are distributed among the replication compartments, we used a two-color fluorescent in situ hybridization assay. Most viral replication compartments in the nucleus occupy unique nuclear territories, implying that they arose from single genomes. Our experiments suggest a correlation between the small number of expressed viral genomes and the limited number of replication compartments.
IMPORTANCE
Herpesviruses use nuclear factors and architecture to replicate their DNA genomes in the host nuclei. Viral replication compartments are distinct nuclear foci that appear during productive infection. We have recently developed a method that uses three viral recombinants (each expressing a different fluorescent protein) to quantify the number of incoming viral genomes that are expressed and replicated in each cell. We found that fewer than seven herpesvirus genomes can be expressed and replicated. Here we have expanded and improved upon our method and demonstrated that the phenomenon of limited genome expression is independent of the recombinants used. We correlated the small number of genomes expressed to the limited number of replication compartments by demonstrating that most replication compartments originate with a single genome. The distinction among replication compartments is maintained even when most of the nucleus is filled with viral DNA, implying that nuclear architecture constrains the compartments.
Introduction
The family Herpesviridae comprises a set of large DNA viruses that replicate in the nucleus of the cell and form similar virion structures. The alphaherpesvirus subfamily shares a common genome organization and the ability to establish lifelong quiescent (latent) infections in neurons. This subfamily contains important human and agricultural pathogens, including herpes simplex 1 and 2 (HSV-1 and HSV-2), varicella zoster virus (VZV), and pseudorabies virus (PRV) (1).
Viral infection begins with the attachment of viral particles to the host cellular membrane, where the nucleocapsids are released into the cytoplasm and transported toward the cell nuclei. Viral genomes enter the cell nuclei at the nuclear pores and begin to express immediate-early proteins, and these in turn allow expression of the early proteins. The early genes initiate viral genome replication in distinct foci known as replication compartments or replication centers (RCs) (2). Late gene transcription occurs after viral DNA replication commences (3, 4). The structural capsid proteins are late gene products that form distinct foci in the nuclei, known as assemblons, where newly synthesized viral genomes are packaged into nucleocapsids (5).
The structure and distribution of RCs in the nucleus are driven by interactions of viral DNA with viral and host proteins (6). Some of the host proteins are derived from nuclear domain 10 (ND10) complexes (7, 8). Although ND10 proteins have a role in silencing foreign DNA, viral genomes associated with ND10 proteins preferentially progress to form viral RCs (9). Both the HSV-1 immediate-early protein ICP0 and its PRV early protein homolog, EP0, inactivate host silencing mechanisms and induce formation of active RCs (10, 11).
Originally, RCs were visualized using indirect immunofluorescence with antibodies directed to viral immediate early proteins (2, 12). Later, other methods, including in situ hybridization to viral DNA (8, 13, 14), fluorescence-tagged proteins (15), and incorporation of labeled nucleotides (16), were also described. Studies with HSV-1 suggested that the number of RCs early after infection is fewer than the number of infectious units added per cell (multiplicity of infection [MOI]) and these early RCs are distributed as distinct foci within the nucleus (13–16). At later times postinfection, these compartments coalesce to form a single large replication body occupying most of the nuclear space (15, 17). Early replication sites also colocalize with transcription sites (16), suggesting that only distinct areas inside the nucleus allow initiation of viral replication and expression.
Discrete nuclear structures containing proteins of both mature and immature capsids were identified as assemblons (5). These structures are located adjacent to the RCs and are involved in capsid maturation and viral genome packaging. The construction of capsid proteins tagged with fluorescent proteins (FPs) allowed live visualization of the formation of assemblons (18, 19). These structures may be related to the accumulation of capsids in crystal-like arrays within the nucleus that were recognized by electron microscopy for many herpesviruses (20).
PRV is a swine alphaherpesvirus commonly used for studies of the molecular biology and pathogenesis of alphaherpesviruses (21). Recently we used three isogenic PRV recombinants, each expressing a different fluorescent protein from the immediate-early cytomegalovirus (CMV) promoter, to estimate the number of incoming viral genomes actively participating in the infection process (22). By monitoring the color distribution of infected cells, we found that only a limited number of incoming genomes are expressed per cell and that in individual cells the expressed genomes are the same genomes that are replicated and packaged. We developed a mathematical model to calculate that on average fewer than seven incoming genomes are replicated per cell (22).
To determine if our results are influenced by the promoter used (CMV immediate-early promoter) or by the insertion site (the gG locus), we constructed a separate set of three isogenic recombinants. These PRV Becker recombinants express unique capsid fusion proteins, each with different fluorescent tags: PRV180 (monomeric red fluorescent protein [RFP]) (23), PRV443 (enhanced green fluorescent protein [EGFP]) (24), and PRV543 (cyan fluorescent protein [CFP]) (unpublished results). All the FPs are expressed as VP26-XFP fusion proteins. These hybrid genes replace the normal VP26 open reading frame and are expressed from the VP26 promoter. VP26 is a small capsid protein present in 900 copies per capsid (24). Using this set of recombinants, we verified our previous findings that only a limited number of viral genomes are expressed per cell.
We also tested the hypothesis that each RC originates from a single genome, as was suggested previously (9). We established a dual-color fluorescent in situ hybridization (FISH) assay for visualizing PRV RCs during coinfection with the two PRV Becker recombinants PRV151 (25) and PRV BeBlu (26), which differ only in a ~3,000-bp insert (egfp from the CMV promoter or lacZ from the native gG promoter, respectively) at an identical locus (gG) in the viral genome. This assay enabled us to directly image the RCs from these two different strains in the same cell. We demonstrate that only a single genome was present in most RCs, suggesting that early RC formation initiates from a single incoming viral genome.
RESULTS
Growth properties of the three PRV recombinants expressing VP26 XFP fusion proteins.
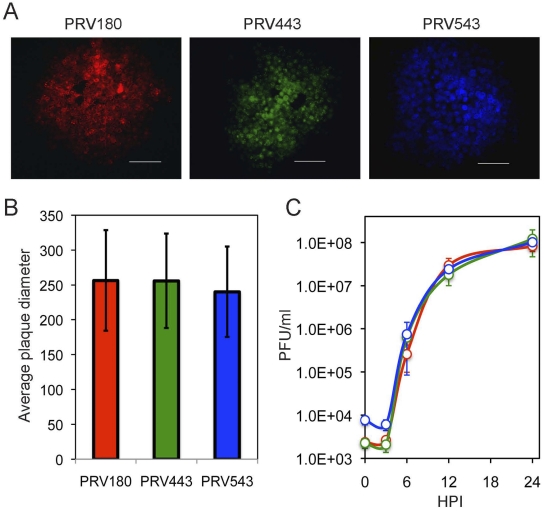
We have shown that only a limited number of incoming viral genomes are expressed and replicated in a single infected cell (22). To ensure that our results were not due to the specific recombinant strains used in the prior study, we constructed three different isogenic recombinants (the differences between the two systems are summarized in Table 1). These recombinants were previously constructed and characterized in our lab: PRV180, PRV443, and PRV543 (Fig. 1A). All strains grew equally well in PK15 cells. There was no statistically significant difference in plaque size (Fig. 1A and B), and the single-step growth curves of the three recombinants in PK15 cells were comparable (Fig. 1C). These results suggest that there are minimal growth differences among the three recombinants
TABLE 1 .
Comparison of the two systems used for quantifying the average number of incoming genomes expressed per cella
| System | Diffusible FPsb | VP26-XFPc |
|---|---|---|
| Promoter | Immediate-early CMV promoter | PRV late VP26 promoter |
| Location of FP genes in genome | In gG gene (US) | Fused to VP26 coding region (UL) |
| FP used | RFP, YFP and CFP | RFP, GFP, and CFP |
| Cell localization | Diffuse | Punctate in nucleus |
A short summary of the major differences between the system used in this article and the system previously used (22). US and UL represent the unique short and unique long regions of the PRV genome, respectively (34).
See reference 22.
See this article.
FIG 1 .
Growth properties of three PRV recombinants expressing VP26-XFP fusion proteins. (A) Representative images of plaques of PRV180, PRV443, and PRV543 were taken 24 hpi. Scale bars, 100 µm. (B) Comparison of the average diameters of 24-hpi plaques (in µm) for the three viruses. At least 100 plaques per well and three wells per virus were measured. Error bars show standard deviations. (C) Single-step growth curve for PRV180, PRV443, and PRV543, marked with red, green, and blue symbols, respectively. Each point is an average of viral titers obtained from three technical replicates. Error bars show standard deviations.
All assemblons in a single nucleus share the same distinct color composition.
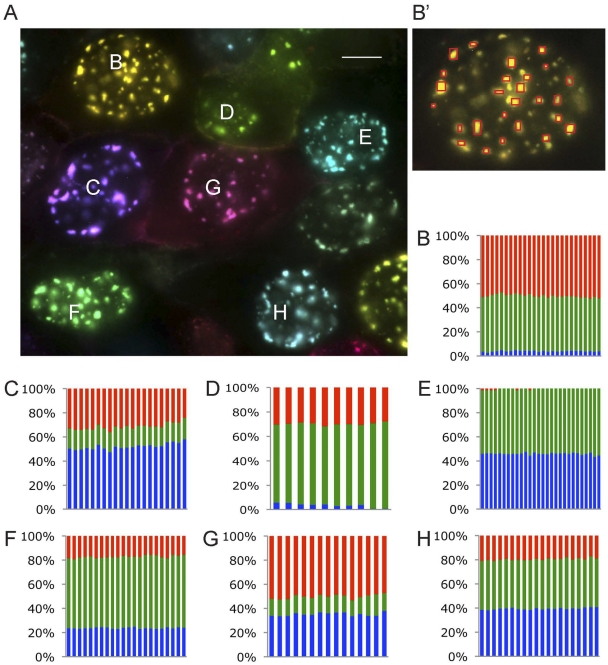
The variability in color spectrum of cells infected with 3 different viruses (each expressing a different FP) negatively correlated with the number of incoming viral genomes expressed in each cell (low variability in color reflects higher numbers of genomes expressed). A stock of an equal number of infectious units from each individual VP26-XFP recombinant (a 1:1:1 mixture based on titer on PK15 cells) was prepared. We infected cells at an MOI of 10 with this mixture (Fig. 2A; see also Movie S1 in the supplemental material). The color spectrum of infected cells was highly variable, corroborating our previous finding that only a limited number of viral genomes are expressed per cell (22). Each graph in Fig. 2B to H represents one nucleus, and the different columns represent the relative ratios of colors in different fluorescent nuclear foci (assemblons), as shown for cell B in Fig. 2B′. In each nucleus, all assemblons detected were the same color, suggesting that viral proteins are uniformly distributed among assemblons in a given nucleus. Moreover, the color of each assemblon remained constant during the time course of infection (see Movie S1).
FIG 2 .
Color analysis of nuclear assemblons. (A) Representative images of PK15 cells infected with a mixture of PRV180, PRV443, and PRV543 at 9 hpi with a MOI of 10. In cells (marked from B to H), individual assemblons were assayed for their fluorescence profiles. Scale bar, 10 µm. (B′) The marking of individual assemblons assayed for cell B are shown. (B to H) The graphs represent the normalized fluorescence composition of individual assemblons in a single nucleus. Each bar corresponds to a single assemblon, and each graph represents a single cell, as indicated by the letters.
Estimating the number of expressed viral genomes using VP26-XFP fusion proteins.
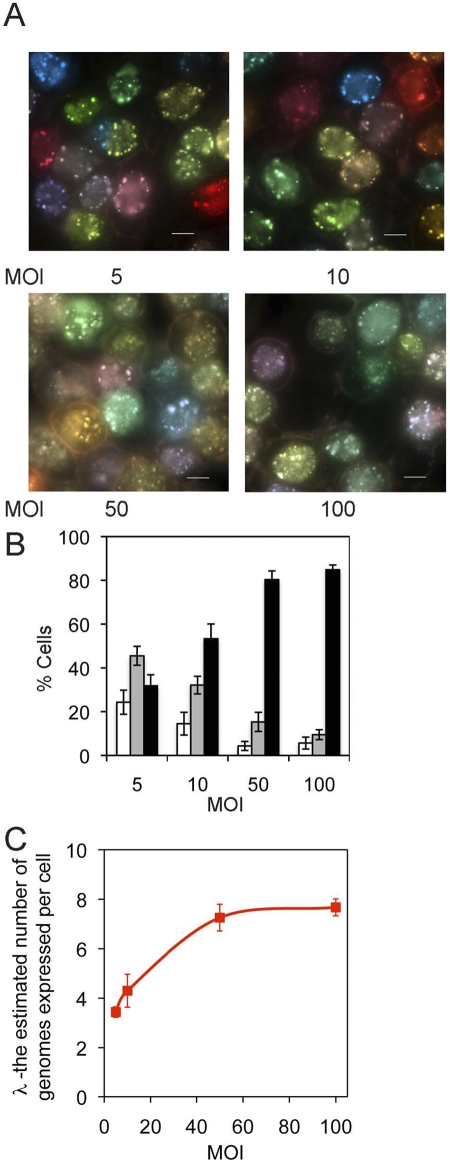
The VP26-XFP fusion protein genes replace the normal VP26 gene and are expressed from the natural VP26 promoter. This promoter functions only after DNA replication commences (a classical late promoter) (27). We used an equal mixture of three isogenic recombinants, PRV180, PRV443, and PRV543, and infected cells at various MOIs (Fig. 3A). Even at the highest MOI tested (MOI = 100), about 5% of the cells expressed a single color (Fig. 3B), and the variability in the color spectrum among cells remained high. We used our model to estimate the number of genomes expressed in each infected cell based on color diversity (22). Briefly, we define λ as the average number of genomes expressed in each cell. We assume that λ is best represented as a Poisson random variable. By determining the color of >600 cells per well (2 technical replicates and 3 biological replicates for each MOI), we estimated λ by maximum-likelihood analysis, according to the following function:
FIG 3 .
A limited number of incoming genomes are expressed and replicated in a newly infected cell. (A) PK15 cells infected with mixture of PRV180, PRV443, and PRV543 at different MOI, as indicated. Imaging of the infected cells was initiated 6 hpi. Scale bar, 10 µm. (B) Percentages of cells that are one color (white), two colors (gray), and three colors (black) were plotted for different MOI as indicated. For each MOI, an average of five random areas were imaged from one well. From each area, 120 cells were analyzed for their color content. Error bars show standard deviations between the areas. (C) The calculated λ values for mixed infection by PRV180, PRV443, and PRV543 are plotted as a function of the MOI. Each point is an average for 6 different replicates. The standard error is presented for each point.
In which r1, r2, and r3 represent the numbers of cells that are one-color, two-color, and three-color, respectively, and n represents the total number of colored cells that were analyzed. We then plotted λ (average for six wells per condition) as a function of the MOI (Fig. 3C).
Our findings confirm that a limited number of PRV genomes are expressed in each cell. Even at an MOI of 100, fewer than eight genomes contribute to the color of the cell. This number is slightly larger than our earlier estimate (fewer than six genomes) using PRV Becker recombinants expressing soluble XFPs from the CMV immediate-early promoter (22). Because the change in the number of expressed genomes as a function of MOI follows the trend reported previously, the small difference in λ may simply reflect experimental variation. We conclude that this method, using cell color to count expressed genomes, does not depend on the promoter or insertion sites used to construct the test recombinants.
Detection of PRV replication compartments using two-color FISH.
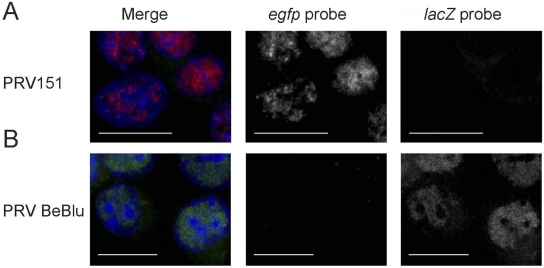
HSV-1 has been shown to form a limited number of RCs (15, 16). Taken together with our finding that a limited number of genomes is expressed per cell, we predicted that each RC would originate with a single genome as was previously suggested (9). To visualize PRV RCs, we used fluorescent in situ hybridization (FISH) probes to detect two PRV recombinants, PRV BeBlu and PRV151, each carrying a unique ~3,000-bp insertion, the lacZ gene or the EGFP gene, respectively. To visualize PRV RCs, we synthesized a unique set of PCR product probes directed to the unique sequence (egfp probes for PRV151 and lacZ probes for PRV BeBlu). At 4 h postinfection (hpi), PRV BeBlu- or PRV151-infected cells were fixed and hybridized with the two probes (Fig. 4). There was no cross-reactivity of the probe with the other viral DNA, and we could distinguish between the two recombinants by FISH. We were not able to detect any RC at earlier time points (2 hpi) or in the presence of the viral DNA synthesis inhibitor phosphonoacetic acid (PAA) (data not shown), suggesting that only replicating genomes are detected by our method. Our results also indicate that the sizes of the RCs detected vary among cells but are comparable for the two viruses at this time point (Fig. 4).
FIG 4 .
Similar viral replication centers after infection with two PRV recombinants. PK15 cells were infected by PRV151 (A) or PRV BaBlu (B) at an MOI of 10. The cells were then fixed at 4 hpi and hybridized with an egfp probe labeled with Alexa Fluor 647 (labeled in red) and a lacZ probe labeled with Alexa Fluor 488 (labeled in green). DAPI-labeled nuclei are in blue. Images are a maximal projection of 5 slices (0.5 µm apart) from a confocal microscope. Scale bar, 20 µm.
PRV151 and PRV BeBlu form distinct RCs in the nucleus.
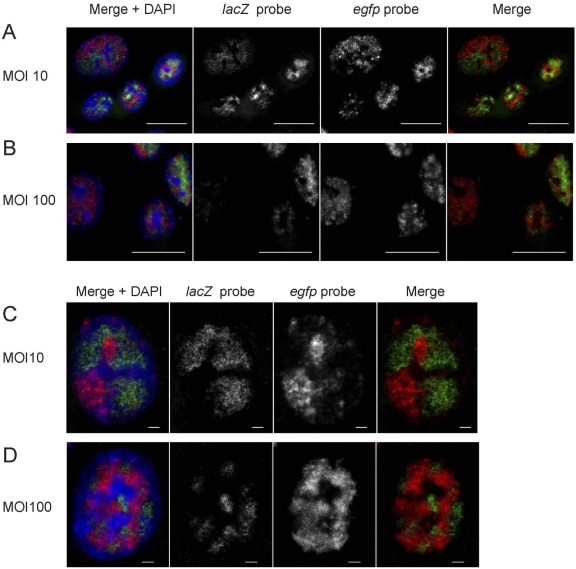
The average number of HSV-1 RCs per cell has been shown to be lower than the number of added infectious units (16). One explanation may be that several viral genomes are needed to establish a functional RC. To test how many viral genomes are found in a single RC, we infected cells with mixtures of PRV151 and PRV BeBlu containing both in equal proportions. Following infection, the cells were fixed 4 hpi and hybridized with both fluorescent DNA probes. As expected, the majority of cells were infected with both viruses (in experiments with both MOI of 10 and 100). The distribution of FISH signal for each probe was restricted to discrete foci with little colocalization of the two probes, suggesting that most RCs contained only a single type of genome (Fig. 5A to D). Even in cells where the RCs coalesce and fill most of the nuclear space, distinct territories, containing a single type of genome with little overlap between the territories, are obvious, as clearly seen in a single confocal slice (Fig. 5C and D).
FIG 5 .
Formation of distinct replication compartments by PRV151 and PRV BeBlu. PK15 cells were infected with PRV151 and PRV BeBlu at an MOI of 10 (A and C) or 100 (B and D), fixed at 4 hpi, and hybridized with an egfp probe labeled with Alexa Fluor 647 (labeled in red) and a lacZ probe labeled with Alexa Fluor 488 (labeled in green). DAPI-labeled nuclei are in blue. Images A and B are a maximal projection of 5 slices (0.5 µm apart) from a confocal microscope, and images C and D are a single slice from a confocal microscope. Scale bars are 20 µm or 2 µm for images A and B or C and D, respectively.
In this experimental setting, if two genomes were involved in each RC, one would expect that 50% of the RCs would harbor both genomes (the percentage will be higher if more genomes were involved). We rarely observed both genomes in a single RC and concluded that most RCs originate with a single viral genome.
DISCUSSION
We were initially surprised to find that even at high multiplicities of infection, only a small number of PRV genomes contributed to the pool of expressed and replicated genomes (22). Here we have corroborated these findings using a different set of recombinant viruses that express different fluorescent fusion proteins from a different promoter that is expressed at a different time after infection.
The VP26 fusion recombinants described in this article provide several advantages for our method. First, the concentration of the FPs in the nuclear assemblons results in a higher signal-to-noise ratio, allowing better detection of all the colors in a single cell. Second, using a native late promoter for expression of the FPs facilitates the detection of replicating genomes rather than any transcriptionally active genome (3, 4). The slightly higher average number of genome reported here (eight) than in the previous study (greater than six) (22) probably reflects the better signal-to-noise ratio.
We have extended our observations showing that even at high MOIs, cells coinfected with PRV151 and PRV BeBlu formed genome-specific RCs. Since both viruses are isogenic except for the lacZ or egfp insert and form comparable numbers of RCs when infected alone, it seems likely that the only difference between the RCs is the genome that is replicated. Our data strongly suggest that most RCs are seeded by a single incoming viral genome. Preliminary evidence of a single source for each RC was obtained by Sourvinos et al. for HSV-1 amplicons (9).
By following HSV-1 ICP8 fused to GFP, Taylor et al. suggested a two-step model for the formation of RCs (15). The first step includes movement of smaller compartments to be next to each other, and the second step is growth by accumulation and fusion of the smaller structures (see Fig. 12 in reference 15). While several RCs come into contact with each other, it is hard to detect mixing of these RCs even at the protein level (17). Our results suggest that each RC forms from a single genome. Remarkably, even when the nucleus is filled with viral DNA, the separation between different RCs is still easily observed (Fig. 5), suggesting that each RC maintains its own territory. This finding is reminiscent of the architecture attributed to chromosomal territories (recently reviewed in reference 28), where different areas of the nucleus serve different functions in transcription, replication, and RNA processing. In agreement with this hypothesis, Chang et al. have shown recently that early HSV-1 RCs move toward and coalesce at nuclear speckles (17). As a result of this movement, nuclear speckles should be found between viral territories, as suggested for chromosomal territories (28).
While most RCs arise from one viral genome, we did identify a few RCs with both genomes. Dual RCs may be the sites for viral recombination, which is well known to occur in the herpesviruses (recently reviewed in reference 29). These dual RCs may represent a population that originated with two (or more) individual genomes. Another explanation is that these dual RCs originated with a single genome, but in the process of replication, a nearby viral genome that was not expressed recombined with the replicating genome. As a result, both genomes now would be detected in the same RC. A third possibility is that dual RCs result from merging of independently formed RCs, as suggested by Taylor et al. (15). To distinguish these and other models for formation of mixed RCs, it will be important to develop real-time live visualization of different viral genomes in single nuclei.
Our findings correlate the known limited number of RCs with the restriction of the number of expressed viral genomes. The limited number of RCs may also result from the restriction of genome expression, since this restriction can be detected even with immediate-early promoters (22). These results also support the finding that not all viral genomes establish RCs (9, 30). We speculate that the limited expression of incoming viral genomes reflects either reduced levels of essential cellular factors needed for initiation of expression or the action of viral proteins, such as VP16, EP0 (ICP0), and IE180 (ICP4), which are known to be involved in early replication and are detected in viral particles (31, 32).
MATERIALS AND METHODS
Viruses and cells.
PRV Becker recombinants PRV180, PRV443, PRV151, and PRV BeBlu were described previously (23–26). PRV543 was constructed by insertion of the CFP gene fused to the VP26 gene, similar to the construction of PRV180 and PRV443 (unpublished data). All viruses were propagated and titers were determined in PK15 (porcine kidney epithelial) cells. The multiplicity of infection (MOI) was calculated as the number of PFU per cell.
VP26-XFP image acquisition and analysis.
To estimate the number of PRV genomes expressed in each infected cell, we first obtained images of cells infected with equal amounts of the three recombinants (PRV180, PRV443, and PRV543) using a Nikon Eclipse Ti-E epifluorescence inverted microscope. Each MOI condition was replicated in two wells, and the experiment was performed three times. From an individual well, five random areas were imaged. From each image, on average 120 cells were analyzed for their color content.
To define the average number of incoming genomes being expressed, we used the mathematical model developed previously (22). Cells were infected for 6 h at a high MOI (>5), allowing synchronized infection. We did not observe any evidence of exclusion after high-MOI infection. Therefore, we concluded that each virus can independently infect a cell. Given these facts, we assumed that the number of incoming viral genomes can be represented as a Poisson random variable (λ).
We then computed the different probabilities for the number of colors (that is, zero, one, two or three) expressed in an infected cell. By combining these probabilities, we estimated the model parameter λ, using maximum-likelihood analysis.
Fluorescence in situ hybridization.
PK15 cells were grown on poly-l-lysine-coated 8-well Lab-Tek chamber slide systems. To synchronize the infections, cell culture slides were inoculated with virus and then kept on ice for 45 min before transfer to 37°C for an additional 15 min. The inoculum was removed, and the infected cells were incubated at 37°C for 4 h. After incubation, the cells were fixed by 100% methanol at −20°C. The methanol was then removed, and the slides were air dried and stored in 4°C until further use.
The probes for in situ hybridization were PCR fragments of two regions each in the egfp and lacZ genes amplified with Advantage2 polymerase (Clontech) from viral DNA. The template DNA was derived from viral stock (either PRV151 or PRV BeBlu) that was boiled for 9 min at 95°C. Primers were designed using the Primer3 oligonucleotide design software program according to the given guidelines (33). To create a variety of fragments differing in length, 3 primer mixtures for the egfp cassette or 5 primer mixtures for the lacZ gene were used to amplify each region, as detailed in Table 2. The three PCRs for egfp were combined as the egfp probe, and the 5 reaction mixtures were combined as the lacZ probe. The purified-mix PCR products were labeled using Ulysis nucleic acid labeling kits (Invitrogen), according to the manufacturer’s protocol, with Alexa Fluor 647 or Alexa Fluor 488, and the labeled probes were purified using Micro Bio-Spin P-30 SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) chromatography columns from Bio-Rad.
TABLE 2 .
Primers used for making FISH probesa
| PCR mix | Primer name | Orientation | Starting codon | Sequence from 5′ |
|---|---|---|---|---|
| egfp 1 | egfp-L31 | Forward | 1 | ATGGTGATGCGGTTTTGG |
| egfp 1 | egfp-R263 | Reverse | 232 | TAGCGGATCTGACGGTTC |
| egfp 1 | egfp-L287 | Forward | 256 | GCTCAAGCTTCGAATTCTGC |
| egfp 1 | egfp-R595 | Reverse | 564 | TCGTGCTGCTTCATGTGG |
| egfp 1 | egfp-L598 | Forward | 567 | TCTTCAAGTCCGCCATGC |
| egfp 1 | egfp-R757 | Reverse | 726 | ATGTTGCCGTCCTCCTTG |
| egfp 2 | egfp-L807 | Forward | 776 | ATGGCCGACAAGCAGAAG |
| egfp 2 | egfp-R985 | Reverse | 954 | TTGGGGTCTTTGCTCAGG |
| egfp 2 | egfp-L1035 | Forward | 1004 | ATCACTCTCGGCATGGAC |
| egfp 2 | egfp-L1146 | Forward | 1115 | TCCCCCTGAACCTGAAAC |
| egfp 2 | egfp-R1297 | Forward | 1266 | GAGTTTGGACAAACCACAAC |
| egfp 2 | egfp-R1403 | Reverse | 1372 | GCCGATTTCGGCCTATTG |
| egfp 3 | egfp-L1393 | Forward | 1362 | CCGAAATCGGCAAAATC |
| egfp 3 | egfp-R1555 | Reverse | 1524 | GGTGATGGTTCACGTAGTGG |
| egfp 3 | egfp-L1566 | Forward | 1535 | TTTTTGGGGTCGAGGTG |
| egfp 3 | egfp-R1766 | Reverse | 1735 | CTGTAGCGGCGCATTAAG |
| egfp 3 | egfp-L1783 | Forward | 1752 | CTGTAGCGGCGCATTAAG |
| egfp 3 | egfp-R1923 | Reverse | 1892 | TGGTTCTTTCCGCCTCAG |
| lacZ 1 | lacZ-L54 | Forward | 1 | ATTCCGCCGATACTGACG |
| lacZ 1 | lacZ-R172 | Reverse | 118 | ATTCCGCCGATACTGACG |
| lacZ 1 | lacZ-L189 | Forward | 135 | AGCGGCTGATGTTGAACTG |
| lacZ 1 | lacZ-R342 | Reverse | 288 | ACTGCCGCCTGTTTTGAC |
| lacZ 1 | lacZ-L361 | Forward | 307 | TTCTTGCGGCCCTAATCC |
| lacZ 1 | lacZ-R547 | Reverse | 493 | GCAGCATCAGGGGAAAAC |
| lacZ 2 | lacZ-L690 | Forward | 636 | CGCCAATGTCGTTATCCAG |
| lacZ 2 | lacZ-R910 | Reverse | 856 | TCTGGCGGAAAACCTCAG |
| lacZ 2 | lacZ-L1048 | Forward | 994 | TTGTGGAGCGACATCCAG |
| lacZ 2 | lacZ-R1226 | Reverse | 1172 | CGCTGACGGAAGCAAAAC |
| lacZ 2 | lacZ-L1286 | Forward | 1232 | GGCGTATCGCCAAAATCAC |
| lacZ 2 | lacZ-R1442 | Reverse | 1388 | ACAGTCTTGGCGGTTTCG |
| lacZ 3 | lacZ-L1453 | Forward | 1399 | GTGGGCGTATTCGCAAAG |
| lacZ 3 | lacZ-R1656 | Reverse | 1602 | CGCTGGATCAAATCTGTCG |
| lacZ 3 | lacZ-L1659 | Forward | 1605 | ACAGCGCGTCGTGATTAG |
| lacZ 3 | lacZ-R1816 | Reverse | 1762 | CATGGTGCCAATGAATCG |
| lacZ 3 | lacZ-L1893 | Forward | 1839 | TCGGATAATGCGAACAGC |
| lacZ 3 | lacZ-R2071 | Reverse | 2017 | GGTGCGGATTGAAAATGG |
| lacZ 4 | lacZ-L2086 | Forward | 2032 | ATCGCAGGCTTCTGCTTC |
| lacZ 4 | lacZ-R2252 | Reverse | 2198 | TCGGCGGTGAAATTATCG |
| lacZ 4 | lacZ-L2278 | Forward | 2224 | CGTTTCACCCTGCCATAAAG |
| lacZ 4 | lacZ-R2448 | Reverse | 2394 | TTCCGTGACGTCTCGTTG |
| lacZ 4 | lacZ-L2457 | Forward | 2403 | TCATCCGCCACATATCCTG |
| lacZ 4 | lacZ-R2556 | Reverse | 2502 | CTGAGCGCATTTTTACGC |
| lacZ 5 | lacZ-L2564 | Forward | 2510 | TCAGACGGCAAACGACTG |
| lacZ 5 | lacZ-R2734 | Reverse | 2680 | TGTTCCCACGGAGAATCC |
| lacZ 5 | lacZ-L2739 | Forward | 2685 | GCGGATTGACCGTAATGG |
| lacZ 5 | lacZ-R2917 | Reverse | 2863 | CAGCCTGAATGGCGAATG |
| lacZ 5 | lacZ-L2916 | Forward | 2862 | TGCGCAACTGTTGGGAAG |
| lacZ 5 | lacZ-R3057 | Reverse | 3003 | TCACTGGCCGTCGTTTTAC |
The sequences of the PCR primers used for generating the FISH probes are listed. The position and orientation of each probe relative to the first probe for each gene are given.
Hybridization was performed using a protocol adapted from the method of Everett et al. (8). The cells were prehybridized by incubation for 30 min at 37°C with hybridization buffer (50% formamide, 10% dextran sulfate, and 4× SSC) in a humidified chamber. RNase digestion with 20 µg/ml RNase A (Boehringer Mannheim Corp.) for 2 h at room temperature had no effect on the results (data not shown). The slides were removed from the chamber and blotted dry. Labeled fragments against each gene region were added to the hybridization buffer to a concentration of 1 ng/µl, after which the slides were incubated in the humidified chamber at 95°C for 2 min to denature the probe and the sample. Hybridization was continued overnight at 37°C. The cells were then washed twice for 10 min at 60°C and once at 37°C with 2× SSC. The slides were counterstained with SlowFade gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) from Invitrogen and imaged in a Leica SP5 confocal microscope.
SUPPLEMENTAL MATERIAL
PK15 cells infected with a mixture of PRV180, PRV443, and PRV543 at a MOI of 10. Imaging was initiated at 3 hpi, and frames were collected every 10 min. Download Movie S1, MOV file, 1.7 MB.
ACKNOWLEDGMENTS
We thank Roger Everett for sharing the FISH protocol. We thank all Enquist lab members for their comments.
O.K. is funded by the International Human Frontier Science Program. L.W.E. acknowledges support from NIH grants 1RC1NS068414 and P40 RR 018604.
Footnotes
Citation Kobiler O, Brodersen P, Taylor MP, Ludmir EB, Enquist LW. 2011. Herpesvirus replication compartments originate with single incoming viral genomes. mBio 2(6):e00278-11. doi:10.1128/mBio.00278-11.
REFERENCES
- 1. Flint SJ, Enquist LW, Racaniello VR, Skalka AM. 2009. Principles of virology, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 2. Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868 [DOI] [PubMed] [Google Scholar]
- 3. Holland LE, Anderson KP, Shipman C, Jr, Wagner EK. 1980. Viral DNA synthesis is required for the efficient expression of specific herpes simplex virus type 1 mRNA species. Virology 101:10–24 [DOI] [PubMed] [Google Scholar]
- 4. Jones PC, Roizman B. 1979. Regulation of herpesvirus macromolecular synthesis. VIII. The transcription program consists of three phases during which both extent of transcription and accumulation of RNA in the cytoplasm are regulated. J. Virol. 31:299–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward PL, Ogle WO, Roizman B. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maul GG. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660–667 [DOI] [PubMed] [Google Scholar]
- 8. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sourvinos G, Everett RD. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Everett RD. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365–374 [DOI] [PubMed] [Google Scholar]
- 11. Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J. Virol. 84:3476–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knipe DM, Senechek D, Rice SA, Smith JL. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maul GG, Ishov AM, Everett RD. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67–75 [DOI] [PubMed] [Google Scholar]
- 15. Taylor TJ, McNamee EE, Day C, Knipe DM. 2003. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 309:232–247 [DOI] [PubMed] [Google Scholar]
- 16. Phelan A, Clements JB. 1997. Functional domains within the nucleus of a cell infected with HSV-1. Rev. Med. Virol. 7:229–237 [DOI] [PubMed] [Google Scholar]
- 17. Chang L, et al. 2011. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. U. S. A. 108:E136–E144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desai P, Person S. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Oliveira AP, et al. 2008. Live visualization of herpes simplex virus type 1 compartment dynamics. J. Virol. 82:4974–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyamoto K. 1971. Mechanism of intranuclear crystal formation of herpes simplex virus as revealed by the negative staining of thin sections. J. Virol. 8:534–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pomeranz LE, Reynolds AE, Hengartner CJ. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. 2010. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat. Commun. 1:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. del Rio T, Ch’ng TH, Flood EA, Gross SP, Enquist LW. 2005. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J. Virol. 79:3903–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith GA, Gross SP, Enquist LW. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 98:3466–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith BN, et al. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. U. S. A. 97:9264–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banfield BW, Yap GS, Knapp AC, Enquist LW. 1998. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J. Virol. 72:4580–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McNabb DS, Courtney RJ. 1992. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J. Virol. 66:2653–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2:a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muylaert I, Tang KW, Elias P. 2011. Replication and recombination of herpes simplex virus DNA. J. Biol. Chem. 286:15619–15624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Everett RD, Zafiropoulos A. 2004. Visualization by live-cell microscopy of disruption of ND10 during herpes simplex virus type 1 infection. J. Virol. 78:11411–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85:6427–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michael K, Klupp BG, Mettenleiter TC, Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 34. Szpara ML, et al. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 7:e1002282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PK15 cells infected with a mixture of PRV180, PRV443, and PRV543 at a MOI of 10. Imaging was initiated at 3 hpi, and frames were collected every 10 min. Download Movie S1, MOV file, 1.7 MB.