Abstract
Background
The role of cholesteryl ester transfer protein (CETP) in the metabolism of HDL cholesterol (HDL-C) is well studied but still controversial. More recently, GWAS and metaanalyses reported the association of a promoter variant (rs3764261) with HDL-C in Caucasians and other ethnic groups. In this study, we have examined the role of genetic variation in the promoter region of CETP with HDL-C, CETP activity, coronary artery disease (CAD), CAD risk factors, and the interaction of genetic factors with environment in a unique diabetic cohort of Asian Indian Sikhs.
Methods and Results
We genotyped four variants; three tagSNPs from promoter (rs3764261, rs12447924, rs4783961) and one intronic variant (rs708272 Taq1B) on 2,431 individuals from the Sikh Diabetes Study. Two variants (rs3764261 and rs708272) exhibited a strong associations with HDL-C in both normo-glycemic (NG) controls (β= 0.12; p= 9.35 ×10−7 for rs3764261; β= 0.10, p= 0.002 for rs708272) and diabetic cases (β= 0.07, p= 0.016 for rs3764261; β= 0.08, p= 0.005 for rs708272) with increased levels among minor homozygous ‘AA’ carriers. In addition, the same ‘A’ allele carriers in rs376426 showed a significant decrease in systolic blood pressure (β= −0.08, p= 0.002) in NG controls. Haplotype analysis of rs3764261, rs12447924, rs4783961, and rs708272 further revealed a significant association of ‘ATAA’ haplotype with increased HDL-C (β= 2.71, p= 6.38 ×10−5) and ‘CTAG’ haplotype with decreased HDL–C levels (β= −1.78, p= 2.5×10−2). Although there was no direct association of CETP activity and CETP polymorphisms, low CETP activity was associated with increased risk to CAD (age, BMI and gender adjusted odds ratio 2.2 95% CI (1.4–3.4, p= 0.001) in this study. Our data revealed a strong interaction of rs3764261 and rs708272 for affecting the association between CETP activity and HDL–C levels; p= 2.2 × 10−6, and p= 4.4 × 10−4, respectively.
Conclusions
Our results, in conjunction with earlier reports confirm low CETP activity to be associated with higher CAD risk. Although there was no direct association of CETP activity with CETP polymorphisms, our findings revealed a significant interaction between CETP SNPs and CETP activity for affecting HDL-C levels. These results urge a deeper evaluation of the individual genetic variation in the CETP before implementing pharmaceutical intervention of blocking CETP for preventing CAD events.
Dyslipidemia with low serum high-density lipoprotein cholesterol (HDL-C) and elevated low-density cholesterol (LDL-C) levels is a well established risk factor for coronary artery disease (CAD) and a leading cause of mortality in individuals with type II diabetes (T2D). In the past decade, decreasing LDL-C has been the major goal in primary and secondary prevention of CAD through treatment with HMG-CoA reductase inhibitors (statins). However, a large body of data suggests that while statin therapy can reduce CAD events by ~30%, the mortality rate due to CAD remains elevated particularly in the patients with metabolic disease and insulin resistance 1. Decreased HDL-C has been suggested to be a strong, independent, predictor of increased risk for CAD by several epidemiological studies 2. Although, hormonal, environmental, and cultural factors determine HDL-C levels within ethnic populations, a genetic component accounts up to 76% of the variation in HDL-C 3. High heritability of HDL-C and HDL-associated lipid traits provide a strong rationale for identifying genetic loci that may help uncover novel pathways crucial for HDL-C regulation and eventually for treatment or early prevention of CAD.
The role of CETP in metabolism of HDL-C is well studied but still controversial. CETP mediates the exchange of lipids between lipoproteins resulting in the net transfer of cholesteryl ester from HDL-C to other lipoproteins and subsequent uptake of cholesterol by hepatocytes through reverse-cholesterol transport 4. Genetic variation in rs708272 (also called Taq1B) in CETP gene has been extensively studied for association with variation in HDL-C in different populations 4, 5. One large meta-analysis study using 147,000 individuals from published studies (between 1970 to January 2008) reported CETP genotypes to be associated with moderate inhibition of CETP activity (with modestly increased HDL-C) and inverse association with CAD 6. However, some other studies have seen greater CAD risk associated with low CETP activity in individuals with genetic deficiency 7, 8. A recent prospective investigation on a moderate size community-based sample from Framingham Heart Study also reported greater CAD risk associated with low CETP activity 9. Most of these genetic studies have been focused on rs708272 (Taq1B) and could not clearly specify the effect of this or other variants on variation in CETP activity or CAD risk. A clear understanding of how genetic variation in CETP affects HDL-C and other risk factors associated with CAD in interaction with environmental factors is still lacking especially in ethnic groups at high risk for T2D and premature CAD. More recently, GWAS studies reported the association of a promoter variant −2568 (rs3764261) with HDL-C variation in Caucasians and has been confirmed in several large meta-analysis studies including different ethnic groups 10, 11.
Based on the premise that increased CETP activity decreased HDL-C levels, a new class of drugs (including torcetrapib) were developed with the intent to raise HDL-C levels through inhibition of CETP activity. However, a failure of the trial of torcetrapib, due to an unacceptable increase in CAD mortality (25%) along with 60% increase in all-cause mortality questioned the logic of CETP inhibition and urged re-evaluation of the role of CETP in possibly preventing CAD events 12. Additionally, recently published DEFINE trial of anacetrapib showing robust effect on lowering LDL-C and increasing HDL-C by CETP inhibition in a moderate size Caucasian populations (n=1,623)13 urges further investigations in other high risk ethnic population (such as this) for ensuring safe inhibition of CETP to lower CAD risk.
In this context, we examined the role of three SNPs (rs3764261 [−2568], rs12447924 [−1700], and rs4783961 [−998]) located in the promoter region and a non-coding SNP (rs708272/Taq1B) located in first intron of the CETP gene. These SNPs were genotyped in a sample of 2,431 participants drawn from our unique Sikh population of Northern India (Punjab) known to have a high prevalence of T2D and CAD14, 15.
METHODS
Study Participants
The study participants are part of our ongoing Sikh Diabetes Study (SDS) 16. DNA and serum samples were drawn from 2,431 participants: 1,307 T2D cases [682 males 56 ± 11 yrs (mean ± SD) and 625 females 55 ± 11 yrs] and 1,124 NG controls [595 males 51 ± 15 yrs and 529 females 50 ± 13 yrs]. The diagnostic criteria used for classifying participants as T2D case, NG control, or impaired glucose tolerant (IGT) have been described in detail elsewhere 17. Briefly, clinical records including medication histories were used to determine the individual’s initial T2D status. Since this study was focused on Punjabi community from Northern India, individuals of South, East, and Central Indian origin were excluded. Individuals with type 1 diabetes (T1D), maturity-onset diabetes of the young (MODY), secondary diabetes due to hemochromatosis, cystic fibrosis, or pancreatitis or individuals with a family member with T1D were excluded from the study. If the individual was eligible for the study, a full clinical evaluation was performed to confirm the diagnosis measuring fasting glucose levels following the guidelines of American Diabetes Association 18. IGT was defined as a fasting blood glucose (FBG) level > 100.8 mg/dL but < 126.1 mg/dL or a 2 hour oral glucose tolerance test (2h OGTT) > 141.0 mg/dL but ≤ 200 mg/dL. The 2h OGTTs were performed following the criteria of the World Health Organization (WHO) (75 g oral load of glucose). BMI was calculated as weight (kg)/[height (m)2]. The NG participants were recruited from the same Punjabi community and from the same geographic location as the T2D participants 14. These were selected on the basis of a fasting glycemia <100.8 mg/dL or a 2h glucose <141.0 mg/dL. Participants with IGT were excluded when data were analyzed for association of variants with T2D.
Questionnaires provided information about the participant’s age, sex and the disease status. In addition to demographic information, the questionnaires collected information on diet, physical activity, smoking, alcohol, medication, and geographic and family health history 14. In general, Punjabi Sikhs do not smoke for religious and cultural reasons. About 50% of participants were life-long vegetarians, and the vast majority of Punjabi females do not drink alcohol 14. Alcohol consumption was scored on a 0–4 scale: 0- for no alcohol, 1– 50 to 100 mL/day, 2– 100 to 400 mL/day, 3– 400 to 1000mL (1L)/day, 4- >1L/day.
Information regarding CAD, date of coronary artery bypass graft or angioplasty, and medication usage was obtained from patient records. CAD was considered if there was use of nitrate medication (nitroglycerine), electrocardiographic evidence of angina pain, coronary angiographic evidence of severe (greater than 50%) stenosis, or echocardiographic evidence of myocardial infarction. In this study, about 15% of participants had CAD.
All participants provided a written informed consent for investigations. The study was reviewed and approved by the University of Oklahoma Health Sciences Center’s Institutional Review Board, as well as the Human Subject Protection Committees at the participating hospitals and institutes in India.
Metabolic Assays
All blood samples were obtained at the baseline visit after having fasted for at least eight hours. Insulin was measured by radio-immuno assay (Diagnostic Products, Cypress, USA). HOMA IR was calculated as (fasting glucose × fasting insulin)/22.5 and HOMA B (fasting insulin × 20/fasting glucose −3.5), as described 19. Quantitation of lipids [total cholesterol, LDL-C, HDL-C, very low-density lipoprotein (VLDL-C), triglycerides (TG)] were quantified by using standard enzymatic methods following manufacturer’s instructions using Hitachi 902 auto-analyzer (Roche, Basel, Switzerland).
CETP Activity
CETP activity was assayed using the Roar CETP Activity Assay Kit. (Roar Biomedical Incorporated, New York, USA) on frozen stored plasma samples of NG controls following manufacturer’s instructions. The assay buffer provided in the kit was diluted 1:10 before use, 5mL 10X buffer in 50mL dH2O. The samples were incubated in a 37°C water bath and ran on a Tecan Infinite™ F200 series using Magellan software (Version 6.5). CETP activity was recorded 1.5 hours. All samples were run in duplication to ensure quality control. Intra- and inter-assay coefficients of variation was less than or equal to 3%.
SNP Genotyping
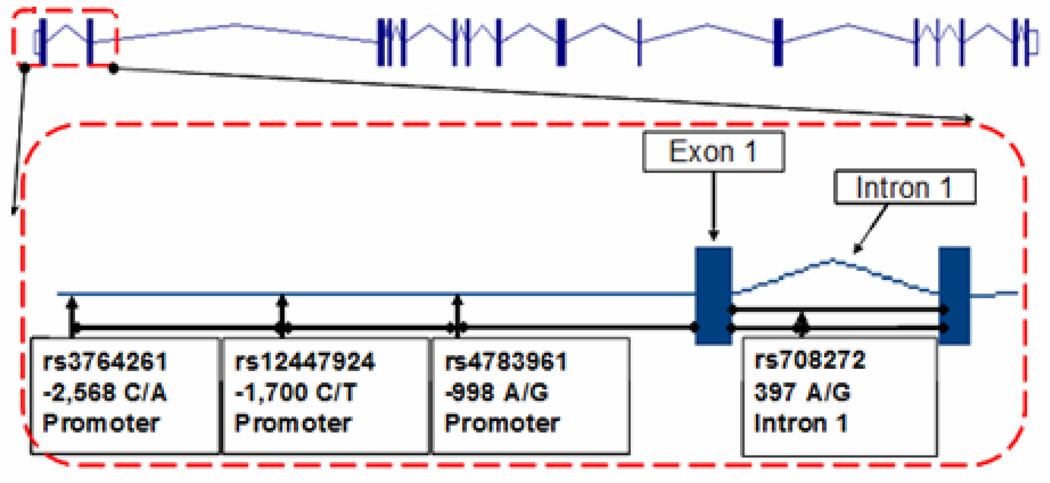
Physical locations of the investigated SNPs in the CETP gene are shown in Figure 1. From the HapMap tagSNP database (which does not include North Indians), the HapMap CEU population sample was chosen as providing the closest SNP coverage in and around the 3 kb region of the 5’ UTR, and a list of maximally informative SNPs was generated 20. TagSNPs with minor allele frequency greater than 10% and that passed Hardy-Weinberg equilibrium (HWE) test were chosen using a pairwise r2 cutoff of 0.8. Three tagSNPs (−2568 C/A rs3764261, −1700 C/T rs12447924, −998 A/G rs4783961) in the promoter region were selected to be genotyped. Additionally, rs708272 (Taq1B) G/A from intron 1 was chosen for its proximity and its extensively reported association with HDL-C in other studies.
Figure 1. Structure of CETP Gene.
Figure 1 denotes structure of CETP locus (3Kb) showing the position of SNPs genotyped for this study. Vertical rectangles in the figure represent exons and horizontal lines denote non-coding sequences (introns). Position of each SNP is marked by arrows.
Interestingly, despite the absence of LD reported between these SNPs in the HapMap database, these SNPs revealed the presence of strong LD in the SDS cohort. With the exception of rs4783961 and rs708272 (D’=0.17 r2=0.02), all other SNPS were in strong LD with D’ ranging from 0.74 to 0.97 and r2 from 0.10 to 0.35. Moreover, rs708272 (Taq1B) was also in strong LD (r2> 0.80) with another extensively reported promoter variant at −629 (rs1800775). The LD and correlation are illustrated in online Figure 1S.
Genotyping for all four SNPs on 2,431 DNA samples was performed using TaqMan pre-designed or TaqMan made-to-order SNP genotyping assays from Applied Biosystems Inc. (ABI, Foster City, USA). Genotyping reactions were performed on an ABI 7900 genetic analyzer using 2 uL of genomic DNA (10 ng/uL), following manufacturers’ instructions. For quality control, 8–10% replicative controls and 4–8 negative controls were included in each 384 well plate. The discrepancy rate of duplicate genotyping was <0.2%. Genotype calling rate was 96% or higher for all of the SNPs genotyped.
Statistical Analysis
Data quality for SNP genotyping was checked by establishing reproducibility of control DNA samples. Departure from HWE in NG controls was tested using the Pearson Chi-square goodness-of-fit test. The genotype and allele frequencies in T2D cases were compared to those in NG control participants using the Chi-square test. Statistical evaluation of genetic effects on T2D risk used multivariate logistic regression analysis with adjustments for age, gender, and other covariates. Continuous traits with skewed sampling distributions (e.g. FBG, 2h glucose, fasting insulin, HDL-C, LDL-C, TG and CETP activity) were log-transformed before statistical analysis. Untransformed measures were reported as arithmetic means while arithmetic means of the transformed variables were re-transformed into the original measurement scale and reported as geometric means. General linear models were used to test the impact of genetic variants on transformed continuous traits (FBG, 2h glucose, HDL-C and LDL-C) in T2D cases, NG controls, and the entire cohort after adjusting for the effects of covariates. Significant covariates for each dependent trait were identified by using Spearman’s correlation and step-wise multiple linear regression with an overall 5% level of significance. These statistical analyses were performed using SPSS for Windows statistical package (version 15.0) (SPSS Inc., Chicago, USA).
Four-site haplotype analysis was performed to determine how the SNPs interacted as one block or haplotype using PLINK (version 1.0.6) (http://pngu.mgh.harvard.edu/~purcell/plink/). To adjust for multiple testing, a Bonferroni’s correction (0.05/ number of observations) was used.
Statistical Power
Statistical power was assessed using the Genetic Power Calculator 21. The general estimates of power in this cohort using additive genetic model at α=0.05, K=0.18 for detecting the effect sizes between 1.11 and 1.30 for T2D, were 56% and 85% when the frequency of T2D risk alleles for rs3764261 and rs12447924 were 0.36 and 0.17, respectively. Our sample had >80% power to calculate the minimum detectable ORs for risk and maximum detectable OR for protective models at α = 0.05 in variants (rs4783961 and rs708272) with allele frequencies ranging from 0.42 to 0.50.
For quantitative traits, assuming the standard threshold of 80% then the power exceeded 90% to detect the inter-genotype difference (e.g. for HDL-C levels), assuming an additive genetic model, (α= 0.05, and Bonferroni’s p= 0.008) at allele frequencies ranging from 0.17–0.50 in using 1,124 NG controls and 1,307 T2D cases. This power was associated with detecting a difference in a quantitative trait, such as HDL-C, of as little as 1 mg/dL and accounts for an effect size of 0.1 which corresponds to detecting significant βs outside of the range of ± 0.05.
RESULTS
Demographic and clinical characteristics of SDS participants are summarized in Table 1. The genotype distributions for all four SNPs were in HWE in NG controls: rs3764261 (p= 0.28), rs12447924 (p= 0.65), rs4783961 (p= 0.74) and rs708272 (p=0.32). As shown in online Figure 1S, there was strong LD between rs3764261, rs12447924, and rs4783961 with D’ ranging from 0.92–0.97, and a strong correlation between rs708272 and rs3764261 (r2=0.30). The genotyping data, including the call rate percentage and number of T2D cases and NG controls for each SNP, is summarized in online Table 1S.
Table 1.
Clinical Characteristics of Study Subjects
| Clinical Characteristics | Gender | NG Controls n=1,124 595 M/529 F |
T2D Cases n=1,307 682 M/625 F |
p values | |
|---|---|---|---|---|---|
| Age (yr) | M | 51.3±15.2 | 55.6±11.4 | 6.1×10−10 | |
| F | 50.2±13.4 | 55.3±10.9 | 7.9×10−11 | ||
| Age at Diagnosis (yr) | M | -- | 46.9±11.2 | -- | |
| F | -- | 48.4±10.9 | -- | ||
| Duration of Diabetes (yr) | M | -- | 8.3±7.2 | -- | |
| F | -- | 7.0±6.3 | -- | ||
| Systolic Blood Pressure (mmHg) | M | 136.0±22.1 | 147.7±22.5 | 2.82×10−18 | |
| F | 135.2±24.3 | 148.1±24.4 | 1.50×10−17 | ||
| Diastolic Blood Pressure (mmHg) | M | 82.1±13.0 | 85.7±12.5 | 1.21×10−6 | |
| F | 81.0±12.5 | 84.9±11.7 | 1.47×10−7 | ||
| BMI (kg/m2) | M | 25.7±4.6 | 26.5±4.4 | 0.002 | |
| F | 26.9±5.8 | 28.2±5.45 | 0.001 | ||
| HDL-C (mg/dL) | M | 35.7±15.1 | 34.3±12.4 | 0.083 | |
| F | 41.0±15.3 | 38.3±12.9 | 0.007 | ||
| LDL-C (mg/dL) | M | 97.4±36.8 | 94.6±38.0 | 0.422 | |
| F | 105.1±36.4 | 106.3±35.9 | 0.077 | ||
| Fasting Blood Glucose (mg/dL) | M | 94.3±12.7 | 162.2±60.9 | 1.8×10−74 | |
| F | 94.3±12.9 | 163.3±64.0 | 1.5×10−59 | ||
| Total Cholesterol (mg/dL) | M | 165.1±55.9 | 167.1±47.4 | 0.705 | |
| F | 181.6±49.2 | 182.1±48.1 | 0.093 | ||
| Triglycerides (mg/dL) | M | 150.5±74.9 | 156.0±83.2 | 0.021 | |
| F | 145.2±68.5 | 160.1±84.5 | 0.015 | ||
| CETP Activity (pmoles/200 µL) | M | 195.6±32.0 | 189.5±33.6 | 0.186 | |
| F | 193.2±33.1 | 206.0±29.4 | 0.004 | ||
| CAD (%) | 11.4 | 16.9 | |||
| Alcohol Consumption (%) | |||||
| No Alcohol | M | 72 | 57 | -- | |
| F | 100 | 100 | -- | ||
| Moderate | M | 22 | 33 | -- | |
| F | 0 | 0 | -- | ||
| High | M | 6 | 10 | -- | |
| F | 0 | 0 | -- | ||
CAD-Coronary Artery Disease
Association of CETP variants with T2D and CAD
Lower serum CETP activity was associated with higher CAD risk in this population. Mean levels of CETP activity varied significantly (p=0.001) among CAD patients and non-CAD patients were (183.6 ± 32.9) and (194.2 ± 33.5), respectively. The odds ratio (OR) for CAD risk associated with CETP activity was 2.34 95%CI (1.51–3.64), p=1.44×10−4 without adjusting for covariates and 2.2 95%CI (1.4–3.4), p=0.001 after controlling for the effects of age, BMI and gender. However, CETP activity was not associated with T2D in this cohort (OR 0.83 95%CI [0.56–1.22], p=0.343).
Association of CETP variants with T2D and CAD
Allelic distributions of all four CETP variants did not differ significantly between T2D cases and NG controls except for the ‘A’ allele of rs4783961 that was moderately associated with T2D (OR 0.81 95% CI (0.7–1.0), p=0.022) after adjusting for age, BMI and gender (Table 2). None of the CETP variants revealed any association with CAD in this cohort.
Table 2.
Genotype Distribution and Association of CETP SNPs with T2D and CAD
| T2D | Allele frequency NG (1,124) |
Allele frequency T2D (1,307) |
Odds Ratios* (OR) (95%CI) |
||
|---|---|---|---|---|---|
| SNP | Allele | p value | |||
| rs3764261 | C/A | 0.64/0.36 | 0.61/0.39 | 1.10 (1.0–1.3) | 0.121 |
| rs12447924 | C/T | 0.83/0.17 | 0.82/0.18 | 1.28 (0.8–2.0) | 0.295 |
| rs4783961 | A/G | 0.58/0.42 | 0.62/0.38 | 0.81 (0.7–1.0) | 0.022£ |
| rs708272 | A/G | 0.5/0.5 | 0.52/0.48 | 0.89 (0.8–1.0) | 0.054 |
| CAD | Allele frequency Control (2,082) |
Allele frequency CAD (349) |
Odds Ratios* (OR) (95%CI) |
||
| SNP | Allele | p value | |||
| rs3764261 | C/A | 0.62/0.38 | 0.62/0.38 | 0.98 (0.8–1.2) | 0.840 |
| rs12447924 | C/T | 0.82/0.18 | 0.81/0.19 | 0.83 (0.4–1.6) | 0.574 |
| rs4783961 | A/G | 0.60/0.40 | 0.61/0.39 | 0.90 (0.7–1.2) | 0.412 |
| rs708272 | A/G | 0.51/0.49 | 0.53/0.47 | 0.84 (0.6–1.1) | 0.244 |
ORs were adjusted for age and BMI;
nonsignificant after Bonferroni’s correction
Association of CETP variants with quantitative traits of obesity, T2D, and serum lipids
There was no evidence of association between CETP variants and traits related to obesity (waist circumference, weight, BMI), FBG, or 2h glucose. However, multiple linear regression analysis of lipids revealed a strong association of variant ‘A’ allele of rs3764261 and ‘A’ allele of rs708272 (Taq1B) with HDL-C in both NG controls (β = 0.123; p= 9.35 ×10−7 for rs3764261; β= 0.10 p= 0.002 for rs708272) and T2D cases (β= 0.07, p= 0.016 for rs3764261; β= 0.08, p=0.005 for rs708272) after adjusting for the effects of age, sex, and BMI (Table 3). None of these SNPs revealed any consistent association with other lipid traits except same alleles were associated with significantly decreased triglyceride levels in this cohort (rs 3764261 β = −0.07, p=0.002; rs708272 β = −0.07, p=0.003) (Table 3)
Table 3.
Association of CETP SNPs genotypes with quantitative traits in NG Controls, T2D Cases, and Combined Cohorts
| NG Controls | T2D Cases | Combined Cohort | ||||
|---|---|---|---|---|---|---|
| Beta | P-value | Beta | P-value | Beta | P-value | |
| rs3764261 | ||||||
| Total Cholesterol mg/dL | 0.08 | 0.025 | −0.01 | 0.507 | 0.01 | 0.223 |
| HDL-C mg/dL | 0.12 | 9.35×10−7 | 0.07 | 0.016 | 0.11 | 9.60×10−7 |
| LDL-C mg/dL | 0.02 | 0.380 | −0.002 | 0.905 | 0.01 | 0.580 |
| TG mg/dL | −0.01 | 0.582 | −0.09 | 3.64×10−4 | −0.07 | 0.002 |
| rs12447924 | ||||||
| Total Cholesterol mg/dL | −0.002 | 0.913 | −0.008 | 0.636 | −0.01 | 0.713 |
| HDL-C mg/dL | −0.03 | 0.177 | −0.008 | 0.662 | −0.02 | 0.154 |
| LDL-C mg/dL | 0.09 | 0.006 | 0.002 | 0.939 | 0.01 | 0.478 |
| TG mg/dL | −0.02 | 0.433 | 0.02 | 0.538 | 0.001 | 0.963 |
| rs4783961 | ||||||
| Total Cholesterol mg/dL | −0.02 | 0.113 | 0.02 | 0.177 | −0.002 | 0.865 |
| HDL-C mg/dL | −0.09 | 0.006 | 0.02 | 0.411 | −0.01 | 0.321 |
| LDL-C mg/dL | −0.01 | 0.475 | 0.01 | 0.660 | −0.001 | 0.951 |
| TG mg/dL | −0.01 | 0.545 | 0.04 | 0.093 | 0.01 | 0.469 |
| rs708272 | ||||||
| Total Cholesterol mg/dL | 0.02 | 0.161 | 0.02 | 0.147 | 0.05 | 0.037 |
| HDL-C mg/dL | 0.10 | 0.002 | 0.08 | 0.005 | 0.09 | 1.69×10−5 |
| LDL-C mg/dL | −0.001 | 0.966 | 0.02 | 0.129 | 0.02 | 0.204 |
| TG mg/dL | −0.07 | 0.043 | −0.07 | 0.004 | −0.07 | 0.003 |
Analysis were adjusted for age, gender, and BMI and alcohol in NG and T2D and age, gender, and BMI and alcohol and disease in combined cohort.
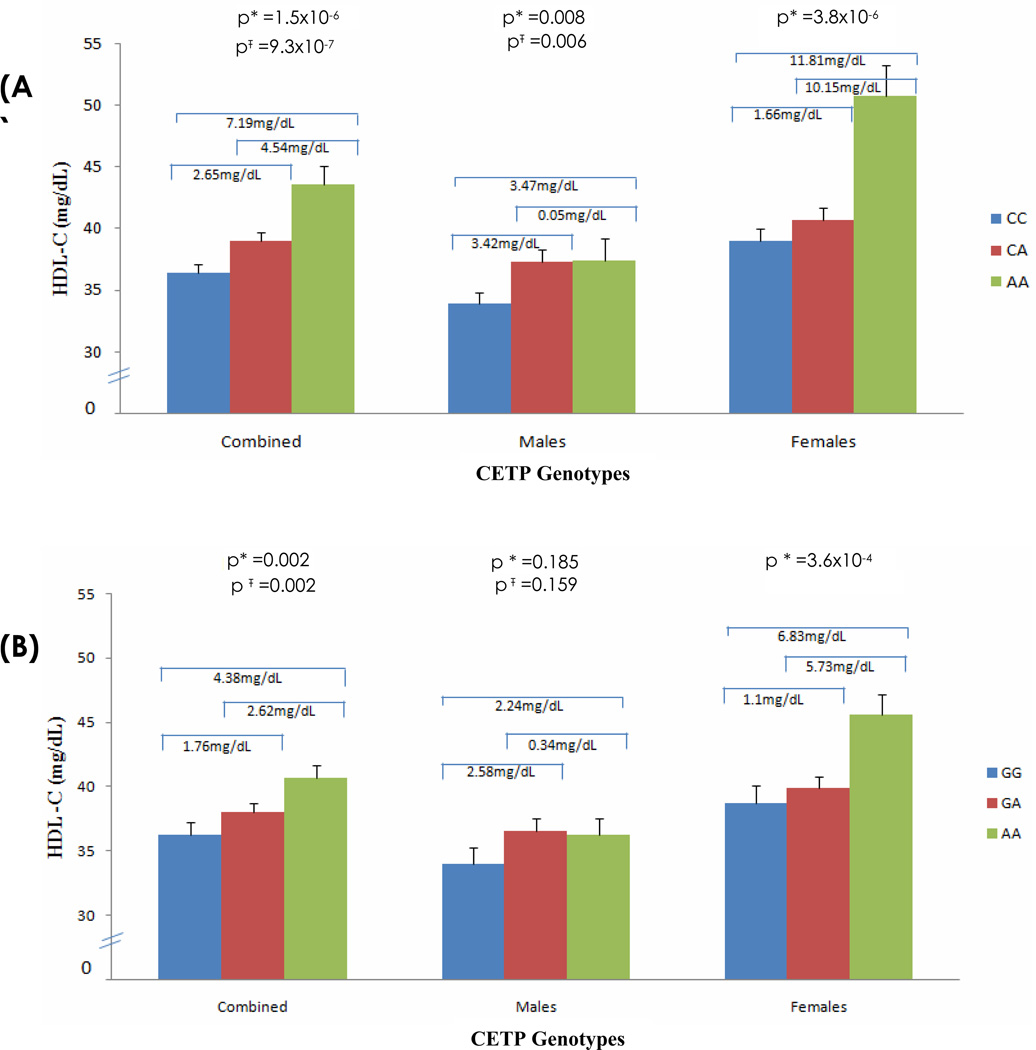
In addition to age and BMI, alcohol consumption was a significant covariate affecting HDL-C levels in males. Since the vast majority of females in this community do not drink alcohol, analysis adjusting for the effect of alcohol on CETP genotypes was performed in the data stratified by gender. For rs3764261 the overall p-values improved from 0.008 to 0.006 in males and 1.5×10−6 to 9.3×10−7 in the combined sample after adjusting for alcohol consumption (Figure 2A). However, alcohol consumption did not modify association of rs708272 (Taq1B) with HDL-C levels in males (p=0.159) or in the combined cohort (p=0.002) (Figure 2B). Similarly, adjusting for alcohol consumption did not modify the association with the other SNPs (data not shown).
Figure 2. Effect of CETP Genotypes rs3764261 and rs708272 on HDL-C Levels in Data Stratified by Gender.
Bar plots in Figure 2 (A) and (B) show association of CETP (rs3764261)(A) and (rs708272)(B) genotypes with HDL-C using multiple linear regression analysis. * In Model 1, analysis was controlled for age, sex, and BMI; Ŧ In Model 2, analysis was controlled for age, sex, BMI, and alcohol.
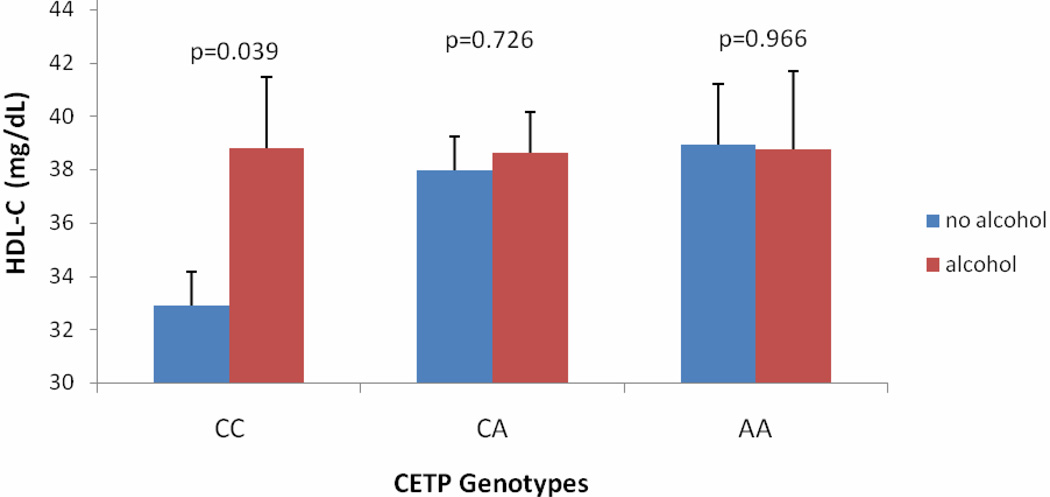
In females, HDL-C levels were significantly increased (3.8×10−6) by 11.8 mg/dL in rs3764261 AA genotype carriers compared to CC genotype carriers and by 6.8 mg/dL in rs708272 AA genotype carriers compared to GG genotype carriers (Figure 2A and 2B). When we further stratified male cohort by alcohol drinkers vs. non-drinkers, the low HDL-C-associated rs3764261 CC genotype carriers showed a moderate increase in HDL-C levels among drinkers (38.8 ± 17.0 mg/dL compared to non-drinkers (32.9 ± 15.1 mg/dL), p=0.039). However, this effect was not seen among ‘favorable’ AA genotype carriers (Figure 3).
Figure 3. Effect of Alcohol on HDL-C in Males (rs3764261).
Figure 3 shows that alcohol was a significant modifier for affecting HDL-C in males. Note that the low HDL-C-associated ‘CC’ genotype carriers had a moderate increase in HDL-C levels among alcohol drinkers (32.9 ± 15.1 mg/dL), compared to non-drinkers (38.8 ± 17.0 mg/dL; p=0.039). However, this effect was not noticed among ‘favorable’ ‘AA’ genotype carriers.
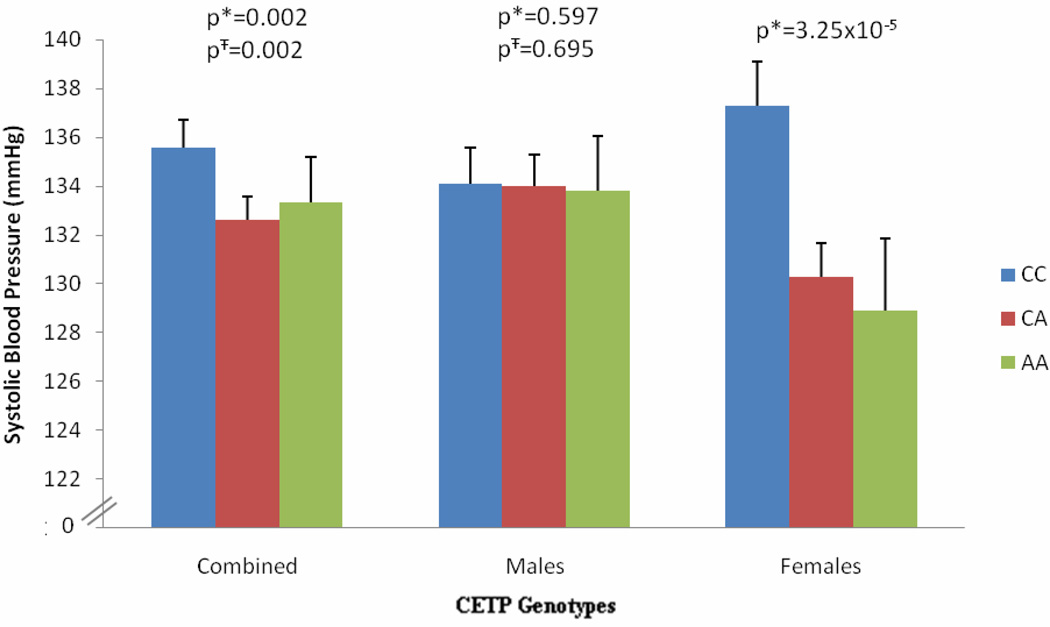
Furthermore, the rs3764261, AA genotype carriers showed significantly lower systolic blood pressure compared to CC genotype carriers in combined NG controls with and without controlling for the effect of alcohol (p=0.002). Interestingly, in gender stratified cohort, AA genotype-associated decrease was highly significant (p=3.25×10−5) in females and no such effect was seen in males either with or without adjustment for alcohol consumption (Figure 4). No other variant in the CETP gene was significantly associated with systolic blood pressure variation in this population.
Figure 4. Effect of CETP Genotypes (rs3764261) on Systolic Blood Pressure.
Figure 4 shows distribution of means for systolic blood pressure by genotypes. * In Model 1, analysis was controlled for age, sex, and BMI; Ŧ In Model 2, analysis was controlled for age, sex, BMI, and alcohol.
Haplotype Analysis
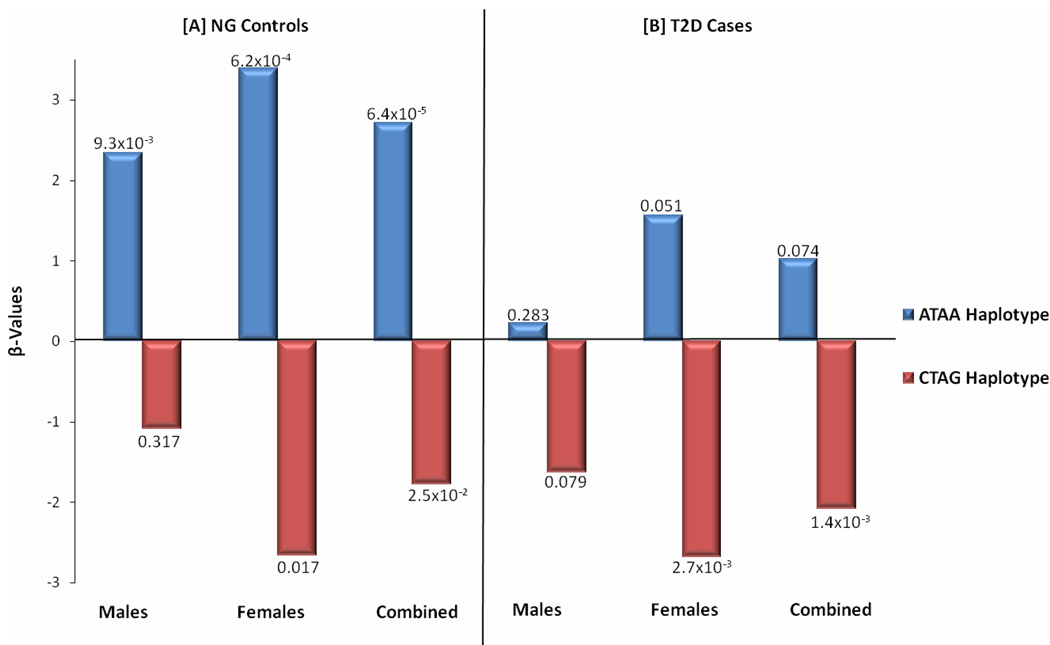
Haplotype analysis using all four investigated SNPs (rs3764261, rs12447924, rs4783961, and rs708272) revealed a strong association of the ‘ATAA’ haplotype with increased HDL-C (β= 2.71, p=6.38×10−5) in NG controls and the ‘CTAG’ haplotype with significantly decreased HDL-C (β= −1.78, p=2.5×10−2); and β= −2.09 p=1.4×10−3) in NG controls and T2D cases, respectively, after adjusting for the effects of age, BMI, and gender. In gender-stratified analysis, the haplotype effect remained highly statistically significant in females (increased HDL-C in the ‘ATAA’ haplotype: β=3.39, p=6.2×10−4; decreased HDL-C in the ‘CTAG’ haplotype: β=−2.67, p=0.017) but was slightly muted in males (increased HDL-C in the ‘ATAA’ haplotype: β=2.34, p=9.3×10−3; non-significant decreased HDL-C in the ‘CTAG’ haplotype: β= −1.09, p=0.317) (Figure 5). Less significant but similar trend associated with increased HDL-C level for the ‘ATAA’ haplotype but significant decreased in HDL-C levels for the ‘CTAG’ haplotype especially in females (β= −2.69, p= 2.7×10−3) and combined sample (β=−2.09, p= 1.4 ×10−3) was seen in T2D cases (Figure 5). None of these haplotypes was associated with other lipid, obesity, or glucose-related traits.
Figure 5. Effect of CETP Haplotypes rs3764261, rs12447924, rs4783961, and rs708272 on HDL-C.
Figure 5 show regression coefficients (β) by ATAA and CTAG haplotypes in NG Controls and T2D cases.
Association of CETP variants with CETP activity and HDL-C levels
In linear regression analysis, none of the investigated SNPs revealed association with CETP activity. As shown in online Figure 2S, the mean levels of CETP activity did not differ significantly across genotypes for either rs3764261 or rs708272. CETP activity also did not differ between T2D cases and NG controls. However, there was a strong positive correlation between HDL-C levels and CETP activity (r=0.12, p=3.1×10−4). Interestingly, as shown in Table 4, we also observed a strong interaction effect between CETP activity and CETP SNPs rs3764261 (p=3.8×10−6), rs708272 (p=3.7×10−4), and rs4783961 (p=0.002) in affecting HDL–C levels. As previously seen, in the data stratified gender, these interaction effects were more pronounced in females for all three SNPs (rs3764261, p=2.95×10−6; rs708272, p=9.49×10−5; and rs4783961, p=0.004). Only rs3764261 showed a significant interaction effect with CETP activity on HDL-C levels in males (p=0.004) (Table 4).
Table 4.
Interaction between Individual SNPs and CETP Activity for affecting HDL-C Levels in Controls
| Combined | Males | Females | ||||
|---|---|---|---|---|---|---|
| Interaction | p* | p Ŧ | p * | p Ŧ | p * | p Ŧ |
| rs3764261*CETP Activity | 3.8×10−6 | 2.2×10−6 | 0.004 | 0.004 | 2.9×10−6 | ----- |
| rs708272*CETP Activity | 3.7×10−4 | 4.4×10−4 | 0.126 | 0.247 | 9.5×10−5 | ----- |
| rs4783961*CETP Activity | 0.002 | 0.001 | 0.166 | 0.177 | 0.004 | ----- |
Dependent variable: HDL-C;
P* analysis was adjusted for age, sex, and BMI;
PŦ analysis was adjusted for age, sex, BMI, and alcohol.
DISCUSSION
Several studies have been undertaken to examine the associations of CETP gene polymorphisms on plasma lipid concentrations 11. The role of Taq1B (rs708272) polymorphism occurring in the first intron of CETP gene has been most extensively studied and is associated with variations in HDL-C concentrations 2. However, the putative correlation of CETP polymorphisms with CETP activity, HDL-C levels and other CAD risk factors is still unclear 22.
As the allelic distributions of Punjab population of North India are similar to European ancestries 23, tagSNPs from upstream of the 5’UTR were chosen using the CEU HapMap data. A much tighter LD (D’= 0.74 −0. 97 and r2= 0.10 – 0.35) among these tagSNPs including the Taq1B (rs708272) (Figure 1S) was found whereas little LD is reported in this region for the CEU sample. Evidently, these findings suggest that the LD structure in this population is different from Euro-Caucasians. Furthermore, variation at three of these four SNPs correlated strongly with variation in HDL-C levels despite the fact that none of these SNPs are known to affect a functional change in CETP; possibly this promoter region may harbor a yet to be discovered functional variant which is in strong LD with these SNPs.
The males in this Punjabi cohort have inherently lower levels of HDL-C compared to females (35.7 ± 15.1 (males) vs. 41.0 ± 15.3 (females), p=9.9×10−8), and these were even lower in diabetics (34.3 ± 12.4 (males) vs. 38.3 ± 12.9 (females), p=4.22×10−8) (Figure 3S). Also, lower HDL-C levels (<40 mg/dl) were more frequently associated in patients with CAD (56% CAD patients had HDL-C <40mg/dL). In general, men are characterized by a less favorable lipoprotein–lipid profile, which is accompanied by smaller HDL-C particle size compared to women 24. Also, the higher HDL-C in women is attributed to higher endogenous estrogen levels and the cardioprotective mechanisms of estrogen and progesterone operating in women 25. However, despite this difference, variant alleles of rs3764261 and Taq1B (rs708272) were directly correlated with increased HDL-C levels in both men and women, and with the magnitude of effect being stronger in women. In rs3764261, HDL-C levels were increased by 7.2 mg/dL (p=1.5×10−6) and in rs708272 by 4.4 mg/dL (p=0.002) in AA genotype carriers vs. CC genotype carriers among NG controls. Notably, the same ‘A’ allele of rs3764261 that significantly increased HDL-C levels also revealed a significant decrease in mean levels of systolic blood pressure from 135.8 ± 1.16 mmHg from CC genotype carriers to 133.2 ± 1.85 mmHg in AA genotype carriers in NG controls (β= −0.10, p=0.002). Interestingly, when separated by gender, this significant association was confined to females (β= −0.17, p= 3.25× 10−5) while no such association was seen in males (β=−0.02, p=0.597) (Figure 4). Increased blood pressure is a risk factor of CAD and has been associated with decreased HDL-C levels 24. Thus, AA individuals of rs3764261 carry a more favorable profile with increased HDL-C levels and decreased systolic blood pressure which was predominantly seen in females. Another study reported gender-specific risk reduction in metabolic syndrome component by Taq1B polymorphism in CETP which was more pronounced in males 26. Further studies on a larger size sample are needed to confirm these findings and to understand the gender specific association of CETP in relation to CAD risk.
Alcohol consumption showed an attenuating effect on the inter-genotype differences of rs3764261 for impacting HDL-C levels in this study. For instance, in rs3764261 among non-alcoholic NG women, the HDL-C levels increased ~30% among ‘AA’ carriers over ‘CC’ (the majority) carrier (Figure 2). In non-drinking NG men the HDL-C levels increased ~18 % in ‘AA’ over the ‘CC’, but in drinking NG men it was increased only ~5% among ‘AA’ carriers. On the other hand, there was a paradoxical increase in improved HDL-C levels in response to alcohol among less favorable ‘CC’ genotype carriers by 5.9 mg/dL (Figure 3). These results suggest a mode of action for both the CETP genetic variation (associated with rs3764261) and its modulation by alcohol consumption. Therefore, it can be speculated that ‘CC’ carriers can be benefitted with moderate alcohol intake. It is also possible that the alcohol consumption disrupts inhibitory mechanism in ‘CC’ carriers so that ‘CC’ carriers behave more like ‘AA’ carriers. However, these findings are speculative till validated and the possibility of other genetic or non genetic factors modulating this effect can also not be ruled out.
In haplotype analysis, in all four investigated SNPs with HDL-C revealed a strong association of the ‘ATAA’ haplotype with increased HDL-C levels (β= 2.71, p=6.38×10−5) and ‘CTAG’ haplotype with decreased HDL-C levels (β= −1.78, p=2.5×10−2) in NG controls after controlling for age, sex, and disease status. When analyzed by gender, the haplotype association was more pronounced in females compared to males (e.g. ‘ATAA’ females β=3.39, p=6.2×10−4, β= 2.34, p=9.34×10−3) (Figure 5).
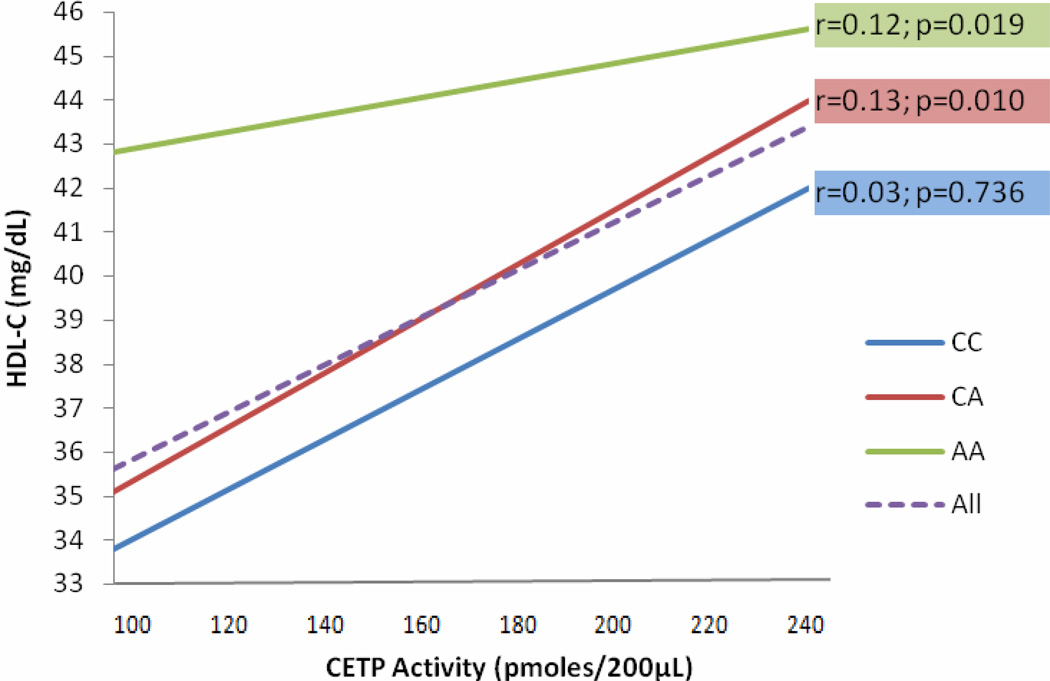
The most interesting part of this investigation was the presence of significant positive correlation of CETP activity and HDL-C levels (r= 0.12 p= 3.1×10−4) which was independent of genetic variation in CETP, in contrast to previous studies 2. Not only that, our results also showed significant increase CAD risk among subjects carrying low CETP activity (OR 2.2 95%CI (1.4–3.4, p=0.001). Although there was no direct association of CETP activity and CETP polymorphisms (Figure 2S), our findings revealed a significant interaction between CETP SNPs and CETP activity for affecting HDL-C levels in rs3764261 and Taq1B (rs708272), and even rs4783961 (Table 4). After including CETP activity in the model, the significant association with HDL-C remained significant in both rs3764261 and rs708272; single SNP analysis (p=9.3 × 10−7 and p= 0.002) and interaction analysis (p=2.2 × 10−6, and p= 4.4 × 10−4) (Tables 3 & 4), respectively. There is little relationship between HDL-C and CETP activity in the ‘AA’ homozygote carriers (rs3764261) as shown in horizontal slop in (Figure 6). From this one could opine that treating ‘AA’ carriers with a CETP inhibitor would contribute minimally to improving HDL-C levels. These inconsistencies between our study and previous studies (supporting high CETP activity linked to CAD risk) suggest the possibility of other factors affecting HDL-C levels and CETP activity. Our results concur with the findings of a prospective investigation from the Framingham Heart Study where low plasma CETP activity was associated with greater CAD risk 9. Taken together with the Framingham Heart Study findings, our investigation argues against inhibiting CETP for improving cardiovascular outcomes and urges a deeper evaluation of the individual genetic variation in the CETP before implementing pharmaceutical intervention of blocking CETP for preventing CAD events 27. These data suggest that, inhibiting CETP in the absence of knowledge of genetic variation we may inhibit the protective effects of CETP (increased HDL-C and decreased systolic blood pressure) in some people (specifically rs3764261 ‘AA’ carriers); where a more personalized medical approach is needed. Our results may partially provide the reason of failure of these drugs for decreasing CAD mortality and may explain why CETP inhibition has not been a full success. Perhaps the off target effects of torcetrapib were associated with genetic variation controlling blood pressure and gender differences modulated by lifestyle and other factors. Although, the overall genetic variance explained by common variants in the CETP locus is ~5–8%, it is likely that the ‘missing heritability’ lies in less common or rare variants in this locus which can be detected by deep sequencing 28. It appears that there are more complex aspects of CETP in the cholesterol metabolism pathway which are currently unknown and further genetic and functional studies can delineate its role in cardiovascular functions.
Figure 6. SNP (rs3764261) CETP Activity Interaction for Affecting HDL-C.
Figure 6 shows the relationships between CETP (rs3764261), HDL-C, and CETP activity. Dashed line (purple) slop shows correlation of HDL-C and CETP activity in combined rs3764261 (CC, CA, AA) genotypes (r= 0.12 p= 3.1×10−4). Three complete lines represent the correlation between HDL-C and CETP activity by each genotype. The uppermost green slop clearly shows that the increase in HDL-C levels in homozygous ‘AA’ carriers is independent of CETP activity.
Nevertheless, our investigation suggests that genetic variation in CETP affects serum HDL-C levels and may play an important role in the risk assessment for CAD. Although, the genetic variation in this locus is not associated with increased risk to T2D or CAD, the variant allele especially in CETP promoter (rs3764261) showed a strong association with increased HDL-C, decreased blood pressure. Low serum CETP activity is associated with greater CAD risk. Although, none of the investigated SNPs is associated with serum CETP activity, through interaction analysis, it is evident that genetic polymorphism and CETP activity significantly affects HDL-C and systolic blood pressure; in addition, genetic effects are significantly modulated by environmental factors (alcohol consumption). Individuals carrying protective genotypes in CETP can be benefitted by improving HDL-C and lowering systolic blood pressure without inhibition of CETP activity. Therefore, genetic and functional studies on a large size datasets from different ethnicities will be required before using CETP inhibitors on every CAD patient.
Supplementary Material
ACKNOWLEDGEMENTS
The study was supported by NIH grants (K01TW006087 and R01DK082766) funded by the Fogarty International Center and National Institute of Diabetes and Digestive and Kidney Diseases, and a seed grant from University of Oklahoma Health Sciences Center, Oklahoma City, OK. Authors are also thankful to Mr. Jagtar S. Sanghera for his help in subject recruitment. Authors also thank all the study participants and are grateful for their contribution in this study.
Abbreviations
- CETP
Cholesteryl Ester Transfer Protein
- T2D
Type 2 diabetes
- NG
Normoglycemic
- CAD
Coronary artery disease
- GWAS
Genome-wide association study
- HOMA-IR
Homeostasis model assessment for insulin resistance
- HOMA-B
Homeostasis model assessment for β cell function
- SNP
Single nucleotide polymorphism
- FBG
Fasting blood glucose
- BMI
Body mass index
- WHR
Waist-to-hip ratio
- MAF
Minor allele frequency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST DISCLOSURE
We declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
REFERENCES
- 1.Libby P. The forgotten majority: unfinished business in cardiovascular risk reduction. J Am Coll Cardiol. 2005;46(7):1225–1228. doi: 10.1016/j.jacc.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Hassanzadeh T, Firoozrai M, Zonouz AE, Zavarehee A, Paoli M. Taq1B polymorphism of cholesteryl ester transfer protein (CETP) gene in primary combined hyperlipidaemia. Indian J Med Res. 2009;129(3):293–298. [PubMed] [Google Scholar]
- 3.Snieder H, van Doornen LJ, Boomsma DI. Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arterioscler Thromb Vasc Biol. 1999;19(12):2826–2834. doi: 10.1161/01.atv.19.12.2826. [DOI] [PubMed] [Google Scholar]
- 4.Yilmaz H, Isbir T, Agachan B, Karaali ZE. Effects of cholesterol ester transfer protein Taq1B gene polymorphism on serum lipoprotein levels in Turkish coronary artery disease patients. Cell Biochem Funct. 2005;23(1):23–28. doi: 10.1002/cbf.1124. [DOI] [PubMed] [Google Scholar]
- 5.Padmaja N, Ravindra Kumar M, Soya SS, Adithan C. Common variants of Cholesteryl ester transfer protein gene and their association with lipid parameters in healthy volunteers of Tamilian population. Clin Chim Acta. 2007;375(1–2):140–146. doi: 10.1016/j.cca.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 6.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299(23):2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 7.Bruce C, Sharp DS, Tall AR. Relationship of HDL and coronary heart disease to a common amino acid polymorphism in the cholesteryl ester transfer protein in men with and without hypertriglyceridemia. J Lipid Res. 1998;39(5):1071–1078. [PubMed] [Google Scholar]
- 8.Agerholm-Larsen B, Nordestgaard BG, Steffensen R, Jensen G, Tybjaerg-Hansen A. Elevated HDL cholesterol is a risk factor for ischemic heart disease in white women when caused by a common mutation in the cholesteryl ester transfer protein gene. Circulation. 2000;101(16):1907–1912. doi: 10.1161/01.cir.101.16.1907. [DOI] [PubMed] [Google Scholar]
- 9.Vasan RS, Pencina MJ, Robins SJ, Zachariah JP, Kaur G, D'Agostino RB, Ordovas JM. Association of circulating cholesteryl ester transfer protein activity with incidence of cardiovascular disease in the community. Circulation. 2009;120(24):2414–2420. doi: 10.1161/CIRCULATIONAHA.109.872705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10(2):109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 12.Poss J, Custodis F, Werner C, Weingartner O, Bohm M, Laufs U. Cardiovascular Disease and Dyslipidemia: Beyond LDL. Curr Pharm Des. 2011;17(9):861–870. doi: 10.2174/138161211795428858. [DOI] [PubMed] [Google Scholar]
- 13.Cannon CP, Shah S, Dansky HM, Davidson M, Brinton EA, Gotto AM, Stepanavage M, Liu SX, Gibbons P, Ashraf TB, Zafarino J, Mitchel Y, Barter P. Safety of anacetrapib in patients with or at high risk for coronary heart disease. N Engl J Med. 363(25):2406–2415. doi: 10.1056/NEJMoa1009744. [DOI] [PubMed] [Google Scholar]
- 14.Sanghera DK, Bhatti JS, Bhatti GK, Ralhan SK, Wander GS, Singh JR, Bunker CH, Weeks DE, Kamboh MI, Ferrell RE. The Khatri Sikh Diabetes Study (SDS): study design, methodology, sample collection, and initial results. Hum Biol. 2006;78(1):43–63. doi: 10.1353/hub.2006.0027. [DOI] [PubMed] [Google Scholar]
- 15.Sanghera DK, Been LF, Ralhan S, Wander GS, Mehra NK, Singh JR, Ferrell RE, Kamboh MI, Aston CE. Genome-wide linkage scan to identify Loci associated with type 2 diabetes and blood lipid phenotypes in the sikh diabetes study. PLoS One. 6(6):e21188. doi: 10.1371/journal.pone.0021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanghera DK, Demirci FY, Been L, Ortega L, Ralhan S, Wander GS, Mehra NK, Singh J, Aston CE, Mulvihill JJ, Kamboh IM. PPARG and ADIPOQ gene polymorphisms increase type 2 diabetes mellitus risk in Asian Indian Sikhs: Pro12Ala still remains as the strongest predictor. Metabolism: clinical and experimental. 2009 doi: 10.1016/j.metabol.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, Kamboh MI. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27 Suppl 1:S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 19.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 20.Halldorsson BV, Istrail S, De La Vega FM. Optimal selection of SNP markers for disease association studies. Hum Hered. 2004;58(3–4):190–202. doi: 10.1159/000083546. [DOI] [PubMed] [Google Scholar]
- 21.Purcell SCS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 22.Freeman DJ, Griffin BA, Holmes AP, Lindsay GM, Gaffney D, Packard CJ, Shepherd J. Regulation of plasma HDL cholesterol and subfraction distribution by genetic and environmental factors. Associations between the TaqI B RFLP in the CETP gene and smoking and obesity. Arterioscler Thromb. 1994;14(3):336–344. doi: 10.1161/01.atv.14.3.336. [DOI] [PubMed] [Google Scholar]
- 23.Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11(6):994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascot A, Lemieux I, Bergeron J, Tremblay A, Nadeau A, Prud'homme D, Couillard C, Lamarche B, Despres JP. HDL particle size: a marker of the gender difference in the metabolic risk profile. Atherosclerosis. 2002;160(2):399–406. doi: 10.1016/s0021-9150(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 25.Rossouw JE. Hormones, genetic factors, and gender differences in cardiovascular disease. Cardiovasc Res. 2002;53(3):550–557. doi: 10.1016/s0008-6363(01)00478-3. [DOI] [PubMed] [Google Scholar]
- 26.Sandhofer A, Tatarczyk T, Laimer M, Ritsch A, Kaser S, Paulweber B, Ebenbichler CF, Patsch JR. The Taq1B-variant in the cholesteryl ester-transfer protein gene and the risk of metabolic syndrome. Obesity (Silver Spring) 2008;16(4):919–922. doi: 10.1038/oby.2007.130. [DOI] [PubMed] [Google Scholar]
- 27.McTaggart F, Jones P. Effects of statins on high-density lipoproteins: a potential contribution to cardiovascular benefit. Cardiovasc Drugs Ther. 2008;22(4):321–338. doi: 10.1007/s10557-008-6113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke AJ, Cooper DN. GWAS: heritability missing in action? Eur J Hum Genet. 18(8):859–861. doi: 10.1038/ejhg.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.