Abstract
In the model organism Caenorhabditis elegans, a family of endogenous small molecules, the ascarosides, function as key regulators of developmental timing and behavior that act upstream of conserved signaling pathways. The ascarosides are based on the dideoxysugar ascarylose, which is linked to fatty acid-like side chains of varying lengths derived from peroxisomal β-oxidation. Despite their importance for many aspects of C. elegans biology, knowledge of ascaroside structures, biosynthesis, and homeostasis remains incomplete. We used an MS/MS-based screen to profile ascarosides in C. elegans wild type and mutant metabolomes, which revealed a much greater structural diversity of ascaroside derivatives than previously reported. Comparison of the metabolomes from wild type and a series of peroxisomal β-oxidation mutants showed that the enoyl CoA-hydratase MAOC-1 serves an important role in ascaroside biosynthesis and clarified the functions of two other enzymes, ACOX-1 and DHS-28. We show that following peroxisomal β-oxidation the ascarosides are selectively derivatized with moieties of varied biogenetic origin and that such modifications can dramatically affect biological activity, producing signaling molecules active at low femtomolar concentrations. Based on these results the ascarosides appear as a modular library of small molecule signals, integrating building blocks from three major metabolic pathways; carbohydrate metabolism, peroxisomal β-oxidation of fatty acids, and amino acid catabolism. Our screen further demonstrates that ascaroside biosynthesis is directly affected by nutritional status and that excretion of the final products is highly selective.
1. INTRODUCTION
Several different aspects of the life history of the nematode Caenorhabditis elegans are regulated by ascarosides, glycosides of the dideoxysugar ascarylose (Figure 1).1–5 The ascarosides ascr#1–3 were originally identified as major components of the dauer pheromone, a population-density signal that promotes entry into an alternate larval stage, the non-feeding and highly persistent dauer diapause.1,2,4,5,6 Entry into the dauer stage is mediated by several highly conserved signaling pathways, including insulin/IGF-1 signaling and transforming growth factor beta (TGF-β) signaling,7 which contributed to interest in the ascarosides' structures, their biosynthesis, and mode of action. More recent work showed that specific mixtures of ascarosides including the 4-aminobenzoic acid derivative ascr#8 act as strong male-specific attractants,3,5 whereas ascarosides including a tryptophan-derived indole-3-carboxy moiety function as aggregation signals at femtomolar concentrations.8
Figure 1.
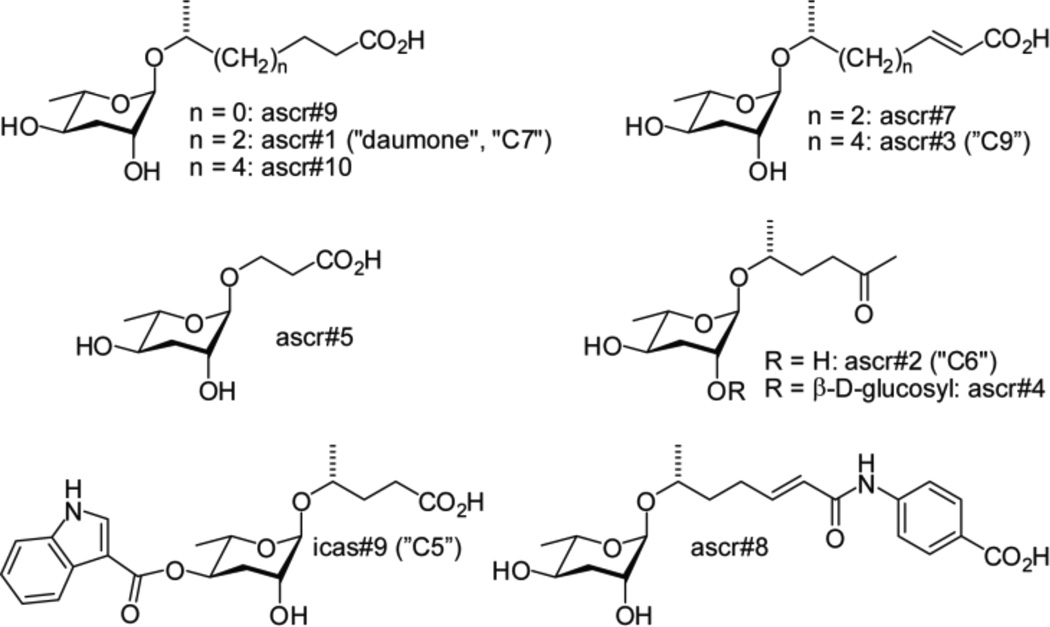
Ascarosides that regulate development and behavior in C. elegans. ascr#1–4, ascr#5 and icas#9 were identified via activity guided fractionation,1,2,3,4,13 ascr#7 and ascr#8 were identified using differential analysis of 2D NMR spectra (DANS),5 and ascr#9 and #10 were detected using mass spectrometric techniques.8
The biosynthetic pathways that control specific assembly of the ascarylose, lipid side-chain and peripheral building blocks are largely unknown. Earlier work showed that worms carrying a mutation in the gene daf-22 are defective in the biosynthesis of both the dauer pheromone and male-attracting signals.3,9,10 daf-22 encodes a protein with strong homology to human sterol carrier protein SCPx and in C. elegans functions in peroxisomal β-oxidation of long-chain fatty acids, producing the 3–9 carbon side chains of the ascarosides.6 Two other components of peroxisomal β-oxidation were shown to participate in ascaroside biosynthesis, the acyl-CoA oxidase ACOX-111 and the β-hydroxyacyl-CoA dehydrogenase DHS-28,6,12 a partial homolog of human multifunctional enzyme type 2 (MFE-2). However, the exact roles of these enzymes in ascaroside biosynthesis remained unclear, because the effects of acox-1 and dhs-28 mutations on ascaroside production were only partially characterized.
The recent discovery of new structural variants with important divergent functions, e.g. icas#3 and icas#9,8,13 suggested that knowledge of ascaroside structures and functions in C. elegans remains incomplete. Results from biological studies further indicate that even small structural differences between ascarosides can be associated with significant functional differences; for example, ascr#3 is a potent male attractant whereas the structurally similar ascr#7 is nearly inactive in this assay.5,14 Correspondingly, several previous studies indicate that ascaroside biosynthesis is tightly controlled by environmental factors such as temperature, nutrient availability, and population density.6,11,15 Therefore, detailed investigation of ascaroside structures and their biosynthetic pathways is essential for many aspects of the biology of this model organism.
In this study, we introduce HPLC-MS/MS-based metabolomics as a tool for ascaroside profiling in C. elegans. Application of this method to wild-type C. elegans revealed that the previously described ascarosides are part of a much larger, structurally diverse library of compounds derived from modular combination of building blocks from three different metabolic pathways. Subsequently we used HPLC-MS/MS of mutant metabolomes to interrogate ascaroside biosynthesis and homeostasis.
2. RESULTS
2.1 LC-MS/MS reveals new ascarosides in wild type C. elegans
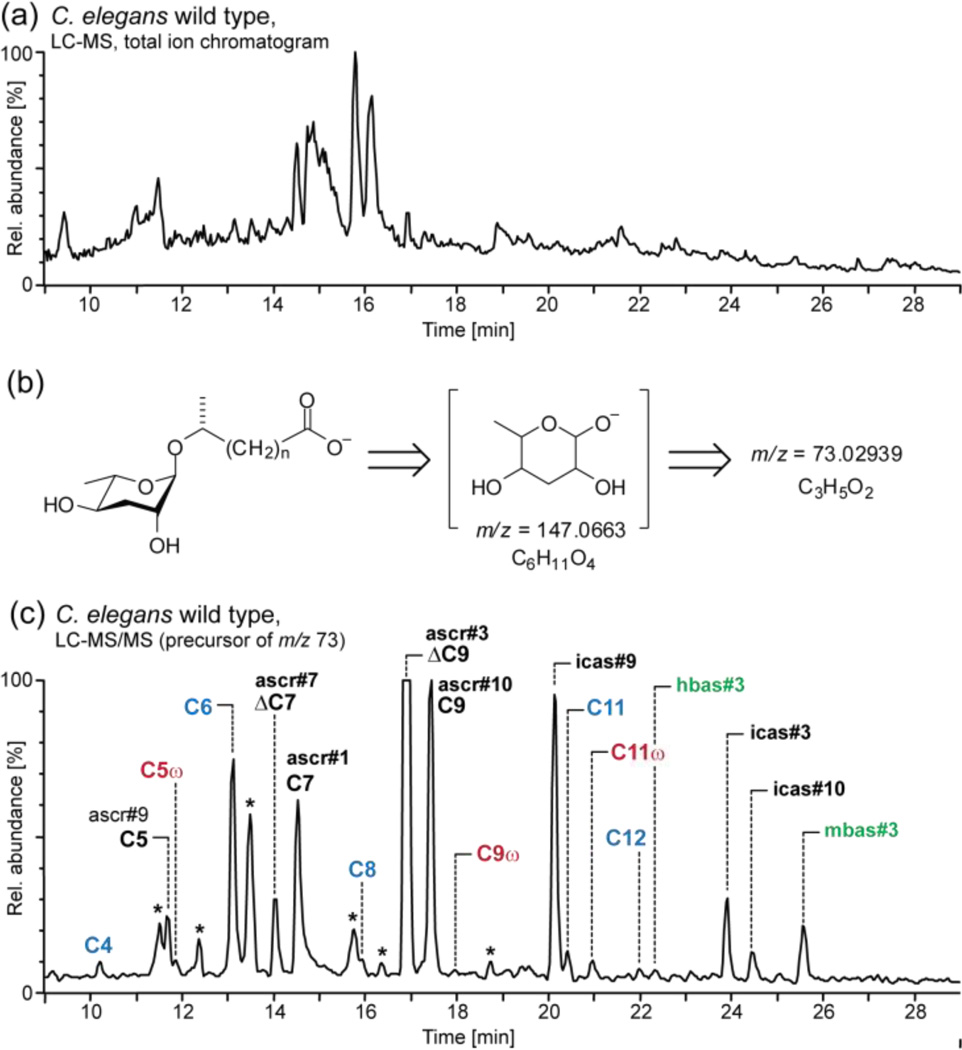
We aimed at developing a method that would (1) facilitate sensitive detection and quantitation of the known ascarosides in the metabolomes of different C. elegans strains and mutants and (2) aid with the discovery of new ascaroside derivatives. Because of the vast complexity of the C. elegans metabolome, HPLC-MS analysis of metabolite extracts results in extremely crowded chromatograms that are difficult to interpret (Figure 2a). However, it seemed likely that structurally related ascarosides would exhibit characteristic MS/MS fragmentation patterns, providing a much more selective and sensitive means for their detection. Therefore we investigated ESI-MS/MS fragmentation of a series of synthetic ascarosides. We found that with negative-ion electrospray ionization (ESI−), ascarosides give rise to an intense and highly diagnostic product ion at m/z 73.02939 [C3H5O2] which originates from the ascarylose unit (Figure 2b and S1). This detection method proved suitable for all known ascarosides, except for ascr#2 and ascr#4 which do not ionize well under ESI− conditions. For detection of ascarosides that ionize only under ESI+ conditions, we monitored neutral loss of 130.0663 amu [C6H10O3] due to cleavage of the ascarylose unit; however, we found that ESI+ MS/MS detection of ascarosides is generally less sensitive than ESI− MS/MS detection as ascarosides fragment less predictably under ESI+ conditions. Next, we tested whether a screen for precursor ions of m/z 73 could be used to detect known as well as yet unidentified ascarosides in the C. elegans wild-type metabolome. For this purpose we used liquid culture metabolite extracts, which contain accumulated excreted metabolites from large numbers of worms (the worm "excretome"). The resulting HPLC-MS/MS chromatograms showed a large number of well-resolved peaks, most of which we found to represent ascarosides, including several families of previously undetected compounds.
Figure 2.
(a) LC-MS total ion current (TIC) chromatogram of wild-type C. elegans excretome (ESI−). (b) MS/MS fragmentation of ascarosides. (c) LC-MS/MS screen (precursors of m/z 73) of wild-type excretome reveals known ascarosides (black), new homologs (blue), new (ω)-oxygenated isomers (red) and new 4’-acylated derivatives (green) (* non-ascarosides). The highly polar ascr#5 elutes at 6.5 min, outside of the shown retention time range.
We first confirmed the identities of the known ascarosides using synthetic standards. In addition we found that the known saturated ascarosides ascr#1, ascr#9, and ascr#10 are accompanied by substantial quantities of homologs with six- to fifteen-carbon side chains (Figure 2c, 3a). Identification of this homologous series was based on high-resolution MS/MS, LC-retention times, and synthesis of representative members (see Supporting Information: Section 1.7 and Figure S2). The LC-MS/MS screen further revealed that ascarosides with side chains of 5 to 11 carbons are accompanied by smaller quantities of slightly less polar isomers. These ascaroside isomers are produced more abundantly by acox-1 mutant worms (vide infra). Several additional peaks in the wild-type MS/MS chromatograms could not be assigned to any of the known ascaroside classes. For two of these compounds, MS/MS product ions at m/z 301.1651 [C15H25O6] suggested that they represent ascr#3 derivatives. The putative ascr#3 derivatives were isolated by preparative HPLC and analyzed using 2D NMR spectroscopy (dqfCOSY, see Figure S3 and S4), which confirmed that these compounds are ascr#3-based and further indicated the presence of a 4-hydroxybenzoyl or (E)-2-methyl-2-butenoyl (tigloyl) moiety attached to the 4-position of the ascarylose (Figure 3c). We corroborated these structural assignments via total synthesis of authentic samples (see Supporting Information). In analogy to the recently reported indole-3-carboxy derivative of ascr#3 ("icas#3"),8 we named the 4-hydroxybenzoyl and (E)-2-methyl-2-butenoyl derivatives of ascr#3 with the Small Molecule IDentifier (SMID)16 "hbas#3" and "mbas#3", respectively. hbas#3 and mbas#3 are the first ascarosides to incorporate 4-hydroxybenzoyl and (E)-2-methyl-2-butenoyl moieties, which in analogy to the indole-3-carboxy moiety in icas#38 could be derived from amino acid precursors. Because of structural similarity to the aggregation-inducing indole ascarosides,8 we tested hbas#3 for its effect on worm behavior. We found that hbas#3 strongly attracts C. elegans at concentrations as low as 10 femto-molar (see Figure S5), which exceeds the potency of any previously known C. elegans small molecule signal.3,8 Low femto molar activity is unusual for small molecule signals in animals, but matched by some classes of peptide hormones.17
Figure 3.
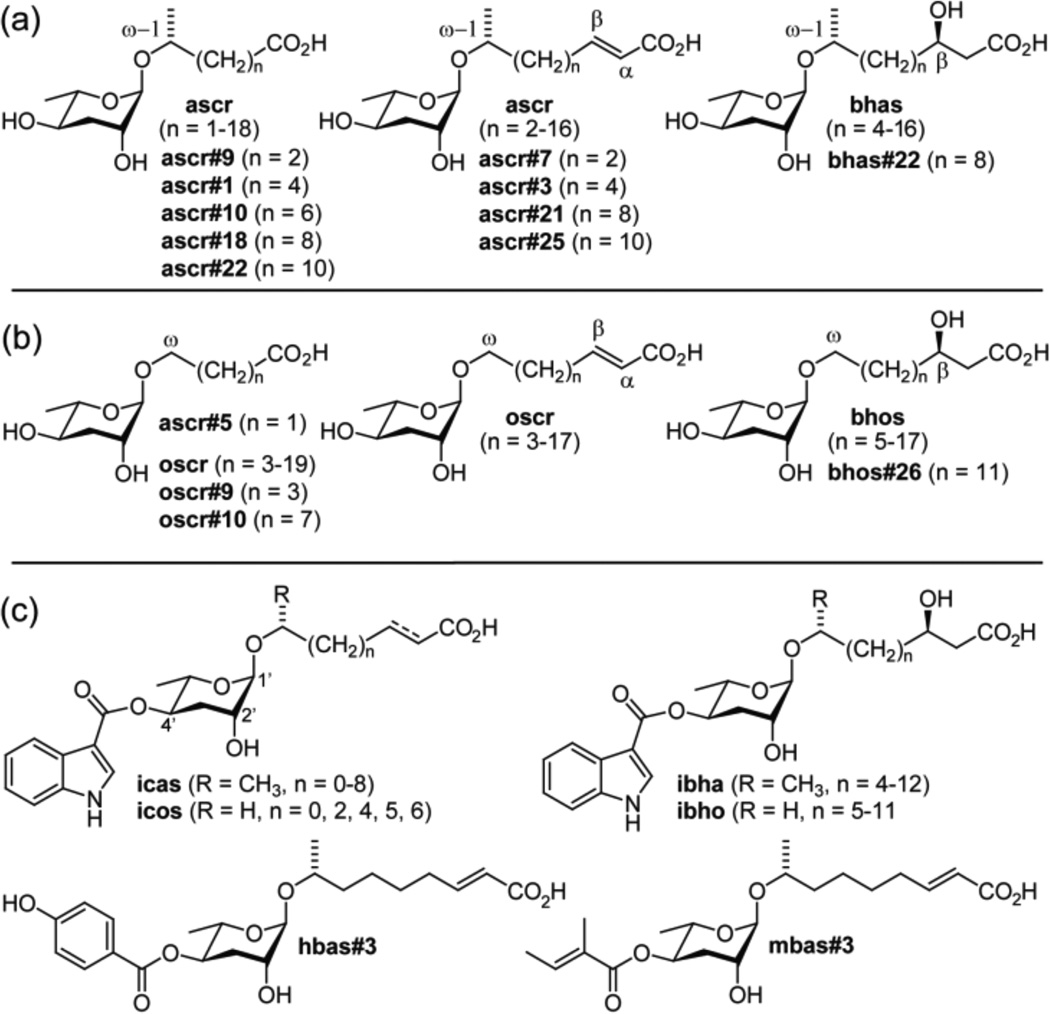
Ascarosides identified in wild-type, acox-1, maoc-1, dhs-28, and daf-22 worms via LC-MS/MS. (a) (ω-1)-oxygenated ascarosides, (b) (ω)-oxygenated ascarosides, and (c) examples for 4'-acylated derivatives. The stereochemistry of compounds that were not synthesized (see Supporting Information for syntheses and a complete list of the 146 characterized ascarosides) was proposed as shown based on analogy and HPLC-MS retention times (Figure S2).
2.2 Peroxisomal β-oxidation in ascaroside biosynthesis
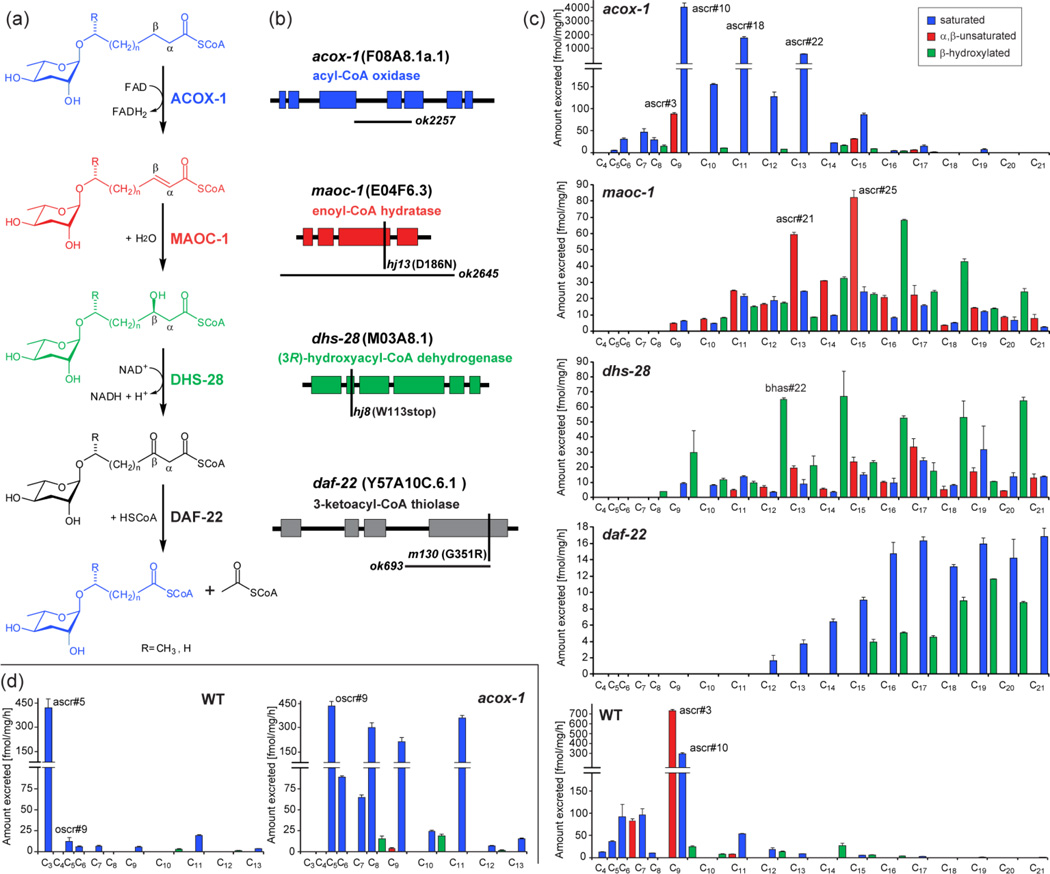
Next we used comparative LC-MS/MS to investigate ascaroside biogenesis. Previous studies suggested that the side chains of the ascarosides are derived from peroxisomal β-oxidation of longer-chained precursors and that the acyl-CoA oxidase ACOX-1 participates in the first step of ascaroside side chain β-oxidation, introducing α,β-unsaturation (Figure 4a, b).11 In vertebrates as well as in Drosophila, the next two steps in peroxisomal β-oxidation, hydration of the double bond and subsequent dehydrogenation to the β-ketoacyl-CoA ester, are catalyzed by one protein, e.g. the multifunctional enzyme type 2 (MFE-2). These two enzymatic functions appear to be separated in C. elegans such that the dehydratase and dehydrogenase are distinct proteins (Figure S6).18 Previous work showed that C. elegans DHS-28, a protein with homology to the (R)-selective β-hydroxyacyl-CoA dehydrogenase domain of human MFE-2, likely participates in converting β-hydroxyacyl-CoA-derivatives into the corresponding β-ketoacyl-CoA intermediates, which are subsequently cleaved by the β-ketoacyl-CoA thiolase DAF-22.6,12 However, it remained unclear which enzyme serves as the enoyl-CoA hydratase that catalyzes the essential second step of the β-oxidation cascade. Using our MS/MS-based ascaroside screen we reinvestigated the ascaroside profiles of acox-1(ok2257), dhs-28(hj8), and daf-22(ok693) mutant worms. Additionally, we analyzed the excretomes of several other peroxisomal mutants, including maoc-1(hj13) worms, because a recent study indicated that maoc-1 encodes a peroxisomal 2-enoyl-CoA hydratase,19 which we hypothesized could participate in the hydration step of ascaroside β-oxidation.
Figure 4.
(a) Proposed roles of peroxisomal β-oxidation enzymes ACOX-1, MAOC-1, DHS-28, and DAF-22 in ascaroside biosynthesis. (b) Gene structures of acox-1, maoc-1, dhs-28, and daf-22 mutant alleles, showing point mutations (vertical bars) and deletions (horizontal bars). (c) (ω-1)-oxygenated ascarosides in wild-type and β-oxidation mutants (acox-1, maoc-1, dhs-28, and daf-22) with saturated (blue), α,β-unsaturated (red), and β-hydroxylated (green) side chains. (d) (ω)-oxygenated ascarosides in wild-type and acox-1 mutants (see also Figure S8).
2.3 Side-chain functionalization precedes β-oxidation
LC-MS/MS analysis of the excretome of acox-1(ok2257) mutant worms revealed that abundance of the α,β-unsaturated ascr#3, the dominating component of wild-type media, was greatly reduced (Figure 4c). This decrease in ascr#3 and other α,β-unsaturated ascarosides does not appear to be the result of overall down regulation in ascaroside production, but instead is accompanied by accumulation of a series of saturated ascarosides. For example, ascr#10, the dihydro-derivative and direct precursor of ascr#3, is 13.6 times more abundant in acox-1(ok2257) than in the wild-type excretome. The corresponding homologs with 11 and 13 carbon side chains, ascr#18 and ascr#22, are 29 times and 66 times more abundant in acox-1 than in the wild-type excretome, respectively. The build-up of longer chained saturated ascarosides in the acox-1(ok2257) excretome confirms the importance of ACOX-1 in α,β-dehydrogenation of the ascaroside side chain (Figure 4a). Because ascaroside biosynthesis is not abolished in acox-1(ok2257) worms, it seems likely that other, yet unidentified ACOX-enzymes contribute to peroxisomal β-oxidation of long chain ascaroside precursors. However, LC-MS/MS analysis of excretome of several other peroxisomal acox mutants (see Supporting Information, Figure S7) revealed largely wild-type like ascaroside profiles.
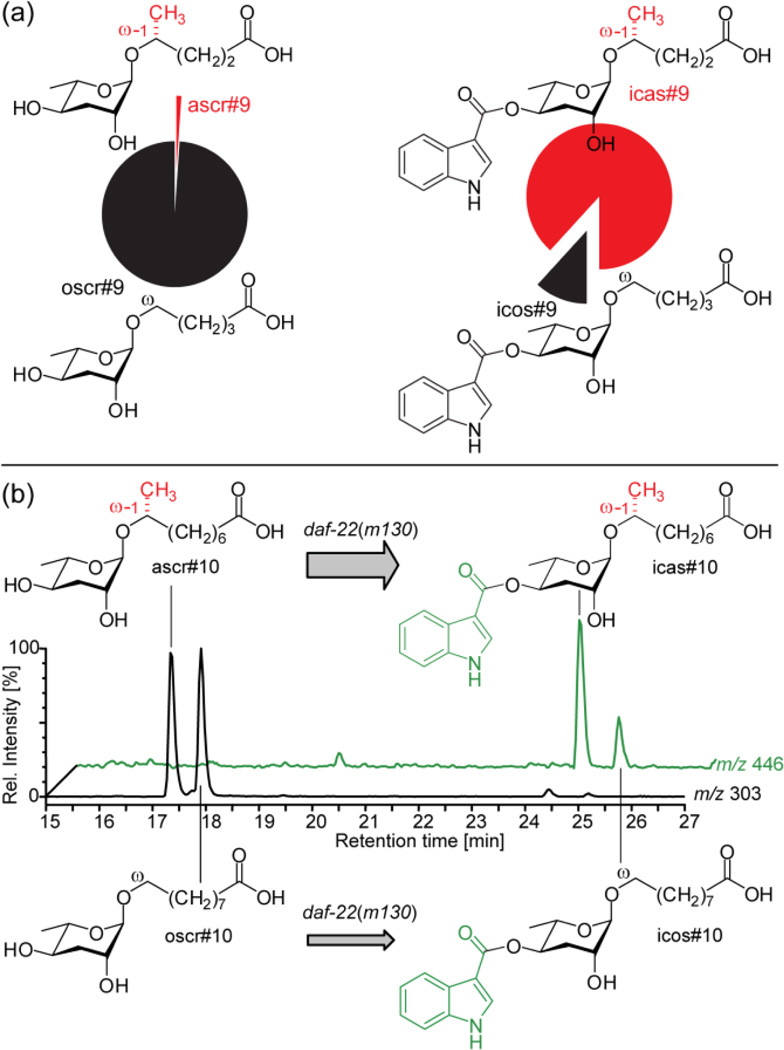
Further analysis of the acox-1(ok2257) excretome revealed the complete absence of ascr#5, one of the major dauer inducing ascarosides produced abundantly in wild type (Figure 4d). Ascr#5 differs from all other previously identified ascarosides in that its side chain is attached to the ascarylose sugar via the terminal carbon ("ω linkage"), and not the penultimate carbon ("ω-1 linkage") (Figure 3b). Instead of ascr#5, we detected large quantities of a new homologous series of saturated ascarosides in acox-1(ok2257), smaller amounts of which were also present in the wild-type excretome (Figure 4d). The most abundant component of this series of isomers was isolated via preparative HPLC and identified by NMR spectroscopy as an ω-linked ascaroside with a C5 side chain (Figure 3b, S9). This suggested that the additional series of compounds observed in acox-1 represents a series of ω-linked saturated ascarosides (Table S2), which we confirmed by synthesis of C5 and C9 ω-linked ascarosides (see Supporting Information). In order to differentiate the (ω)-linked ascarosides from their previously described (ω-1)-linked isomers, we name the newly found (ω)-linked compounds with the SMID “oscr”, e.g. the synthesized (ω)-linked isomers of ascr#9 and ascr#10 were named oscr#9 and oscr#10 (Figure 3).
Thus production of (ω)-linked ascr#5 is abolished in acox-1(ok2257) worms, whereas production of longer chain homologs with 5–13 carbon side chains, e.g. oscr#9, is starkly upregulated (Figure 4d). These results indicate that β-oxidation in acox-1(ok2257) worms is strongly dependent on whether the side chain is (ω-1)- or (ω)-functionalized. Chain shortening of (ω-1)-oxygenated substrates appears to stall at a chain length of 9 carbons as in ascr#10, whereas (ω)-oxygenated substrates are processed for two additional rounds of β-oxidation to afford large quantities of (ω)-oxygenated oscr#9 featuring a 5 carbon side chain. This suggests that sidechain oxygenation precedes peroxisomal β-oxidation. In contrast, the time point of ascarylose attachment seems less certain. The absence of any (ω-1)- or (ω)-hydroxylated fatty acids in the investigated C. elegans mutant metabolome samples suggests a biosynthetic model in which very long-chain ascarosides serve as substrates for peroxisomal β-oxidation; however, the possibility that β-oxidation occurs prior to ascarylose attachment cannot be excluded.
2.4 MAOC-1 and DHS-28 are functional homologs of human MFE-2
In contrast to wild-type and acox-1(ok2257) worms, short chain (< C9) ascarosides were not detected in maoc-1(hj13) and dhs-28(hj8) worms, which instead accumulate several series of (ω-1)- and (ω)-oxygenated medium and long chain ascarosides (≥ C9). The ascaroside profile of the maoc-1(hj13) excretome is dominated by α,β-unsaturated ascarosides such as ascr#21 (C13) and ascr#25 (C15) (Figure 4c), supporting our hypothesis that MAOC-1 functions as an enoyl-CoA hydratase in the ascaroside biosynthetic pathway (Figure 4a). In addition, the maoc-1(hj13) excretome contained smaller amounts of the corresponding saturated ascarosides, along with a third homologous series of compounds, whose HRMS and MS/MS fragmentation suggested that they represent a series of long chain β-hydroxylated ascarosides (Figure 4c, S1, Table S3–4). These structural assignments were confirmed via synthesis of representative members of this series (see Supporting Information), the C9 and C13 β-hydroxylated (ω-1)-ascarosides, bhas#10 and bhas#22, as well as the C15 β-hydroxylated (ω)-ascaroside bhos#26, (SMIDs for (ω-1)- and (ω)-oxygenated β-hydroxylated ascarosides: "bhas" and “bhos”). Since β-hydroxylated ascarosides are putative products of enoyl-CoA hydratases such as MAOC-1, their presence in the excretomes of both studied maoc-1 mutant strains, maoc-1(hj13), which carries a point mutation in the active site (D186N), and the 2110 base pair deletion mutant maoc-1(ok2645) (Figure 4b), suggests that additional enoyl-CoA hydratases may participate in ascaroside biosynthesis.
Similar to our results for maoc-1(hj13), we found that the dhs-28(hj8) ascaroside profile is dominated by compounds with side chains ranging from C9–C21 (Figure 4c). However, in contrast to maoc-1(hj13), saturated and α,β-unsaturated ascarosides constitute a relatively smaller portion of total ascarosides in dhs-28(hj8). Instead, we found (ω-1)- and (ω)-oxygenated β-hydroxy ascarosides (bhas and bhos) with odd numbered side chains from C13–C21 as major components (Figure 4c, S8), which is consistent with the proposed biosynthetic role of DHS-28 as a β-hydroxyacyl-CoA dehydrogenase.6,12 Analysis of the daf-22(ok693) excretome revealed the absence of all ascarosides with side chains shorter than 12 carbons, as reported earlier (Figure 4c, S8).5,6,12 We found that daf-22(ok693) contains small amounts of homologous series of (ω-1)- and (ω)-oxygenated long chain ascarosides featuring saturated (ascr and oscr) and β-hydroxylated side chains (bhas and bhos). In addition the daf-22(ok693) excretome contains very long chain ascarosides (VLCA, >C22) with side chain lengths of up to 33 carbons, as reported earlier.5,20
2.5 Identification of new indole ascarosides
Indole-3-carbonylated ascarosides are much less abundant than the corresponding unfunctionalized ascarosides and have recently been shown to function as highly potent aggregation signals.8 Our LC-MS/MS screen revealed several new types of indole ascarosides (Figure 3c, Table S5–8). Using synthetic samples of icas#3, icas#9, and icas#1,8 we showed that indole ascarosides exhibit a characteristic fragmentation pattern that includes neutral loss of 143 amu [C9H5NO] due to cleavage of the indole-3-carbonyl unit, as well as the ascaroside-diagnostic product ion at m/z 73 (see Figure S1). LC-MS/MS screening for components with this fragmentation pattern revealed that known (ω-1)-oxygenated isomers icas#1, icas#9, and icas#10 in acox-1(ok2257) are accompanied by a series of (ω)-oxygenated indole ascarosides (SMID: icos#1, icos#9, and icos#10), which was confirmed by chemical synthesis of icos#10 as a representative example (see Supporting Information). Peroxisomal β-oxidation mutants that do not produce short chain ascarosides (<9 carbon side chains), for example maoc-1 and dhs-28 worms, also do not produce any of the corresponding indole ascarosides. Instead, we found that the maoc-1 and dhs-28 excretomes contain significant amounts of (ω-1)- and (ω)-oxygenated long chain indole ascarosides (SMIDs icas and icos) and indole β-hydroxy ascarosides (SMIDs ibha and ibho) with side chains from 9–17 carbons (see Table S5–8).
2.6 Indole ascaroside biogenesis
Using application experiments with deuterium labeled tryptophan and axenic in vitro cultures we have previously shown that the indole-3-carbonyl moiety of indole ascarosides originates from L-tryptophan.8 A similar L-tyrosine or L-phenylalanine origin seems likely for the 4-hydroxybenzoyl moiety of hbas#3, whereas the tigloyl group of mbas#3 could be derived from L-isoleucine.21 However, it remained unclear at what stage in ascaroside biosynthesis the indole-3-carbonyl moiety is attached. Comparison of ascaroside and indole ascaroside profiles revealed that indole ascaroside biosynthesis is tightly controlled. For example, we found that in acox-1 mutants the (ω)-ascaroside oscr#9 is over 100 times more abundant than the (ω-1)-ascaroside ascr#9, whereas (ω-1)-indole ascaroside icas#9 is much more prominent than (ω)-indole ascaroside icos#9 (Figure 5a). Similarly, icas#3 and icas#9, the dominating indole ascarosides in the wild type excretome, are produced in roughly equal amounts, whereas ascr#3 is about 20 times more abundant than ascr#9 (Figure S10). This strong dependence of indole ascaroside biosynthesis on side chain length and (ω-1)- vs. (ω)-oxygenation suggested that these compounds originate from specific attachment of a indole-3-carbonyl unit to the corresponding non-indole ascarosides.
Figure 5.
Indole ascaroside biosynthesis. (a) Relative abundance of (ω-1) and (ω)-oxygenated C5-ascarosides ascr#9 and oscr#9 and their corresponding indole ascarosides icas#9 and icos#9 in acox-1 indicates that indole-3-carbonyl attachment is highly specific. (b) LC-MS ion traces of daf-22(m130) excretome following incubation with a 1:1 mixture of ascr#10 and oscr#10, showing a preference for indole-3-carbonyl attachment to the (ω-1)-oxygenated ascr#10.
To test whether non-indole ascarosides serve as precursors for indole ascarosides, we incubated daf-22(m130) worms (which are devoid of all short-chained indole and non-indole ascarosides) with a 1:1 mixture of ascr#10 and oscr#10 for 5 days. Subsequent analysis by LC-MS showed partial conversion into icas#10 and icos#10 (Figure 5b), indicating that non-indole ascarosides serve as specific precursors to their corresponding indole derivatives. Moreover, conversion of (ω-1)-ascaroside ascr#10 was preferred over conversion of (ω)-ascaroside oscr#10, reflecting the ratios of these compounds in wild type and acox-1 mutants. Similarly, we found that daf-22(m130) worms convert added ascr#3 into icas#3. Further conversion of indole or non-indole ascarosides into shorter chain derivatives (e.g. ascr#1 or icas#1) was not observed. These results indicate that attachment of an L-tryptophan derived indole-3-carbonyl unit represents the final step in indole ascaroside biosynthesis.
2.7 Ascaroside excretion is selective
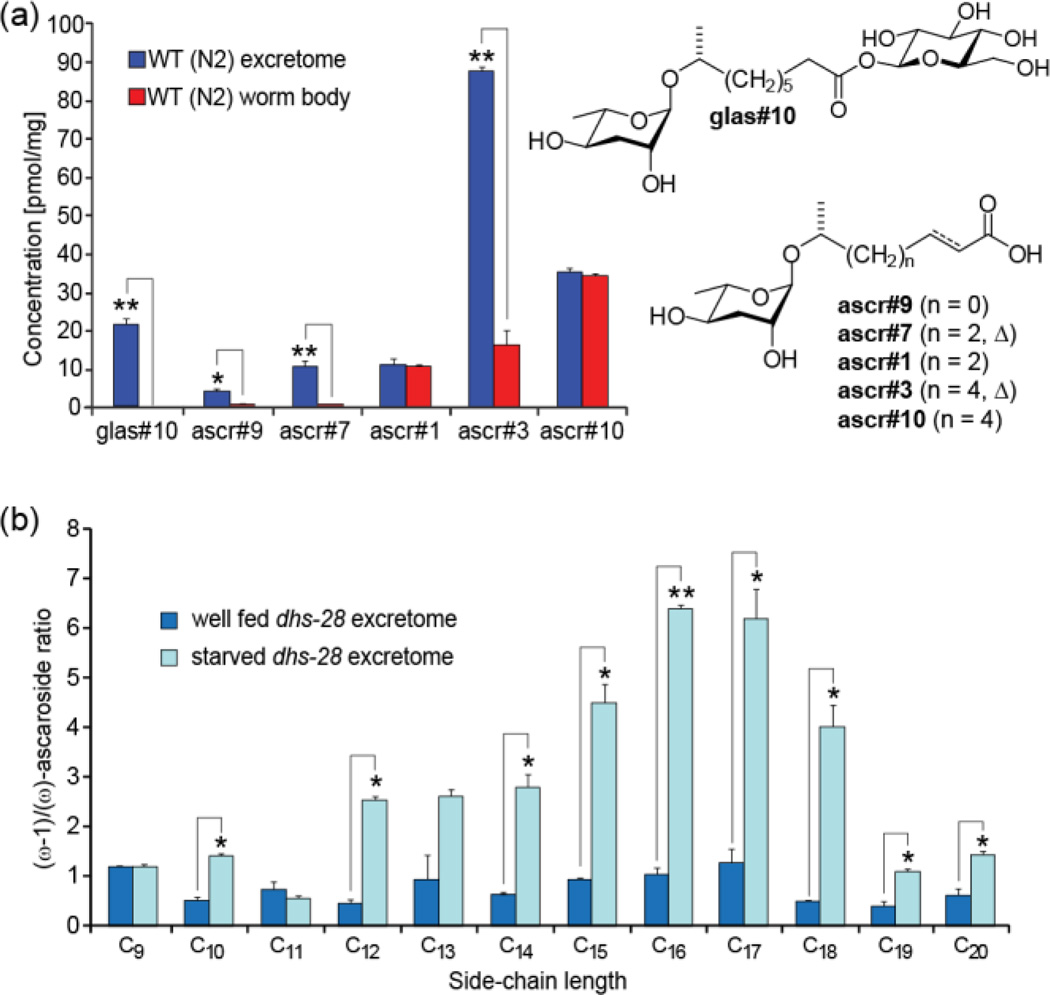
Despite detailed investigations of ascaroside function, little is known about how ascarosides are released and transported from their site of biosynthesis. We compared the ascaroside profiles of the wild-type excretomes (liquid culture supernatant extracts) and worm body metabolomes (worm pellet extracts) to identify possible non-excreted ascaroside derivatives and to determine quantitative differences. We found that ascaroside profiles of worm pellet extracts differ significantly from those excreted into the media, indicating that ascarosides are differentially released by C. elegans (Figure 6a). In the worm pellets, the most abundant ascarosides, for example ascr#3 in wild-type and ascr#10 in acox-1 worms, are accompanied by significantly more polar derivatives, which are absent from the media extracts (Figure S11). MS/MS analysis suggested that these components represent ascaroside O-glycoside esters. Isolation of the putative ascr#10 glycoside from acox-1(ok2257) worm pellet extracts and subsequent NMR spectroscopic analysis (Figure S12) indicated esterification of ascr#10 with the anomeric hydroxy group of β-glucose, which was subsequently confirmed via synthesis (SMID for 1-β-D-glucosyl ascr#10: glas#10, Figure 6a). The fact that large quantities of highly polar glas#10 and other ascaroside glucosides (see Table S9) are retained in the worm bodies but not excreted suggests that they represent transport or storage forms of the ultimately excreted signaling molecules.
Figure 6.
(a) Analysis of worm body ascaroside profiles reveals ascaroside glucosides (e.g. glas#10) and indicates preferential excretion of unsaturated ascarosides. (b) Ratio of (ω-1) to (ω)-linked saturated ascarosides in dhs-28 mutant excretome shows strong dependence on nutritional conditions (for β-hydroxy ascarosides see Figure S14), reflecting starvation dependence of ascr#3/ascr#5 ratio in wild-type C. elegans (see Figure S15) (Welch’s t-test, *: P<0.05; ** P<0.001).
In addition we found that saturated ascarosides are retained in the worm bodies to a much greater extent than their α,β-unsaturated derivatives (Figure 6a). Differential release was also observed for (ω)-oxygenated components (Figure S13). Therefore it appears that C. elegans exhibits remarkable control over the release of ascaroside signals.
2.8 Nutritional state affects ascaroside biosynthesis
Ascaroside biosynthesis has been reported to depend on various environmental factors including food availability,6 developmental stage,15 and temperature.11 We used LC-MS to investigate the effect of nutritional state on ascaroside biosynthesis by comparing the excretomes of well fed and starved cultures of wild-type and mutant worms. We found that the ratio of (ω-1) to (ω)-linked ascarosides strongly depends on nutritional state. Production of long-chain (ω-1)-oxygenated ascarosides in starved cultures of dhs-28 was about 5 times higher than those of well fed cultures (Figure 6b, S14). Similarly, starved wild-type worms excrete significantly larger amounts of the (ω-1)-linked ascr#3 than well-fed worms, relative to the amounts of (ω)-linked ascr#5 (Figure S15). Both ascr#5 and ascr#3 are major components of the dauer pheromone; however, they affect worm behavior differently.1,3,4,22
Therefore, it appears that ascaroside signaling is actively regulated in response to changes in nutrient availability via modulation of (ω-1) and (ω)-functionalization of very long-chain fatty acids upstream of peroxisomal β-oxidation. Together with the recent finding that (ω)- and (ω-1)-functionalized ascarosides are sensed by different families of G-protein coupled receptors,23 these results suggest that (ω)- and (ω-1)-functionalized ascarosides target separate downstream signaling pathways.
3. DISCUSSION
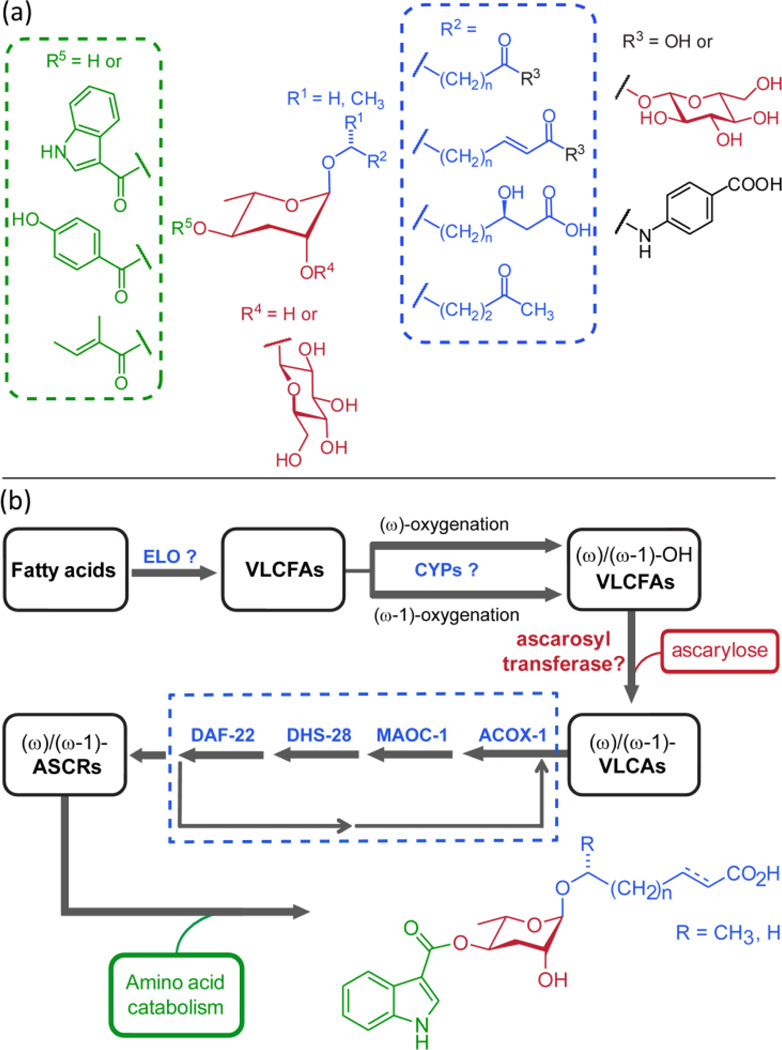
Ascarosides play important roles for several different aspects of C. elegans biology. This functional diversity is paralleled by corresponding structural diversity and a complex biosynthetic pathway. Our MS/MS-based study revealed 146 ascarosides, including 124 previously unreported analogs, which show several unexpected features including (ω)-oxygenation of the fatty acid-derived side chains, 4-hydroxybenzoylation or (E)-2-methyl-2-butenoylation of the ascarylose unit, and glucosyl esters. Most ascarosides occur as members of homologous series of compounds with (ω-1)- or (ω)-linked saturated, α,β-unsaturated, or β-hydroxylated side chains ranging from 3 to 21 (occasionally more) carbons. Importantly, only a few members of each series are produced abundantly in wild type, and incorporation of specific structural features such as modification in position 4 of the ascarylose (e.g. as in indole ascarosides) or the incorporation of α,β-unsaturation appears to be tightly controlled. Given their assembly from carbohydrate, lipid, and amino acid-derived building blocks, the ascarosides appear as a modular library of small molecule signals that integrate inputs from three basic metabolic pathways (Figure 7a). The ascarosides then transduce input from these pathways via their diverse signaling functions in C. elegans’ behavior and development, including dauer formation, mate attraction, hermaphrodite repulsion, and aggregation.8,14 Their specific biosyntheses suggest that many of the newly identified ascarosides also contribute to known or as yet undetermined functions in C. elegans. As an example, we show that hbas#3 acts as an attraction signal whose potency exceeds that of all previously known small molecules in this model organism. More comprehensive biological evaluation will require consideration of synergistic activities (i.e. testing of compound mixtures), which, given the very large number of compounds, may necessitate high-throughput approaches, for example based on microfluidic devices.24
Figure 7.
(a) Modular assembly of ascarosides from amino acid ( ), fatty acid (
), fatty acid ( ) and carbohydrate (
) and carbohydrate ( ) building blocks. (b) Model for ascaroside biogenesis. Chain elongation of fatty acids (by putative elongase homologs elo-1–925) is followed by (ω-1)- or (ω)-oxygenation of VLCFAs and ascarylose attachment. The resulting VLCAs enter peroxisomal β-oxidation via ACOX-1, MAOC-1, DHS-28, and DAF-22 producing short chain ascarosides, which are linked to amino acid-derived moieties and other building blocks.
) building blocks. (b) Model for ascaroside biogenesis. Chain elongation of fatty acids (by putative elongase homologs elo-1–925) is followed by (ω-1)- or (ω)-oxygenation of VLCFAs and ascarylose attachment. The resulting VLCAs enter peroxisomal β-oxidation via ACOX-1, MAOC-1, DHS-28, and DAF-22 producing short chain ascarosides, which are linked to amino acid-derived moieties and other building blocks.
Our results allow proposing a working model for ascaroside biogenesis (Figure 7b). The finding that ascarosides are members of several homologous series with side chains up to 21 (and more) carbons suggests their origin from peroxisomal β-oxidation of very long chain precursors. Previous studies reported the presence of very long chain ascarosides (VLCA) with C29 and C31 side chains in wild-type and daf-22 mutants, which could represent precursors or intermediates in ascaroside biosynthesis.5,20 Alternatively, very long chain fatty acids (VLCFAs) could undergo peroxisomal β-oxidation prior to (ω-1)- or (ω)-functionalization and subsequent attachment of the ascarylose. Our observation that the acox-1 mutation affects (ω-1)- and (ω)-oxygenated ascarosides differently suggests that (ω-1)- and (ω)-functionalization of VLCFA precursors occurs upstream of their breakdown by peroxisomal β-oxidation. Given the large range in side chain lengths it seems likely that resulting (ω-1)- and (ω)-hydroxy VLCFAs are linked to ascarylose prior to entering the β-oxidation pathway, though the presence of a promiscuous ascarosyl transferase cannot be excluded (Figure 7b).
Chain shortening of VLCA then progresses via repetitive cycles of peroxisomal β-oxidation. The results from our LC-MS/MS screen allowed us to propose precise roles for enzymes participating in each of the four-step β-oxidation cycle: the acyl-CoA oxidase ACOX-1, enoyl-CoA hydratase MAOC-1, β-hydroxyacyl-CoA dehydrogenase DHS-28, and β-ketoacyl-CoA thiolase DAF-22. We show that mutations in acox-1, maoc-1, and dhs-28 result in specific changes of the corresponding ascaroside profiles, in agreement with their proposed functions.
The acyl-CoA oxidase ACOX-1 has been subject of a previous study which suggested that mutations in acox-1 primarily affect the biosynthesis of ascr#2 and ascr#3, but not of ascr#1.11 However, our results indicate that acox-1(ok2257) mutants have a reduced ability to process C9 (ω-1)-functionalized ascarosides, resulting in diminished production of all shorter-chained ascarosides and build-up of C9 and longer chained saturated ascarosides. Mutations of maoc-1 (as well as dhs-28 and daf-22) have been shown to result in expansion of intestinal lipid droplets and cause an increase in fasting- and lipolysis-resistant triglycerides.19 Our results show that MAOC-1 participates in ascaroside biosynthesis, acting as the previously unidentified enoyl-CoA hydratase. These findings further demonstrate that hydration of enoyl-CoAs and dehydrogenation of β-hydroxyacyl-CoAs in C. elegans are catalyzed by two distinct enzymes, MAOC-1 and DHS-28, as had been suggested based on their homology to separate functional domains of human MFE-2.19
We further show that attachment of the tryptophan-derived indole-3-carbonyl unit in indole ascarosides likely represents the last step in their biosynthesis, and that this step is highly specific. As attachment of an indole-3-carbonyl group to ascarosides can dramatically alter their biological function, such tight regulation makes sense. For example, indole-3-carbonyl addition to the dauer inducing and strongly repulsive signal ascr#3 results in the potent hermaphrodite attractant icas#3.8 Therefore, identification of the enzymes that attach indole-3-carbonyl and other functional groups to the ascarosides will be of great interest.
The biosynthesis of ascarylose in C. elegans has not been investigated, but detection of ascarosides in axenic C. elegans cultures demonstrated that C. elegans produce ascarylose endogenously.8 Ascarylose biosynthesis in bacteria is well understood, and the C. elegans genome includes several homologs of bacterial genes in this pathway, for example ascE from Yersinia pseudotuberculosis (Figure S16),26 providing potential entry points for the study of ascarylose biosynthesis and its regulation in nematodes. In addition, the oxidases catalyzing (ω-1)- or (ω)-functionalization of VLCFA precursors remain to be identified. Our finding that the ratio of (ω-1)/(ω)-oxygenated ascarosides in wild type and peroxisomal β-oxidation mutants is strongly affected by starvation suggests that this step may encode information about nutritional state. Finally, we demonstrate that ascaroside excretion is surprisingly specific. Given the high sensitivity and selectivity of LC-MS/MS, we believe that ascaroside profiling using this method will aid identifying additional genes and environmental factors that participate in ascaroside biosynthesis and homeostasis.
Supplementary Material
ACKNOWLEDGMENT
We thank Arthur Edison (University of Florida, Gainesville) for helpful suggestions, the Caenorhabditis Genetics Center, Ho Yi Mak (Stowers Institute), and Shohei Mitani (Tokyo Women’s Medical University) for providing C. elegans mutant strains, and Maciej Kukula (BTI Mass Spectrometry Facility) and Wei Chen (Proteomics and Mass Spectrometry Core Facility, Cornell University) for assistance with HR-MS. This work was supported in part by the National Institutes of Health (GM088290, GM085285, and T32GM008500), and the Cornell/Rockefeller/Sloan-Kettering Training Program in Chemical Biology.
ABBREVIATIONS
- SMID
Small Molecule Identifier
- VLCA
very long chain ascaroside
- VLCFA
very long chain fatty acid
Footnotes
ASSOCIATED CONTENT
Supporting Information. Detailed experimental procedures including the synthesis of reference compounds, a full list of identified ascarosides, and their LC-MS and NMR spectroscopic data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Jeong PY, Jung M, Yim YH, Kim H, Park M, et al. Nature. 2005;433:541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 2.Butcher RA, Fujita M, Schroeder FC, Clardy J. Nat. Chem. Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW, Schroeder FC. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butcher RA, Ragains JR, Kim E, Clardy J. Proc. Natl. Acad. Sci. USA. 2008;105:14288–14292. doi: 10.1073/pnas.0806676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg P, Schroeder FC. Proc. Natl. Acad. Sci. USA. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Proc. Natl. Acad. Sci. USA. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]; (b) Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]; (c) Schroeder FC. ACS Chem. Biol. 2006;1:198–200. doi: 10.1021/cb600173t. [DOI] [PubMed] [Google Scholar]; (d) Fielenbach N, Antebi A. Genes. Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Sommer RJ, Ogawa A. Curr. Biol. 2011;21:R758–R766. doi: 10.1016/j.cub.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho M, O’Doherty O, Edison A, Sternberg P, Schroeder FC. PLoS Biol. 2011 doi: 10.1371/journal.pbio.1001237. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golden JW, Riddle DL. Mol. Gen. Genet. 1985;198:534–536. doi: 10.1007/BF00332953. [DOI] [PubMed] [Google Scholar]
- 10.White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, Jorgensen EM. Curr. Biol. 2007;17:1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Joo HJ, Kim KY, Yim YH, Jin YX, Kim H, Kim MY, Paik YK. J. Biol. Chem. 2010;285:29319–29325. doi: 10.1074/jbc.M110.122663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joo HJ, Yim YH, Jeong PY, Jin YX, Lee JE, Kim H, Jeong SK, Chitwood DJ, Paik YK. Biochem. J. 2009;422:61–71. doi: 10.1042/BJ20090513. [DOI] [PubMed] [Google Scholar]
- 13.Butcher RA, Ragains JR, Clardy J. Org. Lett. 2009;11:3100–3103. doi: 10.1021/ol901011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edison AS. Curr. Opin. Neurobiol. 2009;19:378–388. doi: 10.1016/j.conb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan F, Srinivasan J, Mahanti P, Ajredini R, Durak O, Nimalendran R, Sternberg PW, Teal PEA, Schroeder FC, Edison AE, Alborn HT. PLoS ONE. 2011;6:e17804. doi: 10.1371/journal.pone.0017804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SMID: Small Molecule IDentifier for small molecules identified from C. elegans and other nematodes. The SMID database (www.smid-db org) is an electronic resource maintained by Frank C. Schroeder and Lukas Mueller at the Boyce Thompson Institute in collaboration with Wormbase (www.wormbase.org). The purpose of this database is to introduce searchable, gene-style identifiers, "SMIDs", for all small molecules newly identified from C. elegans and other nematodes.
- 17.(a) Rittschof D, Cohen JH. Peptides. 2004;25:1503–1516. doi: 10.1016/j.peptides.2003.10.024. [DOI] [PubMed] [Google Scholar]; (b) Gozes I, Morimoto BH, Tiong J, Fox A, Sutherland K, Dangoor D, Holser-Cochav M, Vered K, Newton P, Aisen PS, Matsuoka Y, van Dyck CH, Thal L. CNS Drug Rev. 2005;11:353–368. doi: 10.1111/j.1527-3458.2005.tb00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haataja TJK, Koski MK, Hiltunen JK, Glumoff T. Biochem. J. 2011;435:771–781. doi: 10.1042/BJ20101661. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. Proc. Natl. Acad. Sci. USA. 2010;10:4640–4645. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zagoriy V, Matyash V, Kurzchalia T. Chem. Biodivers. 2010;7:2016–2022. doi: 10.1002/cbdv.201000012. [DOI] [PubMed] [Google Scholar]
- 21.Attygalle AB, Wu X, Will KW. J. Chem. Ecol. 2007;33:963–970. doi: 10.1007/s10886-007-9276-3. [DOI] [PubMed] [Google Scholar]
- 22.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K, Sengupta P. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Lab. Chip. 2011;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agbaga MP, Brush RS, Mandal NA, Henry K, Elliott MH, Anderson RE. Proc. Natl. Acad. Sci. USA. 2008;105:12843–12848. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorson JS, Lo SF, Olivier P, He X, Liu HW. J. Bacteriol. 1994;176:5483–5493. doi: 10.1128/jb.176.17.5483-5493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.