Abstract
Primary fatty acid amides (PFAM) are important signaling molecules in the mammalian nervous system, binding to many drug receptors and demonstrating control over sleep, locomotion, angiogenesis, and many other processes. Oleamide is the best-studied of the primary fatty acid amides, whereas the other known PFAMs are significantly less studied. Herein, quantitative assays were used to examine the endogenous amounts of a panel of PFAMs, as well as the amounts produced after incubation of mouse neuroblastoma N18TG2 and sheep choroid plexus (SCP) cells with the corresponding fatty acids or N-tridecanoylethanolamine. Although five endogenous primary amides were discovered in the N18TG2 and SCP cells, a different pattern of relative amounts were found between the two cell lines. Higher amounts of primary amides were found in SCP cells, and the conversion of N-tridecanoylethanolamine to tridecanamide was observed in the two cell lines. The data reported here show that the N18TG2 and SCP cells are excellent model systems for the study of PFAM metabolism. Furthermore, the data support a role for the N-acylethanolamines as precursors for the PFAMs and provide valuable new kinetic results useful in modeling the metabolic flux through the pathways for PFAM biosynthesis and degradation.
Keywords: N-acylethanolamine as PFAM precursors, choroid plexus cells, oleamide metabolism, N-fatty acylglycine oxidation
Fatty acid amides have a long biological history, dating back over a century to the pioneering studies of the ceramides and sphingolipids (1, 2). Primary fatty acid amides (PFAM), R-CO-NH2 with R being a long-chain fatty acid, were first identified from a biological source in 1989 with the identification of palmitamide, palmitoleamide, oleamide, elaidamide, and linoleamide in luteal phase plasma (3). At the time of their discovery, the biological function of these mammalian PFAMs was unknown. Interest in the PFAMs dramatically intensified with the discoveries that oleamide accumulated in the cerebrospinal fluid (CSF) of sleep-deprived cats, that it is a natural component of the CSF in the cat, rat, and human, and that the administration of synthetic oleamide induced physiological sleep in rats (4). Intriguingly, later studies found that oleamide levels in the brain of the ground squirrel were ∼2.5-fold higher in hibernating animals relative to those found in nonhibernating animals (5). Other functions ascribed to oleamide, since its discovery as a sleep-inducing PFAM, include the ability to modulate gap junction communication in glial cells (6, 7), tracheal epithelial cells (8), seminiferous tubule cells (9), and fibroblasts (10); to allosterically activate the GABAA receptors and specific subtypes of the serotonin receptor (11–13); to affect memory processes (14); to increase food intake (15); to reduce anxiety and pain (16, 17); to depress body temperature and locomotor activity (17, 18); to stimulate Ca(II) release (19); and to relax blood vessels (20, 21).
Although much of the research regarding the PFAMs has focused on oleamide, studies show that some of the other known PFAMs are bioactive. Palmitamide is weakly anticonvulsant (22); linoleamide regulates Ca(II) flux (23) and inhibits the erg current (24); and erucamide stimulates angiogenesis (25) and controls water balance (26). In addition, both mammalian phospholipase A2 (PLA2) and epoxide hydrolase (EH) are inhibited by series of PFAMs, with arachidonamide and γ-linolenamide being the most potent for the inhibition of PLA2 (27) and elaidamide being the most potent for the inhibition of EH (28). The physiological significance of the reported functions of oleamide and the other PFAMs is not completely clear as relatively high concentrations are sometimes used to elicit the indicated responses. Nonetheless, the PFAMs have emerged as an important class of mammalian cell signaling lipids. For recent reviews of PFAM metabolism, see Farrell and Merkler (29) and Ezzili et al. (30).
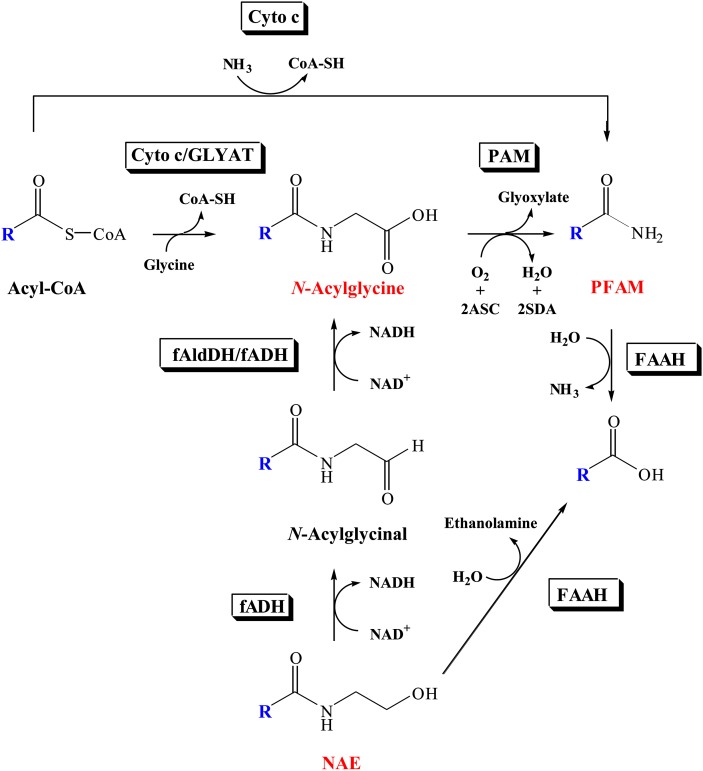
The major route for PFAM degradation in vivo is the hydrolysis to fatty acid and ammonia, a reaction catalyzed by fatty acid amide hydrolase (FAAH) (31, 32). Less is definitively known about PFAM biosynthesis. One proposed route is the ammonolysis of fatty acyl-CoA thioesters (33), whereas a second proposed route involves the oxidative cleavage of N-fatty acylglycines (34, 35). Mouse neuroblastoma N18TG2 cells are known to produce oleamide (36) and, thus, must possess the enzymatic machinery necessary for oleamide biosynthesis. N-Oleoylglycine was characterized after growth of the N18TG2 cells in the presence of inhibitor of peptidylglycine α-amidating monooxygenase (PAM) (37), supporting the hypothesis that the N-fatty acylglycines serve as PFAM precursors. PAM, an enzyme with a well-defined role in α-amidated peptide biosynthesis (38), has been suggested to catalyze N-fatty acylglycine cleavage in vivo. A number of N-fatty acylglycines have been identified from mammalian sources (37, 39), and there is evidence that these members of the fatty acid amide family are also cell signaling lipids (40–42). The biosynthesis of these lipids is also unclear; suggested pathways include glycine conjugation of the fatty acyl-CoA thioesters (34, 43) and sequential oxidation of the N-acylethanolamines (44, 45) (Fig. 1). The N-acylethanolamines (NAE) are another branch of the fatty acid amide family of cell signaling lipids, with N-arachidonoylethanolamine (anandamide) being the most studied of these group of molecules. Given there are multiple pathways known for the biosynthesis of the NAEs (44–46), it is likely that there are also multiple routes for the in vivo production of the N-fatty acylglycines and the PFAMs (29, 30). In fact, Bradshaw et al. (46) have shown that the N-fatty acylglycines are produced in the C6 glioma cells via the glycine-dependent and NAE-dependent reactions shown in Fig. 1.
Fig. 1.
Proposed pathways for the biosynthesis and degradation of the N-fatty acylglycines and the PFAMs. Other possible biosynthetic and degradative reactions are discussed in the review by Farrell and Merkler (29). ASC, ascorbate; Cyto c, chytochrome c; fADH, fatty alcohol dehydrogenase; fAldDH, fatty aldehyde dehydrogenase; GLYAT, glycine N-acyltransferase; PAM, peptidylglycine α-amidating monooxygenase; SDA, semidehydroascorbate.
The N18TG2 cells produce oleamide and are known to express PAM and FAAH (36, 47, 48). Oleamide has been the focus of the PFAM research in the N18TG2 cells, and one goal of the work detailed herein was to broaden the scope of the N18TG2-based work to other PFAMs. It is essential to determine whether the pathways defined for oleamide biosynthesis and degradation could account for the other known PFAMs. Our second goal was to establish choroid plexus cells, the cells that produce the oleamide-containing CSF, as another model for future PFAM metabolic studies.
We report here, for the first time, the N18TG2-mediated conversion of a series of fatty acids to PFAMs, including the production of tridecanamide from N-tridecanoylethanolamine. Using the same set of fatty acids and N-tridecanoylethanolamine, we report also that cultured sheep choroid plexus (SCP) cells produce PFAMs. The PFAM levels in the SCP cells were generally higher than those in the N18TG2 cells, likely resulting from the lack of detectable FAAH expression in the SCP cells. The data reported demonstrate that the N18TG2 and SCP cells are excellent models for the study of PFAM metabolism and provide useful indices for modeling of PFAM metabolism as the biosynthetic and degradative pathways are better defined.
MATERIALS AND METHODS
Materials and chemicals
Linoleic acid, palmitic acid, 6-thioguanine, and fatty acid-free BSA (BSA) were from Sigma; elaidic acid and palmitoleic acid were from Fisher; penicillin/streptomycin, EMEM, and DMEM were supplied by Mediatech Cellgro; fetal bovine serum (FBS) was from Atlanta Biologicals; BSTFA and silica were from Suppelco; [2H33]heptadecanoic acid (margaric acid, C2H3-(C2H2)15-COOH) was from C/D/N isotopes; goat ant-FAAH (V-17) antibody and its blocking peptide were from Santa Cruz Biotech; donkey anti-goat IgG fused to horse radish peroxidase (HRP) was from ICN Biomedical; SuperSignal chemiluminescent HRP substrate was from Bio-Rad; and MagicMark XP molecular weight markers were from Invitrogen. Mouse neuroblastoma N18TG2 cells were purchased from the Deutsche Sammlung von Mikrooganism und Zellkuturen GmBH, and the sheep choroid plexus cells came from the American Type Culture Collection. All other reagents and supplies were purchased from commercial sources at the highest quality available and used without further modification.
Primary fatty acid amide synthesis
PFAMs were synthesized from the acyl chloride essentially as described by Fong et al. (49). The undistilled acyl chloride was added drop-wise to ice-cold concentrated NH4OH at a ratio of 1:6 (v/v) acyl chloride:NH4OH, and the reaction was allowed to proceed until the precipitation of the PFAM visibly ceased. Excess NH4OH was removed from the PFAM crystals by washing with H2O.
For most of the desired PFAMs, the required acyl chloride was available commercially. Unavailable acyl chlorides were synthesized by mixing the free fatty acid and thionyl chloride in a 1:2 molar ratio under N2 (with stirring), and then allowing the reaction to proceed under reflux at 50°C for 30 min. Unreacted thionyl chloride was removed by heating or by reduced pressure until gas evolution ceased.
N-Tridecanoylethanolamine (TDEA) was synthesized by combining triethylamine, acetonitrile, and ethanolamine in a 1.5:1:1 (v/v/v) ratio under N2 at room temperature. Tridecanoyl chloride was dissolved in a minimal volume of acetonitrile, added drop-wise to the ethanolamine mixture, and left to stir overnight at room temperature. The resulting solution was taken to dryness in a vacuum, yielding a yellow solid. The yellow solid was dissolved in a small volume of warm ethanol and recrystalized from cold H2O. After crystallization, the compound was washed with H2O.
Use of BSA as a fatty acid carrier
BSA was used as a carrier for fatty acids in aqueous media. Fatty acids were dissolved in ethanol, converted to the sodium salt with excess NaOH, dried under vacuum, and then dissolved in phosphate-buffered saline (PBS) to make a 25 mM solution. The sample was then heated in 49°C water bath for 5 min before adding 2.5 mM BSA (fatty acid free). The sample was stirred at room temperature for 4 h, sterile filtered, and stored at −20°C. A TDEA solution was made similarly, but an additional sonication step was added prior to addition of BSA.
Cell culture
All cells were grown in 225 cm2 culture dishes. The N18TG2 cells were grown in DMEM supplemented with 100 μM 6-thioguanine. The SCP cells were grown in EMEM supplemented with 100 µM sodium pyruvate. SCP and N18TG2 media had 10% FBS with 100 IU/ml penicillin and 1.0 mg/ml streptomycin. They were incubated at 37°C with 5% CO2 according to instructions from the supplier. Cultures were grown to 80–90% confluency, and then the culture media were removed and replaced with fresh media containing 0.5% FBS and a 2.5 mM fatty acid/0.25 mM BSA mixture. After 12, 24, or 48 h incubation, media was collected, the cells washed with PBS, and then detached from the flask by trypsinization. An aliquot was taken for counting on a hemacytometer with trypan blue. Cells were centrifuged and stored at −80°C. Conditioned medium was centrifuged to remove any cells, and the supernatant was stored at −80°C. The zero (0) time samples were collected without addition of the fatty acid-BSA mixture or 0.5% FBS medium, but they were collected in 10% FBS medium under normal growing conditions. A set of control experiments was carried out using cells grown in 0.5% FBS media.
Organic solvent extraction of the lipid metabolites
Metabolites were extracted from cells using procedures similar to those described by Sultana and Johnson (50). Methanol (4 ml) was added to the cell pellets, and the resulting mixture was sonicated for 15 min at room temperature. The samples were the centrifuged to remove cellular debris, and the supernatant was dried under N2 in a warm water bath at 40°C. The pellet was reextracted with 4 ml 1:1:0.1 (v/v/v) chloroform:methanol:water, sonicated for 10 min, vortexed for 2 min, and centrifuged. This second supernatant was added to the dried supernatant from the first extraction, and mixture was dried under N2 at 40°C. The pellet was extracted for a third time with 4.8 ml of chloroform:methanol 2:1 (v/v) and 800 µl of 0.5 M KCl/0.08 M H3PO4, sonicated for 2 min, vortexed for 2 min, and centrifuged to separate particulates from the liquid phases. The lower lipid phase from this centrifugation step was added to the dried combination of first and second supernatants, and this mixture taken to dryness under N2 at 40°C.
Lipid metabolites were extracted from the conditioned media using two 15 ml washes of chloroform:methanol 2:1 (v/v) (total extraction volume = 30 ml for this step), followed by two washes of a mixture consisting of 15 ml of chloroform:methanol 2:1 (v/v) plus 2.4 ml of 0.5 M KCl/0.08 M H3PO4 (total extraction volume = 34.8 ml for this step). Precipitated protein was condensed between the aqueous and organic layers by centrifugation. The lipid-containing organic phases were combined and dried under N2 at 40–50°C or in a vacuum.
Solid phase extraction of the lipid metabolites
Silica columns (0.5 g) were washed with n-hexane and run essentially as described by Sultana and Johnson (50). The dried lipid extracts, prepared as described above, were redissolved in 100 µl n-hexane and applied to a washed silica column. The mobile phase was run without positive pressure as follows: 4 ml of n-hexane, 1 ml of 99:1 (v/v) hexane:acetic acid, 1 ml of 90:10 (v/v) hexane:ethyl acetate, 1 ml of 80:20 (v/v) hexane:ethyl acetate, 1 ml of 70:30 (v/v) hexane:ethyl acetate, 1.5 ml of 2:1 (v/v) chloroform:isopropanol, and lastly, 0.5 ml of methanol. The eluant from each different organic wash was collected separately. The eluants from the chloroform:isopropanol wash and the methanol wash were combined and taken to dryness under N2 at 40–50°C. An internal standard, 3 nmol of [2H33]heptadecanoic acid, was added, and the mixture dried before derivitization.
To test effectiveness of the extraction methods, cells and media were spiked with each PFAM and subjected to the organic solvent extraction and solid phase extraction procedures before GC-MS quantification. Percentage recoveries ranged from 82% to 101%.
Gas chromatography-mass spectrometry
All separations were performed using a Shimadzu QP-5000 GC-MS. Separations were achieved on a J&W Scientific DB-5 column (0.25 mm × 30 m) in splitless mode. The GC temperature program was 55–150°C at 40°C/min, hold at 150°C for 3.6 min, ramp at 10°C/min to 300°C, and hold for 1 min. The transfer line was held at 280°C and the injection port at 250°C throughout the separation. Helium was used as a carrier gas at a flow rate of 0.9 ml/min. The mass range detected was 35–450 amu with a scan speed of 2,000. The solvent cut time was set to 7 min.
Before injection, dried samples were trimethylsilylated with 100 µl BSTFA [N,O-bis(trimethylsilyl)trifluoroacetamide]. Samples were flushed with dry N2, BSTFA added, flushed briefly again, and allowed to react at 55–60°C for 1 h before GC-MS analysis. The major product of PFAM derivitization was the corresponding acyl nitrile, and the PFAM-TMS was also observed. Samples were injected twice: 33.3% of the sample volume, followed by 50% of the remaining sample volume, so that 1 nmol internal sample was in each sample run. After analyzing a set of experimental samples by GC-MS (0, 12, 24, and 48 h time intervals, each injected twice), one of the 24 or 48 h samples was dried down, spiked with the PFAM of interest, redried, rederivitized with BSTFA, and analyzed again by GC-MS to validate PFAM and nitrile retention times.
Some PFAMs, particularly oleamide and erucamide, are used as slip additives in polyethylenes (51–53). Thus, background PFAM levels must be determined due to potential slip additive contamination. Background PFAM levels were measured by running solvents through the same glassware and plasticware that were used for the cell and media samples, and the collected solvents were subjected to sonication, solid phase extraction, and derivitization. Integration of the GC trace was taken over the same time integral as standard amides and for the same set of ions. An averaged number of cells were used to calculate blank amount of amide per cell in the blank samples. Unconditioned media were also subjected to extraction and SPE to assay for background amides.
In addition to unconditioned media and glass/plasticware, conditioned media and unconditioned media were incubated with various metabolites and tested to see if PFAMs would accumulate over time in the absence of cells. Unconditioned media were incubated with either oleamide, N-oleoylglycine, or N-oleoylethanolamine at 37°C for 48 h. Conditioned media were sterile-filtered with a 0.45 µm filter to remove any cells after the 48 h incubation with desired cells. The conditioned media were then incubated with oleamide, N-oleoylglycine, or N-oleoylethanolamine in BSA at 37°C for an additional 48 h. Oleamide extraction was performed as described above.
Finally, to test for the change in oleamide production, SCP and N18TG2 cells were incubated for 48 h with media containing 0.5% FBS and 0.25 mM BSA carrier but no long chain fatty acyl metabolite. Cells and media were both collected and subjected to PFAM analysis as described above.
Data analysis
Total ion chromatograms (TIC) were collected, and postrun, a set of selected ions unique to the amides and nitriles under examination was overlaid and integrated so that effects of any coeluting compounds could be minimized. These unique ion sets are based on electron-impact (EI) fragmentation of the PFAMs and nitrile standards and can be found in Table 1. Due to the high background in the range of 17.5–20 min in the linoleic acid-incubated media, the amount of amide was calculated using the nitrile only, plotted against a standard curve of linoleonitrile from the linoleamide derivitization reaction. Similarly, endogenous oleic acid and octadecanoic acid coeluted with palmitamide-TMS. This potentially interfering artifact was circumvented by intergrating palmitonitrile only, comparing these data against a standard curve of palmitonitrile. Note that no palmitonitrile was found in cells grown in the presence of exogenously added oleic acid, so palmitamide-TMS was not found in the oleamide spectral and could not interfere with the quantification of oleamide. Representative chromatograms of the endogenous oleic acid from the N18TG2 cells (supplementary Fig. I) and SCP cells (supplementary Fig. II) are provided to clarify the separation and specificity.
TABLE 1.
Selected ions for integration of amides and nitriles
| Compound | Selected m/z Values | Retention Time (min) |
| [ H33]Heptadecanoic acid | 119, 135, 149, 360, 375 | 15.1 |
| Tridecanonitrile | 82, 97, 110, 124, 138, 147, 152, 166, 170, 188, 192, 195 | 9 |
| Tridecanamide-TMS | 93, 100, 128, 131, 158, 170, 186, 200, 213, 270, 285 | 13.4 |
| Palmitoleonitrile | 122, 136, 150, 164, 178, 192, 206, 235 | 12.66 |
| Palmitoleamide-TMS | 59, 116, 128, 131, 144, 184, 198, 200, 240, 253, 310, 325 | 16.07 |
| Palmitonitrilea | 69, 110, 124, 138, 152, 166, 180, 194, 208, 237 | 12.9 |
| Linoleonitrileb | 95, 109, 120, 134, 148, 162, 176, 261 | 14.8 |
| Linoleamide-TMS | 59, 67, 81, 91, 95, 109, 116, 119, 121, 128, 131, 135, 144, 147, 149, 336, 352, 279 | 15.98 |
| Oleo/elaido-nitrile | 83, 97, 110, 122, 136, 150, 164, 190, 206, 220, 234, 263 | 14.9 |
| Ole/elaid-amide-TMS | 59, 86, 112, 116, 122, 126, 128, 131, 136, 140, 144, 154, 158, 170, 184, 186, 198, 200, 226, 238, 264, 281, 338, 353 | 16.05 |
Palmitamide-TMS was not integrated due to interference of relatively high amounts of oleic acid-TMS and octadecanoic acid-TMS that interfered with the selected ion integration. Amounts of palmitamide were determined based only on palmitonitrile, using standard curves of palmitonitrile only.
Linoleamide-TMS was not integrated for a similar reason as for palmitamide-TMS, and only linoleonitrile was integrated, using standard curves of linoleonitrile only.
[2H33]Heptadecanoic acid was spiked into each sample as an internal standard to measure the instrument performance. The amides and nitriles of interest were integrated along with the [2H33]heptadecanoic acid, both were compared with their standard curves, and a correction factor was determined based on the integration of [2H33]heptadecanoic acid. Each extraction sample was run on the GC-MS twice, and each incubation sample of FFA or NAE with SCP and N18TG2 cells was run at least twice.
Western analysis
SDS polyacrylamide gel electrophoresis was run according to the method of Laemmli (54). Transfer from the SDS gels onto a PVDF membrane was carried out at 80 V for 60 min.
To ensure specificity, controls were run with the FAAH antibody preincubated with its blocking peptide prior to Western blot analysis. Antibodies were incubated with a 50-fold molar excess of the blocking peptide for 2.5 h at 36°C with agitation and this was followed by a 2–24 h incubation at 4°C in 500 ml of PBS. The antibodies were then used directly from the PBS solution.
RT-PCR
Cell samples (N18TG2 and SCP) were isolated from culture (one T-75 cm2 flask, grown to ∼90% of confluency) and lysed. mRNA was isolated using the MicroPoly(A) Pure mRNA purification kit from Ambion. cDNA was generated from the isolated mRNA using the RETROscript reverse transcription kit from Ambion. PCR conditions were as follows: an initial 3 min denaturation at 95°C followed by 25–45 cycles of denaturation at 95°C (1 min), annealing at 45°C (1 min), and elongation at 72°C (1 min), with a final elongation cycle of 7 min at 72°C. The PCR product was gel purified using a QIAQuick gel extraction kit (Qiagen), and the product was sequenced to verify accuracy.
RESULTS
Analysis of background PFAMs
Endogenous amounts of the PFAMs were measured to provide a baseline reference level (time = 0), and are shown in Table 2. The levels of linoleamide, oleamide, palmitamide, palmitoleamide, and tridecanamide isolated from unconditioned media generally represent a small percentage of those isolated from conditioned media. The PFAM levels in the unconditioned media blanks were 1–6% of those found in media conditioned by the SCP cells and 1–22% of those found in media conditioned by the N18G2 cells. Linoleamide was an exception, at 44% of that measured from the N18TG2 cell conditioned media.
TABLE 2.
Endogenous PFAM levels in the N18TG2 and SCP cells
| PFAM | N18TG2 Cells | SCP Cells |
| pmoles per10 cells7 | pmoles per10 cells7 | |
| Palmitoleamide | 340 ± 110 | 410 ± 50 |
| Palmitamide | 930 ± 250 | 1200 ± 210 |
| Oleamide | 120 ± 50 | 6400 ± 2300 |
| Linoleamide | 100 ± 60 | 250 ± 180 |
PFAM amounts are reported as the average ± SD for three determinations.
We also measured PFAM levels in solvents, glassware, and plasticware used in the extractions to measure background amounts from slip additives (51–53). PFAM levels in the solvent blanks were low, ranging between 0.5% and 8% of the amounts isolated from the SCP or N18TG2 cells. Tridecanamide was an exception, as the blank value was 18% of that identified from the N18TG2 cells postincubation.
We divided the background PFAM levels by the average number of viable cells per experiment (with all extraction volumes the same) to enable a direct comparison of the background amounts to those we measured in the cells and the cell-conditioned media (Table 3). For example, 90 pmoles of palmitamide divided by the average number of viable N18TG2 cells, 7.9 × 107, at the start of our incubation yields an average background palmitamide level of 12 pmoles per 107 N18TG2 cells (Table 3). This background level of palmitamide is ∼1% of the endogenous level of palmitamide from the N18TG2 cells, 930 pmoles per 107 N18TG2 cells.
TABLE 3.
Background PFAM levels from the solvents and plasticware
| PFAM | Background Amount for N18TG2 Cells | Background Amount for SCP Cells |
| pmoles per10 cells7 | pmoles per10 cells7 | |
| Palmitoleamide | 23 ± 7.0 | 140 ± 46 |
| Palmitamide | 12 ± 2.3 | 76 ± 15 |
| Oleamidea | 8.7 ± 4.1 | 55 ± 26 |
| Linoleamide | 14 ± 6.5 | 89 ± 41 |
| Tridecanamide | 17 ± 19 | 410 ± 450 |
To facilitate comparison of these data to the endogenous PFAM levels in the N18TG2 and SCP cells and to the cellular production data, the moles of amide found in these blanks was divided by an average number of viable cells (1.24 × 107 for SCP or 7.9 × 107 for N18TG2). The PFAM amounts are reported as the average ± SD for four determinations.
Each oleamide value actually represents the sum of the background levels of oleamide and elaidamide. Background levels of elaidamide (trans-9-octadecenic acid) are most likely ∼0 (3).
The methods used to measure the background levels of the PFAMs could not discriminate oleamide (cis-9-octadecenic acid) from elaidamide (trans-9-octadecenic acid). We assumed that the background 18:1 amide reported here is oleamide because oleamide is a common slip additive (51,53).
Analysis of PFAMs produced by the N18TG2 cells
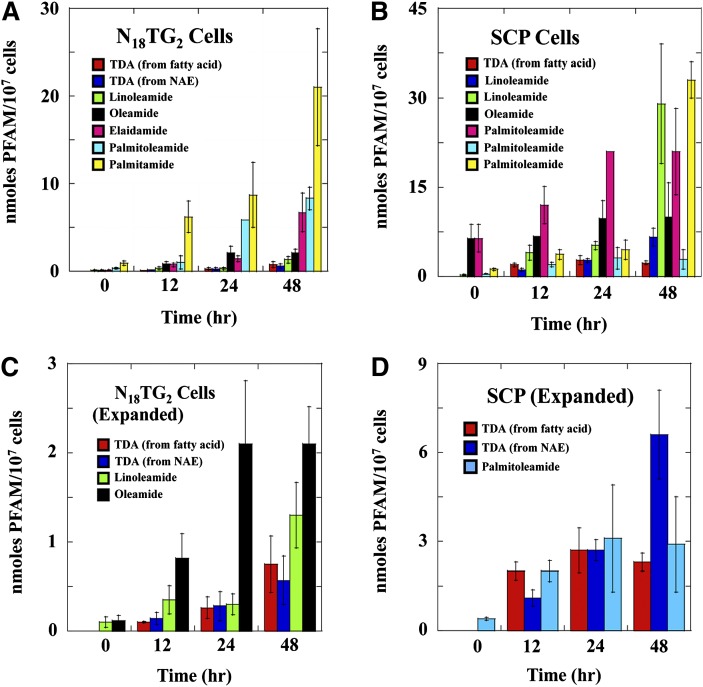
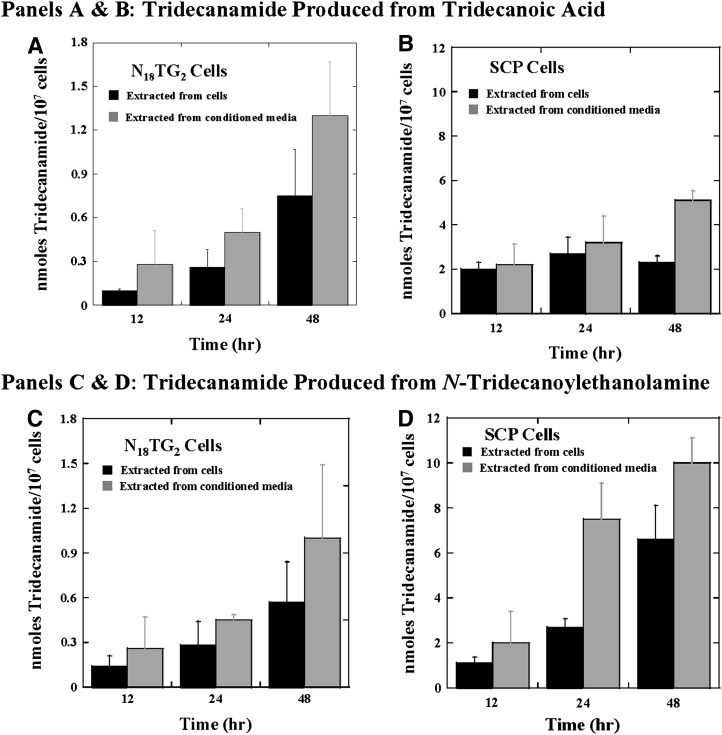
We first measured the amounts of linoleamide, palmitoleamide, palmitamide, and oleamide in the N18TG2 cells before growth in the presence of exogenously added fatty acids. The C18:1 PFAM identified endogenously is assumed to be oleamide rather than elaidamide. The reasons are that oleamide and not elaidamide has been identified in N18TG2 cells (36) and that trans-fatty acids are not produced in non-ruminant animals, but are derived from food (55). The concentrations of the endogenous PFAMs were relatively low, ranging from 100 ± 60 pmoles/107 cells for linoleamide to 930 ± 250 pmoles/107 cells for palmitamide in the N18TG2 cells (Table 2). Upon growth of the N18TG2 cells in the presence of exogenously added fatty acids (using BSA as a carrier), the corresponding PFAM was identified from both the cells and the N18TG2 cell-conditioned media (Table 4). Statistical analysis of the PFAM production data to define the robustness of the measured differences in the PFAM levels is included in supplementary Tables I and II. In most cases, the amount of PFAM found in the conditioned media was similar to the amount isolated from the washed cells. Incubation of the N18TG2 cells for 0, 12, 24, or 48 h in the presence of exogenously added fatty acids led to continued production of the corresponding PFAM (Table 4 and Fig. 2A, C). Tridecanamide was produced by the N18TG2 cells from two precursors added to the cell culture medium, tridecanoate and N-tridecanoyethanolamine (Table 4 and Fig. 3A, C). There is evidence that the NAEs can serve as precursors to the N-fatty acylglycines and the PFAMs (44–46); this is the first evidence that such chemistry can take place in the N18TG2 cells. Palmitamide was the most abundant PFAM after incubation of the N18TG2 cells with palmitic acid. Palmitoleamide was the next most abundant PFAM, followed by elaidamide, oleamide, linoleamide, and tridecanamide after 48 h incubation with the corresponding free fatty acid.
TABLE 4.
PFAMs in cells and media
| PFAM Origin | SCP Cells | SCP Media | N18TG2 Cells | N18TG2 Media |
| Tridecanoic acid | ||||
| 12 h | 2.0 ± 0.31 | 2.2 ± 0.92 | 0.10 ± 0.010 | 0.28 ± 0.23 |
| 24 h | 2.7 ± 0.75 | 3.2 ± 1.2 | 0.26 ± 0.12 | 0.50 ± 0.16 |
| 48 h | 2.3 ± 0.30 | 5.1 ± 0.43 | 0.75 ± 0.32 | 1.3 ± 0.37 |
| N-Tridecanoylethanolamine | ||||
| 12 h | 1.1 ± 0.28 | 2.0 ± 1.4 | 0.14 ± 0.068 | 0.26 ± 0.21 |
| 24 h | 2.7 ± 0.36 | 7.5 ± 1.6 | 0.28 ± 0.16 | 0.45 ± 0.037 |
| 48 h | 6.6 ± 1.5 | 10 ± 1.1 | 0.57 ± 0.27 | 1.0 ± 0.49 |
| Palmitoleic acid | ||||
| 12 h | 2.0 ± 0.36 | 3.8 ± 0.35 | 1.0 ± 0.77 | 1.9 ± 1.6 |
| 24 h | 3.1 ± 1.8 | 5.4 ± 2.1 | 5.8 | 8.5 |
| 48 h | 2.9 ± 1.6 | 7.9 ± 0.76 | 8.3 ± 1.3 | 15 |
| Palmitic acid | ||||
| 12 h | 3.7 ± 0.79 | 5.3 ± 1.4 | 6.2 ± 1.8 | 2.2 ± 1.0 |
| 24 h | 4.5 ± 1.6 | 6.4 ± 0.74 | 8.7 ± 3.7 | 21 ± 8.2 |
| 48 h | 33 ± 3.0 | 25 ± 8.4 | 21 ± 6.7 | 74 ± 27 |
| Oleic acid | ||||
| 12 h | 6.8 | 5.7 ± 3.7 | 0.82 ± 0.27 | 0.78 ± 0.32 |
| 24 h | 9.8 ± 2.9 | 17 ± 2.1 | 2.1 ± 0.71 | 1.4 ± 0.20 |
| 48 h | 10 ± 5.8 | 13 ± 3.9 | 2.1 ± 0.42 | 1.7 ± 0.13 |
| Elaidic acid | ||||
| 12 h | 12 ± 3.1 | 17 | 0.74 ± 0.20 | 1.7 ± 0.20 |
| 24 h | 21 | 51 ± 19 | 1.4 ± 0.32 | 1.9 ± 0.59 |
| 48 h | 21 ± 7.2 | 130 ± 19 | 6.7 ± 2.2 | 11 ± 0.74 |
| Linoleic acid | ||||
| 12 h | 4.0 ± 1.3 | 7.0 ± 1.7 | 0.35 ± 0.16 | 0.39 ± 0.044 |
| 24 h | 5.2 ± 0.73 | 26 ± 8.1 | 0.30 ± 0.12 | 0.41 ± 0.11 |
| 48 h | 29 ± 10 | 54 ± 30 | 1.3 ± 0.37 | 0.85 ± 0.27 |
Amounts are reported as nmoles/107 cells and are the average ± SD for two to three determinations. In a few instances, only a single determination was made. Blank values were subtracted from the amounts found in experimental samples prior to reporting: solvent blank for the cell samples and unconditioned media blank for the media samples. The PFAM amounts were measured after incubation of the indicated cells for 12, 24, or 48 h with media containing the corresponding free fatty acid.
Fig. 2.
PFAMs produced in N18TG2 and SCP cells incubated with the indicated fatty acids. Data here is only for the amount of the PFAMs extracted from the N18TG2 (A, C) and SCP (B, D) cells after the indicated incubation time. The data for the PFAMs from the cell-conditioned media is in Table 4. Panels C and D were included to better reveal the low levels of PFAM production in the N18TG2 and SCP cells.
Fig. 3.
Tridecanamide produced in N18TG2 (A) and SCP (B) cells from N-tridecanoylethanolamine. The amount of tridecamide found in cells (black) and excreted into media (gray) is shown. (A, B) The production of tridecanamide from tridecanoate in the N18TG2 cells and in the SCP cells. (C, D) The production of tridecanamide from N-tridecanoylethanolamine in the N18TG2 cells and the SCP cells.
Analysis of PFAMs produced by the SCP cells
Incubation of the SCP cells with each fatty acid led to the production of corresponding PFAM (Fig. 2B, D), including the nonnaturally occurring tridecanamide. In most cases, the amount of PFAM found in the conditioned media was similar to the amount of PFAM found in the cells themselves (Table 4), as was seen for N18TG2 cells. The amounts of amides found from the SCP cells and in the SCP-conditioned media after 0, 12, 24, or 48 h incubation with corresponding fatty acids can be found in Table 3 and Fig. 2B, D. Overall, higher levels of the PFAMs were identified from the SCP cells and SCP cell-conditioned media relative to those measured for the N18TG2 cells and N18TG2 cell-conditioned media. Note that oleamide was the most abundant PFAM produced by the SCP cells.
As we observed in the N18TG2 cells, we found that tridecanamide was produced by the SCP cells from both tridecanoate and N-tridecanoyethanolamine (Table 4 and Fig. 3B, D). This is the first report of the conversion of an NAE to a PFAM in the SCP cells, similar to what we found in the N18TG2 cells.
Controls for the time-dependent production of oleamide in the N18TG2 and SCP cells
To insure that the increasing amounts of the PFAMs we measured over time (Table 4 and Fig. 2) results from the cellular conversion of the exogenously added fatty acid to the PFAM, we carried out a set of control experiments to determine if the observed increases in oleamide versus time in the N18TG2 cells could be attributed to either a) reactions independent of the oleic acid added to the culture medium or b) reactions that occur in the cell culture medium alone.
To test for oleamide production in the cells grown in the absence of exogenously added oleic acid, both N18TG2 and SCP cells were incubated for 48 h with media containing 0.5% FBS and 0.25 mM BSA carrier alone. There was no significant difference between the amount of oleamide found at time point zero (Table 4) and the amount found in cells incubated for 48 h with BSA and low serum media only (data not shown).
A set of cell-free controls were run to test for the formation of PFAMs in media containing added oleic acid, N-oleoylglycine, or N-oleoylethanolamine. Mueller and Driscoll (56) have identified oleamide-synthesizing activity in FBS, a component of the culture media in used in our experiments. Mueller and Driscoll (56) reported that incubation of oleoyl-CoA and ammonia or glycine in the presence of FBS resulted in the formation of oleamide or N-oleoylglycine, respectively. To test for oleamide-forming activity in the media, cell-free unconditioned and conditioned media were incubated with oleic acid, N-oleoylglycine, or N-oleoylethanolamine for 48 h. We found no significant difference between the amount of oleamide present in the blank controls and media (both conditioned and unconditioned) exposed to oleoyl metabolites.
FAAH expression studies
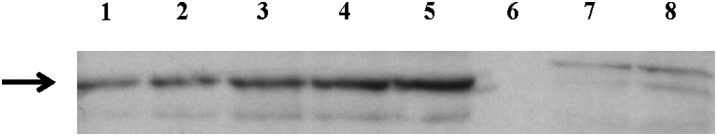
The PFAM levels produced in the SCP cells is generally higher than those found in N18TG2 cells (Table 4). One possible explanation for these results could be the lack of expression of FAAH in the SCP cells. FAAH is likely the main enzyme involved in the PFAM degradation in vivo (31, 32). Western analysis and RT-PCR were employed to interrogate FAAH expression in the SCP cells, N18TG2 cells, and human embryonic kidney (HEK-293) cells. The N18TG2 and HEK-293 cells serve as a controls, as both of these cell lines are known to express FAAH (48, 57). Both Western analysis (Fig. 4) and RT-PCR (data not included) show FAAH expression in the N18TG2 and HEK-293 cells, but we found no detectable FAAH expression in the SCP cells by either method.
Fig. 4.
FAAH expression via Western analysis. Lanes 1 and 2 are from the HEK-293 cells; lanes 3–5 are from the N18TG2 cells; lane 6 is an empty lane; and lanes 7 and 8 are from the SCP cells.
DISCUSSION
Our study is the first report of the conversion of free fatty acids (other than oleic acid) to their respective PFAMs in N18TG2 cells, and it is the first finding of the other endogenous PFAMs in N18TG2 cells. Although oleamide is the best-studied PFAM, it is only one of many known mammalian PFAMs (3). In addition, this is the first report of PFAMs found both endogenously and upon incubation with free fatty acids in SCP cells. Our controls show that the PFAMs produced by the N18TG2 and SCP cells cannot be attributed to contamination or cell-free production by reactions in the culture media.
There are a few discernible patterns when comparing the PFAM quantification data from or between the two cell lines. In general, the levels of the endogenous PFAMs are higher in the SCP cells. This is most evident for oleamide, for which the endogenous level in the SCP cells is ∼50-fold higher than that in the N18TG2 cells (6,400 pmoles per 107 cells versus 120 pmoles per 107 cells; see Table 2). The endogenous PFAM levels reflect a steady-state balance between production, degradation, and secretion, and they are likely influenced by the fatty acids and other fatty acid-containing moieties of the growth media.
As was observed with the endogenous PFAM levels, there are a few obvious patterns in comparing PFAM production data (in cells and excreted into the media) in the two cell lines. Similar to the endogenous PFAM levels, total PFAM production is generally higher for the SCP cells, ranging from ∼4-fold higher for tridecanamide to ∼40-fold higher for linoleamide. Palmitoleamide and palmitamide are exceptions, with ∼2-fold higher amounts produced by the N18TG2 cells relative to the SCP cells. In comparing the amounts of the specific PFAMs that were produced, the elaidamide levels were consistently higher than the oleamide levels, and tridecanamide was always the lowest in the two cell lines (whether produced from the tridecanoate or N-tridecanoylethanolamine). One likely reason for the higher levels of total PFAM production in the SCP cells is the lack of detectable FAAH expression in these cells (Fig. 4 and Ref. 58). These data and higher endogenous PFAM levels in the SCP cells (Table 2) are consistent with published results showing that FAAH is the major enzyme involved in PFAM degradation (31, 32). In fact, the relative endogenous levels of the PFAMs and the steady-state PFAM levels at 48 h in the N18TG2 cells are a reasonable match for the substrate preferences for FAAH (59–62); the PFAMs with a relatively high VMAX/KM value are found in lower amounts, suggesting the FAAH is the dominant PFAM degradative enzyme in these cells.
Because elaidamide is a nonnatural PFAM, it might be a relatively poor substrate for FAAH and other pathways for PFAM degradation. This could explain the high abundance of elaidamide relative to oleamide after incubation with the corresponding fatty acid. Similarly, tridecanoate is a nonnatural fatty acid and might be a relatively poor substrate for the enzyme(s) of PFAM biosynthesis, accounting for the relatively low abundance of tridecanamide. In vitro data on the PAM-catalyzed oxidation of N-fatty acylglycines to the corresponding PFAMs show a preference for longer acyl chain substrates (34, 35). The higher PFAM48/PFAM12 ratio in the N18TG2 cells relative to the SCP cells reflects a greater net rate of PFAM biosynthesis in the N18TG2 cells, a lower net rate of PFAM degradation in the N18TG2 cells, or a combination of both. Given the current uncertainties about the pathways for biosynthesis and degradation of the PFAMs and the transport mechanism into and out of cells, definitive conclusions about the relative abundances of the PFAMs in the N18TG2 and SCP cells are not possible. The data presented here provide key information to evaluate models of PFAM flux as the PFAM biosynthetic and degradative pathways are better defined.
In addition to testing for the conversion of endogenously found fatty acids to their respective primary amides, tridecanoate, a nonnatural fatty acid, and its corresponding NAE were also incubated with the cells to test for their conversion to a PFAM. These experiments were to demonstrate that a model fatty acid with a nonnaturally occurring acyl chain could serve as a substrate for PFAM biosynthesis, to determine whether NAEs could also serve as PFAM substrates in these cells, and to compare the flux of an NAE and its corresponding FFA to PFAM. In vitro data and data from other cells have suggested that NAEs are precursors to the PFAMs (44–46, 63). Our hypothesis was that the nonnaturally occurring N-tridecanoylethanolamine would be a relatively poor FAAH substrate and, thus, result in greater accumulation of tridecanamide. We identified and quantified tridecanamide in the model cell lines after incubation with N-tridecanoylethanolamine (Fig. 3). This is the first report of the conversion of an NAE to the corresponding PFAM in the N18TG2 and SCP cells, providing additional evidence for the NAEs serving as precursors to the PFAMs. The levels of tridecanamide produced from N-tridecanoylethanolamine were higher than those produced by tridecanoate in the SCP cells. The differences in overall production of tridecanamide between the SCP and N18TG2 cells could result from differences in the conversion rates of tridecanoate into phospholipids between the two cells. Nonetheless, it is intriguing that the SCP cells, which do not express FAAH (Fig. 4), produce a higher level of tridecanamide, suggesting that FAAH plays a major role in the cellular degradation of the NAEs.
In conclusion, five endogenous PFAMs were found in N18TG2 and SCP cells, all of which are new findings (except for oleamide in N18TG2). The amount of endogenous PFAMs, particularly oleamide, was much higher in the SCP cells. These data indicate the focus on oleamide within the field of primary fatty acid amides can and should be broadened to other members of this class of fatty acid amides. After incubation with the corresponding fatty acid, PFAMs could be found in both cells and conditioned media, and the profile of PFAM levels was different between the two cell lines. The data presented here provide strong support for the role of FAAH as the major enzyme of PFAM degradation in vivo, demonstrate that the N18TG2 cells are a good model system for the study of PFAM metabolism, show that SCP cells are an important FAAH-independent model for the study of PFAM metabolism, support a role for the NAEs as PFAM precursors, and provide useful kinetic results for modeling the metabolic flux through the pathways of PFAM biosynthesis and degradation.
Supplementary Material
Acknowledgments
The authors would like to thank Milena Ivkovic for synthesizing the N-tridecanoylethanolamine, Lamar Galloway for technical assistance, and Jeremy Gibson for assistance with the statistical analysis of our data.
Footnotes
Abbreviations:
- CSF
- cerebrospinal fluid
- FAAH
- fatty acid amide hydrolase
- NAE
- N-acylethanolamine
- PAM
- peptidylglycine α-amidating monooxygenase
- PFAM
- primary fatty acid amide
- SCP
- sheep choroid plexus
This work was supported in part by National Institutes of Health General Medical Sciences Grant R15-GM-059050; Florida Center for Excellence for Biomolecular Identification and Targeted Therapeutics (FCoE-BITT) Grant GALS020; the Shirley W. and William L. Griffin Foundation, the Gustavus and Louise Pfeiffer Research Foundation, the Wendy Will Case Cancer Fund, the Milheim Foundation for Cancer Research, the Eppley Foundation for Research, and the Alpha Research Foundation, Inc. (D.J.M.); a Graduate Multidisciplinary Scholars (GMS) Award (E.K.F.) through the USF Thrust Life Sciences Program administered by the Florida Center of Excellence for Biomolecular Identification and Targeted Therapeutics. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and two tables.
References
- 1.McIlwain H. 1958. Thudichum and the medical chemistry of the 1860s to 1880s. Proc. R. Soc. Med. 51: 127–132. [PMC free article] [PubMed] [Google Scholar]
- 2.Levene P. A. 1916. Sphingomyelin. III. J. Biol. Chem. 24: 69–89. [Google Scholar]
- 3.Arafat E. S., Trimble J. W., Andersen R. N., Dass C., Desiderio D. M. 1989. Identification of fatty acid amides in human plasma. Life Sci. 45: 1679–1687. [DOI] [PubMed] [Google Scholar]
- 4.Cravatt B. F., Prospero-Garcia O., Siuzdak G., Gilula N. B., Henriksen S. J., Boger D. L., Lerner R. A. 1995. Chemical characterization of a family of brain lipids that induce sleep. Science. 268: 1506–1509. [DOI] [PubMed] [Google Scholar]
- 5.Stewart J. M., Boudreau N. M., Blakely J. A., Storey K. B. 2002. A comparison of oleamide in the brains of hibernating and non-hibernating Richard's ground squirrel (Spermophilus richardsonii) and its inability to bind to brain fatty acid binding protein. J. Therm. Biol. 27: 309–315. [Google Scholar]
- 6.Guan X., Cravatt B. F., Ehring C. R., Hall J. E., Boger D. L., Lerner R. A., Gilula N. B. 1997. The sleep-inducing lipid oleamide deconvolutes gap junction communication and calcium wave transmission in glial cells. J. Cell Biol. 139: 1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boger D. L., Patterson J. E., Guan X., Cravatt B. F., Lerner R. A., Gilula N. B. 1998. Chemical requirements for inhibition of gap junction communication by the biologically active lipid oleamide. Proc. Natl. Acad. Sci. USA. 95: 4810–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boitano S., Evans W. H. 2000. Connexin mimetic peptides reversibly inhibit Ca2+ signaling through gap junctions in airway cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 279: L623–L630. [DOI] [PubMed] [Google Scholar]
- 9.Decrouy X., Gasc J. M., Pointis G., Segretain D. 2004. Functional characterization of Cx43 based gap junctions during spermatogenesis. J. Cell. Physiol. 200: 146–154. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich H. P., Sun B., Saggers G. C., Kromath F. 2006. Gap junction commications influence upon fibroblast synthesis of type I collagen and fibronectin. J. Cell. Biochem. 98: 735–743. [DOI] [PubMed] [Google Scholar]
- 11.Coyne L., Lees G., Nicholson R. A., Zheng J., Neufield K. D. 2002. The sleep hormone oleamide modulates inhibitory ionotropic receptors in mammalian CNS in vitro. Br. J. Pharmacol. 135: 1977–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huidobro-Toro J. P., Harris R. A. 1996. Brain lipids that induce sleep are novel modulators of 5-hydroxytryptamine receptors. Proc. Natl. Acad. Sci. USA. 93: 8078–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedlund P. B., Carson M. J., Sutcliffe J. G., Thomas E. A. 1999. Allosteric regulation by oleamide of the binding properties of 5-hydroxytryptamine7 receptors. Biochem. Pharmacol. 58: 1807–1813. [DOI] [PubMed] [Google Scholar]
- 14.Varvel S. A., Cravatt B. F., Engram A. E., Lichtman A. H. 2006. Fatty acid amide hydrolase(-/-) mice exhibit an increased sensitivity to the disruptive effects of anandamide or oleamide in a working memory water maze task. J. Pharmacol. Exp. Ther. 317: 251–257. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-González D., Bonilla-Jaime H., Morales-Otal A., Henriksen S. J., Velázquez-Moctezuma J., Prospéro-García O. 2004. Oleamide and anandamide effects on food intake and sexual behavior of rats. Neurosci. Lett. 364: 1–6. [DOI] [PubMed] [Google Scholar]
- 16.Akanmu M. A., Adesun S. O., Ilesanmi O. R. 2007. Neuropharmacological effects of oleamide in male and female mice. Behav. Brain Res. 182: 88–94. [DOI] [PubMed] [Google Scholar]
- 17.Fedorova I., Hashimoto A., Fecik R. A., Hedrick M. P., Hanuš L. O., Boger D. L., Rice K. C., Basile A. S. 2001. Behavioral evidence for the interaction of oleamide with multiple neurotransmitter systems. J. Pharmacol. Exp. Ther. 299: 332–342. [PubMed] [Google Scholar]
- 18.Huitrón-Reséndiz S., Gombart L., Cravatt B. F., Henriksen S. J. 2001. Effect of oleamide on sleep and its relationship to blood pressure, body temperature, and locomotor activity in rats. Exp. Neurol. 172: 235–243. [DOI] [PubMed] [Google Scholar]
- 19.Lo Y. K., Tang K. Y., Chang W. N., Lu C. H., Cheng J. S., Lee K. C., Chou K. J., Liu C. P., Chen W. C., Su W., et al. 2001. Effect of oleamide on Ca2+ signaling in human bladder cancer cells. Biochem. Pharmacol. 62: 1363–1369. [DOI] [PubMed] [Google Scholar]
- 20.Hoi P. M., Hiley C. R. 2006. Vasorelaxant effects of oleamide in rat small mesenteric artery indicate action at a novel cannabinoid receptor. Br. J. Pharmacol. 147: 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudhahar V., Shaw S., Imig J. D. 2009. Mechanisms involved in oleamide-induced vasorelaxation in rat mesenteric resistance arteries. Eur. J. Pharmacol. 607: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert D. M., Vandevoorde S., Diependaele G., Govaerts S. J., Robert A. R. 2001. Anticonvulsant activity of N-palmitoylethanolamide, a putative endocannabinoid, in mice. Epilepsia. 42: 321–327. [DOI] [PubMed] [Google Scholar]
- 23.Huang J. K., Jan C. R. 2001. Linoleamide, a brain lipid that induces sleep, increases cytosolic Ca2+ levels in MDCK renal tubular cells. Life Sci. 68: 997–1004. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y. C., Wu S. N. 2003. Block of erg current by linoleoylamide, a sleep-inducing agent, in pituitary GH3 cells. Eur. J. Pharmacol. 458: 37–47. [DOI] [PubMed] [Google Scholar]
- 25.Wakamatsu K., Masaki T., Itoh F., Kondo K., Sudo K. 1990. Isolation of fatty acid amide as an angiogenic principle from bovine mesentery. Biochem. Biophys. Res. Commun. 168: 423–429. [DOI] [PubMed] [Google Scholar]
- 26.Hamberger A., Stenhagen G. 2003. Erucamide as a modulator of water balance: new function of a fatty acid amide. Neurochem. Res. 28: 177–185. [DOI] [PubMed] [Google Scholar]
- 27.Jain M. K., Ghomashchi F., Yu B. Z., Bayburt T., Murphy D., Houck D., Brownell J., Reid J. C., Solowiej J. E., Wong S. M., et al. 1992. Fatty acid amides: scooting mode-based discovery of tight-binding competitive inhibitors of secreted phospholipases A2. J. Med. Chem. 35: 3584–3586. [DOI] [PubMed] [Google Scholar]
- 28.Morisseau C., Newman J. W., Dowdy D. L., Goodrow M. H., Hammock B. D. 2001. Inhibition of microsomal epoxide hydrolases by ureas, amides, and amines. Chem. Res. Toxicol. 14: 409–415. [DOI] [PubMed] [Google Scholar]
- 29.Farrell E. K., Merkler D. J. 2008. Biosynthesis, degradation, and pharmacological importance of the fatty acid amides. Drug Discov. Today. 13: 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezzili C., Otrubova K., Boger D. L. 2010. Fatty acid amide signaling molecules. Bioorg. Med. Chem. Lett. 20: 5959–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cravatt B. F., Lichtman A. H. 2002. The enzymatic inactivation of the fatty acid amide class of signaling lipids. Chem. Phys. Lipids. 121: 135–148. [DOI] [PubMed] [Google Scholar]
- 32.McKinney M. K., Cravatt B. F. 2005. Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 74: 411–432. [DOI] [PubMed] [Google Scholar]
- 33.Driscoll W. J., Chaturvedi S., Mueller G. P. 2007. Oleamide synthesizing activity from rat kidney. Identification as cytochrome c. J. Biol. Chem. 282: 22353–22363. [DOI] [PubMed] [Google Scholar]
- 34.Merkler D. J., Merkler K. A., Stern W., Fleming F. F. 1996. Fatty acid amide biosynthesis: a possible new role for peptidylglycine α-amidating enzyme and acyl-coenzyme A:glycine N-acyltransferase. Arch. Biochem. Biophys. 330: 430–434. [DOI] [PubMed] [Google Scholar]
- 35.Wilcox B. J., Ritenour-Rogers K. J., Asser A. S., Baumgart L. E., Baumgart M. A., Boger D. L., DeBlassio J. E., deLong M. A., Glufke U., Henz M. E., et al. 1999. N-Acylglycine amidation: implications for the biosynthesis of fatty acid primary amides. Biochemistry. 38: 3235–3245. [DOI] [PubMed] [Google Scholar]
- 36.Bisogno T., Sepe N., De Petrocellis L., Mechoulam R., Di Marzo V. 1997. The sleep inducing factor oleamide is produced by mouse neuroblastoma cells. Biochem. Biophys. Res. Commun. 239: 473–479. [DOI] [PubMed] [Google Scholar]
- 37.Merkler D. J., Chew G. H., Gee A. J., Merkler K. A., Sorondo J. P., Johnson M. E. 2004. Oleic acid derived metabolites in mouse neuroblastoma N18TG2 cells. Biochemistry. 43: 12667–12674. [DOI] [PubMed] [Google Scholar]
- 38.Mains R. E., Park L. P., Eipper B. A. 1986. Inhibition of peptide amidation by disulfiram and diethyldithiocarbamate. J. Biol. Chem. 261: 11938–11941. [PubMed] [Google Scholar]
- 39.Bradshaw H. B., Rimmerman N., Hu S. S., Burstein S., Walker J. M. 2009. Novel endogenous N-acyl glycines: identification and characterization. Vitam. Horm. 81: 191–205. [DOI] [PubMed] [Google Scholar]
- 40.Chaturvedi S., Driscoll W. J., Elliot B. M., Farady M. M., Grunberg N. E., Mueller G. P. 2006. In vivo evidence that N-oleoylglycine acts independently of its conversion to oleamide. Prostaglandins Other Lipid Mediat. 81: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimmerman N., Bradshaw H. B., Hughes H. V., Chen J. S., Hu S. S., McHugh D., Vefring E., Jahnsen J. A., Thompson E. L., Masuda K., et al. 2008. N-Palmitoyl glycine, a novel endogenous lipid that acts as a modulator of calcium influx and nitric oxide production in sensory neurons. Mol. Pharmacol. 74: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHugh D., Hu S. S., Rimmerman N., Juknat A., Vogel Z., Walker J. M., Bradshaw H. B. 2010. N-Arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waluk D. P., Schultz N., Hunt M. C. 2010. Identification of glycine N-acyltranferase-like 2 (GLYATL2) as a transferase that produces N-acyl glycines in humans. FASEB J. 24: 2795–2803. [DOI] [PubMed] [Google Scholar]
- 44.Aneetha H., O'Dell D. K., Tan B., Walker J. M., Hurley T. D. 2009. Alcohol dehydrogenase-catalyzed in vitro oxidation of anandamide to N-arachidonoyl glycine, a lipid mediator: synthesis of N-acyl glycinals. Bioorg. Med. Chem. Lett. 19: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivkovic M., Dempsey D. R., Handa S., Hilton J. H., Lowe E. W., Jr, Merkler D. J. 2011. N-Acylethanolamines as a novel alcohol dehydrogenase 3 substrates. Arch. Biochem. Biophys. 506: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradshaw H. B., Rimmerman N., Hu S. S., Benton V. M., Stuart J. M., Masuda K., Cravatt B. F., O'Dell D. K., Walker J. M. 2009. The endocannabinoid anandamide is a precursor for the signaling lipid N-arachidonoyl glycine by two distinct pathways. BMC Biochem. 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritenour-Rodgers K. J., Driscoll W. J., Merkler K. A., Merkler D. J., Mueller G. P. 2000. Induction of peptidylglycine α-amidating monooxygenase in N18TG2 cells: a model for studying oleamide biosynthesis. Biochem. Biophys. Res. Commun. 267: 521–526. [DOI] [PubMed] [Google Scholar]
- 48.Bisogno T., Melck D., De Petrocellis L., Bobrov M. Y., Gretskaya N. M., Bezuglov V. V., Sitachitta N., Gerwick W. H., Di Marzo V. 1998. Arachidonoylserotonin and other novel inhibitors of fatty acid amide hydrolase. Biochem. Biophys. Res. Commun. 248: 515–522. [DOI] [PubMed] [Google Scholar]
- 49.Fong C., Wells D., Krodkiewska I., Hartley P. G., Drummond C. J. 2006. New role for urea as a surfactant headgroup promoting self-assembly in water. Chem. Mater. 18: 594–597. [Google Scholar]
- 50.Sultana T., Johnson M. E. 2006. Sample preparation and gas chromatography of primary fatty acid amides. J. Chromatogr. A. 1101: 278–285. [DOI] [PubMed] [Google Scholar]
- 51.Watson J., Greenough E. B., Leet J. E., Ford M. J., Drexler D. M., Belcastro J. V., Herbst J. J., Chatterjee M., Banks M. 2009. Extraction, identification, and functional characterization of a bioactive substance from automated compound-handling plastic tips. J. Biomol. Screen. 14: 566–572. [DOI] [PubMed] [Google Scholar]
- 52.McDonald G. R., Hudson A. L., Dunn S. M. J., You H., Baker G. B., Whittal R. M., Martin J. W., Jha A., Edmondson D. E., Holt A. 2008. Bioactive contaminants leach from disposable laboratory plasticware. Science. 322: 917. [DOI] [PubMed] [Google Scholar]
- 53.Lv G., Wang L., Li S. 2009. Method for the determination of fatty acid amides in polyethylene packaging materials--gas chromatography/mass spectrometry. J. Chromatogr. A. 1216: 8545–8548. [DOI] [PubMed] [Google Scholar]
- 54.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 55.Sommerfeld M. 1983. Trans unsaturated fatty acids in natural products and processed foods. Prog. Lipid Res. 22: 221–233. [DOI] [PubMed] [Google Scholar]
- 56.Mueller G. P., Driscoll W. J. 2009. Biosynthesis of oleamide. Vitam. Horm. 81: 55–78. [DOI] [PubMed] [Google Scholar]
- 57.De Petrocellis L., Bisogno T., Maccarrone M., Davis J. B., Finazzi-Agrò A., Di Marzo V. 2001. The activity of anandamide at vanilloid VR1 receptors requires facilitated transport across the cell membrane and is limited by intracellular metabolism. J. Biol. Chem. 276: 12856–12863. [DOI] [PubMed] [Google Scholar]
- 58.Farrell E. K. 2009. Biosynthesis of Fatty Acid Amides. PhD Thesis. University of South Florida, Tampa, FL. [Google Scholar]
- 59.Maurelli S., Bisogno T., De Petrocellis L., Di Luccia A., Marino G., Di Marzo V. 1995. Two novel classes of neuroactive fatty acid amides are substrates for mouse neuroblastoma ‘anandamide hydrolase’. FEBS Lett. 377: 82–86. [DOI] [PubMed] [Google Scholar]
- 60.Desarnaud F., Cadas H., Piomelli D. 1995. Anandamide amidohydrolase activity in rat brain microsomes. J. Biol. Chem. 270: 6030–6035. [DOI] [PubMed] [Google Scholar]
- 61.Giang D. K., Cravatt B. F. 1997. Molecular characterization of human and mouse fatty acid amide hydrolases. Proc. Natl. Acad. Sci. USA. 94: 2238–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueda N., Puffenbarger R. A., Yamamoto S., Deutsch D. G. 2000. The fatty acid amide hydrolase (FAAH). Chem. Phys. Lipids. 108: 107–121. [DOI] [PubMed] [Google Scholar]
- 63.Burstein S. H., Rossetti R. G., Yagen B., Zurier R. B. 2000. Oxidative metabolism of anandamide. Prostaglandins Other Lipid Mediat. 61: 29–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.