Abstract
The compartmentation of neutral lipids in plants is mostly associated with seed tissues, where triacylglycerols (TAGs) stored within lipid droplets (LDs) serve as an essential physiological energy and carbon reserve during postgerminative growth. However, some nonseed tissues, such as leaves, flowers and fruits, also synthesize and store TAGs, yet relatively little is known about the formation or function of LDs in these tissues. Characterization of LD-associated proteins, such as oleosins, caleosins, and sterol dehydrogenases (steroleosins), has revealed surprising features of LD function in plants, including stress responses, hormone signaling pathways, and various aspects of plant growth and development. Although oleosin and caleosin proteins are specific to plants, LD-associated sterol dehydrogenases also are present in mammals, and in both plants and mammals these enzymes have been shown to be important in (steroid) hormone metabolism and signaling. In addition, several other proteins known to be important in LD biogenesis in yeasts and mammals are conserved in plants, suggesting that at least some aspects of LD biogenesis and/or function are evolutionarily conserved.
Keywords: endoplasmic reticulum, lipids, membranes, phospholipids, lipid storage, plant
The seeds of plants store significant amounts of neutral lipids, namely triacylglycerols (TAGs), in cytosolic lipid droplets (LDs), most of which are subsequently mobilized immediately after germination in order to fuel the growth and development of the seedling prior to photosynthetic establishment. In developing seeds, TAGs are assembled in the endoplasmic reticulum (ER) from acyl-CoAs and glycerol by the conserved “Kennedy” pathway that operates in all eukaryotes. Recently, however, additional acyl-CoA-independent reactions have been identified in developing seeds and other plant tissues that contribute to the synthesis of TAGs, although the relative contributions of these alternative pathways to TAG accumulation may vary depending on the tissue and/or species (1).
There also has been a growing appreciation in the past few years that the compartmentation of neutral lipids in LDs of plants extends well beyond their role as simply static depots for carbon storage in seeds and, consequently, there is renewed interest in the cellular ontogeny and dynamics of this organelle. For instance, LDs are observed in nearly all cell types in plants, and although the biogenesis of LDs in nonseed tissues is still poorly understood, we now know that they are involved in many unique processes, such as stress response, pathogen resistance, and hormone metabolism. Furthermore, there are highly specialized roles for LDs in anther development, wherein LDs contribute significantly to the formation of the hydrophobic barrier of the pollen coat as tapetal tissues undergo programmed cell death. Interestingly, this process is somewhat similar to the specialized role that LDs play in the formation of the hydrophobic barrier that comprises the outer layer of mammalian skin.
Overall, knowledge of LD function in both seed and nonseed tissues has been greatly enhanced by efforts to characterize the major proteins that specifically associate with these organelles, namely oleosins, caleosins, and sterol dehydrogenases (steroleosins) (Fig. 1). Here, after a brief description of LD biogenesis in plant cells, we review the functional properties of these major LD-associated proteins and discuss how they compare with the properties of their known or potential counterparts in yeasts and mammals. We also describe the specialized role of LDs in pollen coat formation and how this process has some interesting parallels with skin formation in mammals. Finally, we describe approaches for identifying additional proteins involved in LD biogenesis and function in plants, such as the recent identification of the Arabidopsis homolog of the human comparative gene identification-58 (CGI-58) (2, 3), the causative gene in the human neutral lipid storage disorder, Chanarin-Dorfman syndrome. For a comprehensive comparison of LDs in plants, animals and microorgansims, readers are directed to a recent review by Murphy (4).
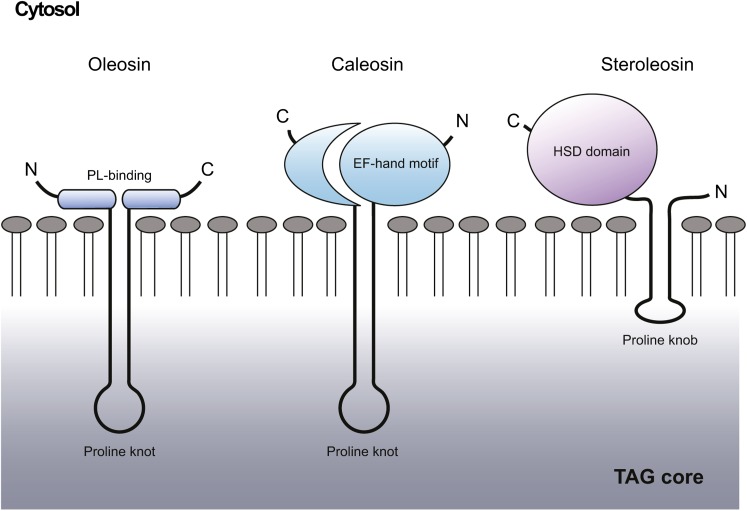
Fig. 1.
Schematic representations of the structures of oleosin, caleosin, and steroleosin at the surface of a TAG-filled LD. Shown are the cytosolic-facing, N- and C-terminal domains for oleosin [including the two regions of the protein proposed to be involved in its interaction with the charged, phospholipid (PL) head groups] (123), caleosin (including its calcium-binding EF-hand motif) and steroleosin (including its HSD domain). Shown also for each protein is its major hydrophobic domain, each of which is depicted as penetrating into the TAG-filled core of the LD and includes a so-called “proline knot” or “proline knob”. Based on illustrations presented in (35, 47, 74).
LD BIOGENESIS IN PLANTS
In plants, as in all other eukaryotes, LDs (referred to also as spherosomes, oil/lipid bodies, and oleosomes) are considered to arise from the ER, with some of the earliest and perhaps most substantial evidence in support of this concept coming from ultrastructural studies of developing seeds, including those showing frequent and remarkably intimate associations between the two organelles (5, 6). Indeed, the now commonly held theory that LD assembly begins with the accumulation of TAGs between the two leaflets of the ER bilayer, followed by the expansion and eventual pinching off of the LD into the cytoplasm, was formulated primarily from ultrastructural studies of developing plant embryos (7, 8). Some of these same studies demonstrated convincingly that LDs consist of a TAG core surrounded by a protein-containing half unit-membrane (9).
There also is support from studies in plants for the generally held idea that LDs form at distinct locations or ‘subdomains’ of the ER. That is, the ER is now well recognized as being a dynamic and intricate network that contains numerous specialized regions involved in unique functions, including LD biogenesis (10–12). In plants, for instance, diacylglycerol acyltransferase (DGAT), a key enzyme responsible for synthesizing the final step in TAG biosynthesis, as well as oleosins, the structural proteins that stabilize LDs (see Oleosins section below), have both been shown to localize within discrete regions of the ER (13–17). These results, as well as those obtained from recent studies with yeasts (18) or mammals (19), have led to the premise that a localized partitioning of DGAT, and/or perhaps its substrate, diacylglycerol (DAG), within the ER serves as the platform by which TAG accumulation and eventually the induction of LD formation takes place. This concept of spatial organization, however, must be reconciled with the essential involvement of acyl-chain modification reactions (e.g., desaturation) of fatty acids for TAG biosynthesis, a process that occurs directly on phosphatidylcholine (PC) as a substrate in plants (20–22), and for which there is no direct evidence for a spatial separation of TAG and PC biosynthesis.
Little is known with respect to the mechanistic details by which LD biogenesis (i.e., the induction, growth, and dissociation from ER subdomains) occurs, although some headway is being made into identifying the protein machinery through genetic screens of LD mutants in yeast (23–25), neutral lipid storage disorders in humans (26), and homology-based searches in plants (see Approaches to identify novel proteins involved in LD biogenesis in plants section below). It is also unclear whether small LDs emerge from the ER and then coalesce to form larger droplets, whether they bud to form their final size, or whether a combination of these processes is involved (Fig. 2). For instance, arguments have been made at the biophysical level that the thermodynamics of lipid demixing promote the formation of small, 12 nm droplets that would bud from the ER surface and then fuse in the cytosol (27), but this may not entirely account for the participation of protein machinery in the process of LD biogenesis. On the other hand, alternative models for LD biogenesis predict that LDs form from COP (coatomer) transport vesicles (28), or that they do not even detach from the ER, but instead remain as an integral part of the ER, growing and shrinking depending on the needs of the cell/organism (29). There is also the likelihood that the process of LD biogenesis is species and even cell-type specific, and may involve different classes of proteins that bind to LD and mediate LD formation, size, and interaction (30–32). Hence, a unified concept of LD biogenesis may draw on several proposed mechanisms that may not be mutually exclusive.
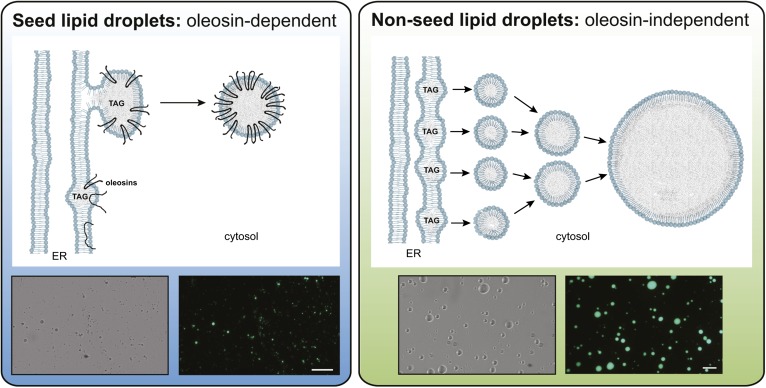
Fig. 2.
Schematic representation of models for oleosin-dependent and oleosin-independent LD formation from the ER in plant cells. In seed tissues (left panel), oleosins are cotranslationally inserted into the ER where they partition into domains in which TAG is accumulating between two leaflets. This promotes orientation of the oleosin proteins with N- and C termini facing the cytosol and the rest of the hydrophobic region of the protein adopting an extended hairpin configuration in the TAG matrix. The LDs in the cytosol are stabilized by oleosins and kept from fusing despite rapid dehydration and rehydration of these tissues during seed desiccation and imbibition. Micrographs are of isolated LDs from Arabidopsis seeds in bright-field (left) or by epifluorescence (right) following staining with Bodipy 493/503, a neutral lipid selective stain. LDs in nonseed tissues (right panel) may form from smaller TAG droplets that initially pinch off from the ER, then fuse to form larger droplets. Oleosins are not present in these LDs and the protein composition of LDs in nonseed tissues remains unknown. Micrographs are of LDs isolated from the oleaginous mesocarp of avocado fruit imaged in bright-field (left) or by Bodipy493/503 fluorescence (right). The white bars represent 50 microns. LDs from seeds tend to be smaller and more uniform compared with those from nonseed tissues, which fuse readily even in solution. Figure prepared by Dr. Charlene Case and Mr. Patrick Horn.
THE ROLE OF LDS IN PLANTS AS REVEALED BY CHARACTERIZATION OF SURFACE-ASSOCIATED PROTEINS
The functions of organelles in eukaryotic cells are determined at least in part by their protein and lipid constituents, both of which can vary in response to environmental and/or physiological cues, or as a result of the cellular differentiation that occurs in higher eukaryotic organisms. In plants, LDs have classically been viewed as inert storage organelles, but there is mounting evidence that these organelles, similar to their counterparts in yeasts and mammals (33), are highly dynamic and actively involved in many different physiological processes. Knowledge of LD protein and lipid composition, as well as mechanisms underlying LD protein targeting, assembly, and regulation, are essential for developing a better understanding of LD function. Below is a summary of the functional and biogenetic aspects of the major LD-associated proteins in plant cells.
Oleosins
Oleosin proteins are small (∼15–30 kDa), abundant proteins in the seeds of plants that bind to the surface of LDs and help prevent their coalescence during seed desiccation (30, 34, 35). Oleosins also are found in pollen grains (36), the vegetative gametophytes of moss (37), and the tapetal cells of some plant species (38), but they are otherwise generally absent from nonseed plant tissues, although oleosins do localize to LDs in leaves or leaf-derived cells when expressed in transgenic cells/tissues (39–41).
Like most membrane-bound proteins, oleosins are initially inserted into the ER in a cotranslational manner (42–44), after which they adopt an orientation that includes their N and C termini facing toward the cytosol and a large hydrophobic domain intercalated within the ER bilayer (45). A conserved, so-called “proline knot” at the middle of the hydrophobic domain is essential for targeting (partitioning) the oleosin protein to LDs (45) (Fig. 1), probably by assisting in the formation of an extended hydrophobic hairpin configuration projecting toward the interior of the droplet that is stabilized as TAGs concomitantly accumulate between the ER leaflet in the growing droplet. Such a concentration of oleosin proteins and TAG “bubbles” would therefore serve as an effective nucleator for LD formation at localized regions of the ER (Fig. 2), a process that might also be aided by the oligomerization of oleosins on the surface of LDs (46), as well as by another protein termed caleosin (see Caleosins section below) that is also localized in ER subdomains and LDs (47).
Oleosins are directly involved in regulating size and stability of LDs in seeds. For instance, RNAi-mediated suppression of the major seed-expressed oleosin gene, OLEO1, in Arabidopsis led to LDs of aberrant size in embryos, as well as dehydration-induced fusion of LDs in mature seeds (48). Knockout mutants (individually and in combination) of the major seed-specific oleosins in Arabidopsis, oleo1, 2, 3, 4, supported similar conclusions, i.e., reduced oleosin levels contributed to larger seed LDs (49). Cold temperatures during rehydration of oleosin-mutant seeds promoted fusion of LDs with reduced oleosin content. In addition, RNAi-mediated suppression of oleosin gene expression in soybean seeds led to large LDs and “micro-inclusions” of lipid in the ER, reinforcing the idea that oleosins are important for ER-mediated LD biogenesis in addition to controlling their stability (32).
Upon imbibition and seed germination, the TAGs housed within LDs are rapidly mobilized to release fatty acids for β-oxidation within glyoxysomes (a specialized class of peroxisomes found in geminated oilseeds and fungi). Lipid mobilization results in the production of sucrose (via the glyoxylate and gluconeogenic pathways), which serves as a carbon and energy reserve for the growing seedling prior to establishment of photosynthetic activity (50, 51). In this regard, oleosins are thought to play an important role in LD breakdown by recruiting lipases and perhaps other enzymes (e.g., phospholipases, proteases) that are involved in storage oil degradation (52). For instance, the rate of lipid mobilization was slowed dramatically in transgenic Arabidopsis seedlings when oleosin gene expression was suppressed, yet the full complement of TAG lipases was intact (48). Although it has been speculated that the reduced surface-to-volume ratio of the larger LDs in these transgenic seedlings may have slowed the access of the lipases and/or other degradative enzymes to the TAG substrate, it may be that oleosins play a critical role in recruiting and/or providing recognition sites for lipase activation.
It is tempting to draw parallels between the oleosin proteins of plant seeds and the PAT family of proteins [named after P erilipin, A dipophilin, and T IP47 (for 47 kDa Tail-Interacting Protein)] from mammals and insects. The PAT proteins are abundant LD surface-associated proteins that are involved in both the formation of LDs as well as recruitment of various LD-related enzymes such as lipases (53, 54). Comparison of oleosin and PAT proteins, however, show almost no primary sequence similarities. Furthermore, there are no obvious homologs for oleosins in mammals or PAT proteins in plants. The mechanisms of PAT and oleosin protein interaction with LDs are also likely to be fundamentally different, because PAT proteins associate mainly with the surface of LDs whereas oleosins have a characteristic hydrophobic domain that penetrates deeper into the lipid core. It is interesting to note, however, that although mammals lack any apparent homologs to oleosins, certain viral proteins have oleosin-like proline knot motifs that mediate their localization to LDs in mammalian cells (55). Furthermore, ectopic expression of plant oleosins in either mammalian or yeast cells results in their proper targeting to LDs in the heterologous system (44, 55). Taken together, these and other observations suggest that even though nonplant organisms lack oleosin-like proteins, the basic mechanisms involved in the targeting of oleosin to LDs (which likely involves biophysical partitioning into favorable lipid environments, as described above) are generally conserved in eukaryotes.
It is noteworthy to mention also that the PAT family of proteins is involved in biogenesis and regulation of LDs in a variety of cell types in mammals, including both lipid-storing and nonlipid-storing tissues. Oleosins, on the other hand, are expressed mainly in seeds. As such, there are likely other proteins present in plants that are involved specifically in the biogenesis and regulation of LDs in nonseed tissues.
Caleosins
Caleosins are a more recently identified group of LD-associated proteins in plants that appear to serve both structural and functional roles in the LD lifecycle. Caleosins were first reported as minor constituents of purified LDs from sesame seeds (56) but have since been found associated with LDs from seeds of many different plant species (57–60). Like oleosins, caleosins contain a long, central hydrophobic hairpin structure that contains a proline knot motif for LD association. The proteins differ, however, in that the caleosins have significantly larger N- and C-terminal regions (Fig. 1). The N-terminal region contains a single calcium-binding site known as a helix-loop-helix EF-hand motif and the C-terminal region contains several conserved protein phosphorylation sites (56). There are also conserved histidine residues in both the N- and C-terminal regions that together coordinate the binding of a heme prosthetic group (61).
Evidence for a role of caleosin in structural maintenance of LDs is based in part on observations that certain lower plant species such as cycads (sago palms) contain caleosin, rather than oleosin, as their major LD-associated protein (59, 62). Caleosins can also bind to and stabilize artificial LDs in vitro (41, 63), and suppression of oleosin genes in soybean (via RNAi) results in a corresponding increase in the amount of caleosin proteins in LDs (32). Furthermore, caleosins are present in fungi and single-celled algae (47, 57), whereas oleosins are present predominantly in higher plant species (37). Collectively, these observations suggest that caleosins may represent a more ancient structural protein for LDs in plants, and that oleosins evolved from caleosins to become more specifically associated with LD formation and maintenance in tissues that are subject to rapid desiccation/rehydration, i.e., in the seeds and pollen of higher plants.
In addition to their structural role, caleosins have unique functional properties. For instance, caleosins can act as peroxygenases (61, 64), which catalyze the hydroperoxide-dependent oxygenation of unsaturated fatty acids to produce epoxy fatty acids. These types of lipid transformations are frequently associated with oxylipin metabolism in plants, which involves the production of various lipophilic hormone-like molecules associated with plant stress response and innate immunity (65). Alternatively, caleosins have been implicated in the degradation of polyunsaturated fatty acids, as the products of peroxidized linoleic acid can accumulate to high levels in germinated seeds, and these products can subsequently be broken down by β-oxidation in glyoxysomes (66, 67). Additional support for a role of caleosins in oil breakdown comes from the analysis of gene knockout mutants in Arabidopsis, where disruption of the major seed-expressed caleosin resulted in a decreased rate of fatty acid degradation (68).
The structural and functional properties of caleosin proteins are sensitive to calcium, hence the name given to this group of plant LD-associated proteins. For instance, addition of calcium ions is known to alter the surface binding properties of caleosin to LDs in vitro (69), and the peroxygenase activity of caleosin is entirely calcium-dependent (61). Caleosin proteins also are known to be N-acetylated, phosphorylated, and, like oleosins, ubiquitinated under certain conditions (70), suggesting that caleosin (and oleosin) activity in plant cells is highly regulated, perhaps even in a coordinated manner.
Functional roles for caleosins in nonseed tissues of plants also have been reported. For instance, there are five caleosin genes in the Arabidopsis genome (AtCLO1-5), and whereas AtCLO-1 is highly expressed in developing seeds, the other four genes are expressed at lower basal levels throughout the plant during normal growth and development (57). Upon exposure of Arabidopsis plants to drought conditions, however, one of the earliest and strongest induced genes based on microarray studies is AtCLO-3 [also referred as RD20 ( R esponse to D ehydration 20 )] (71, 72), and the green-fluorescent-protein-tagged protein product of AtCLO-3 was shown to be localized to LDs in vegetative cells (72). Similarly, the AtCLO-4 gene encodes an LD-associated protein that mediates seed germination potential, as well as whole plant responses to various stresses, including drought, salt, and osmotic changes (73), indicating that caleosins play important signaling roles in both seed or nonseed tissues. How caleosins interact with LDs to generate peroxygenase-derived lipid metabolites or influence other signaling components in plant cells remains to be determined.
Steroleosins
Steroleosins are a third group of proteins that are frequently associated with LDs in plant seeds and, as their other name (sterol dehydrogenases) implies, they share significant sequence similarity with the LD-associated hydroxysteroid dehydrogenase (HSD) family of enzymes in mammals (74). Unlike the oleosin and caleosin proteins, steroleosins have just two main structural domains, including an N-terminal hydrophobic region required for LD association followed by a C-terminal HSD domain (Fig. 1). Although the N-terminal hydrophobic region lacks a proline knot motif, it does contain conserved proline residues that are predicted to form a so-called “proline knob” that functions in association with the LD surface [(74) and Fig. 1].
In mammals, the HSD family of enzymes is involved in modulating the steady-state concentrations of biologically active and less active forms of various steroid hormones, usually through the interconversion of hydroxyl and ketone groups present on the steroid backbone (29, 75, 76). For instance, 17β-estradiol is a potent estrogen-type hormone that contains a hydroxyl group at the 17th position, which can be dehydrogenated by 17 β-HSD 2/4 to produce a less active ketone-containing derivative called estrone (77) (Fig. 3). By contrast, other HSD family members, such as 17 β-HSD 1, can catalyze the reverse reaction, converting the ketone group on estrone back into an alcohol and thus regenerating the active hormone (Fig. 3). Given this scenario, it is often the case that pairs of HSD enzymes exist in a given tissue/cell so that they can fine-tune the amounts of biologically active hormones. The differences in biological potency are also presumably determined by specific hormone receptors, which favor either the hydroxyl- or ketone-containing form of the molecules.
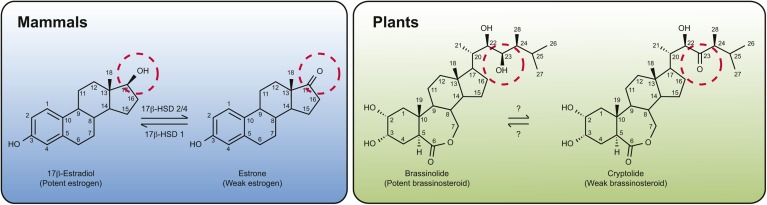
Fig. 3.
Structure and interconversion of steroid hormones in animals and plants. In mammals, 17β-estradiol, a potent estrogen-type hormone that contains a hydroxyl group at the 17th position (circled), is dehydrogenated by 17 β-HSD 2/4 to produce a less active ketone-containing estrone, whereas 17 β-HSD 1 can catalyze the reverse reaction. In an analogous manner, in plants, the 23-hydroxyl group of BL (circled) can be dehydrogenated to produce a ketone-containing derivative called cryptolide. The enzyme(s) responsible for this interconversion, however, is not known.
Given the high sequence similarity of steroleosin proteins to mammalian HSDs, it is not surprising that several groups have demonstrated that the plant proteins can actually function as HSDs in vitro by converting estradiol and to a lesser degree, cortisol, into the ketone-containing products, estrone and cortisone (74, 78). Although the significance is not clear, the steroleosin AtHSD1 from Arabidopsis also was shown to catalyze the reverse reaction, i.e., the ketosteroid reduction of estrone into estradiol (78). None of these compounds naturally exists in plants, however, and the exact substrates and products of these enzymes remain to be determined.
There is mounting evidence, however, that steroleosins play an important role in brassinosteroid (BR) metabolism and signaling in plants. BRs are similar to mammalian steroids in several ways, including some shared aspects of their biosynthesis, as well as having multiple roles in growth, development, fertility, and stress responses (79–81). The BR with highest biological activity is called brassinolide (BL), and there are over 40 BR derivatives known to date, indicating that the metabolic pathways of this plant steroid are complex and that BR modification(s) likely plays an important role in modulating biological activity (82). Interestingly, analogous to the interconversion of estrone and estradiol in animals, dehydrogenation/reduction reactions appear to be important in regulating BL activity, because the 23-hydroxyl group of BL can be dehydrogenated to produce a ketone-containing derivative called cryptolide (82) (Fig. 3). There is little information, however, regarding the genes and enzymes responsible for this BL to cryptolide conversion, or for most other chemical transformations involved in BR metabolism (81, 82).
A role for steroleosins in BR signaling pathways is supported also by recent studies of Arabidopsis plants either lacking or overexpressing the AtHSD1 gene, all of which displayed similar phenotypic traits as plants that are disrupted in or overexpressing known BR biosynthetic genes (83, 84). Furthermore, endogenous AtHSD1 expression is strongly upregulated in various tissues by ectopic application of BRs (84), and AtHSD1 overexpression affects both seed dormancy and germination (85). Taken together, these data suggest that, like their counterparts in mammals, plant steroleosins may influence steroid signaling pathways by regulating the amounts of biologically active hormones through chemical interconversions. Clues to the specific steroidal metabolites generated and acted upon by steroleosins, as well as other potential roles of LDs in this process, will undoubtedly be uncovered through the continued analysis of plants overexpressing or disrupted in specific steroleosin genes.
Other plant LD-associated proteins
There are a number of other proteins that are frequently associated with LDs in the seeds of plants, including proteases, phospholipases, lipoxygenases, and lipases, many of which facilitate the breakdown of LDs and TAGs following seed germination (86). In some cases, proteins of unknown functions have been identified in LDs from seeds [e.g., ref.(87)]. Other recent reviews provide a comprehensive discussion of TAG mobilization from LDs in seed and seedling tissues (50, 52). Recently, a calcium-dependent protein kinase that mediates the innate immune response during pathogen infection (88), was shown to target to both LDs and peroxisomes in vegetative cells of Arabidopsis (suspension-cultured and root cells), supporting the idea that LDs may participate in multiple aspects of stress responses (71–73, 84, 88). It is likely that additional LD-associated enzymes involved in both biotic and/or abiotic stress responses will be identified in the near future.
A ROLE FOR LDS IN ANTHER DEVELOPMENT
LDs in plants also serve important roles in organ development. For instance, the anthers of plants contain a specialized layer of cells called the tapetum, which plays an essential part in development of pollen. Pollen production is a highly coordinated and specialized process that includes both cellular differentiation and structural and morphological changes in the tissues of the anther. For instance, the tapetal cells accumulate high amounts of lipids in two distinct types of subcellular compartments, namely: i) elaioplasts, which are specialized chloroplasts that are largely devoid of thylakoids, but contain abundant LDs enriched in steryl esters; and ii) tapetosomes, which are multivesicular-like bodies located in the cytoplasm of the cell and which include a complex association of ER cisternae and LDs containing high amounts of TAG coated with oleosin proteins (35). During the latter stages of pollen development, the tapetal cells of the pollen sac degenerate to release their lipid/protein contents onto the surface of the pollen grains. Interestingly, most of the TAG is metabolized, whereas the steryl esters, oleosins, and other lipids [including flavonoids and alkanes (38)] are combined to form a complex hydrophobic barrier that helps protect the pollen grains from dehydration and other environmental stresses, such as UV radiation.
Given the scenario discussed above, a number of similarities appear to exist between the process of pollen coat formation in plants and formation of the hydrophobic barrier that comprises the outer layer of mammalian skin [reviewed in (89)]. As the largest organ in mammals, the epidermis plays a critical role in protecting underlying tissues from pathogen invasion and dehydration. Formation of the outermost layer of skin (the stratum corneum) involves the terminal differentiation of keratinocytes into corneocytes, which subsequently become highly crosslinked and embedded within a complex hydrophobic matrix that is secreted from the underlying layer of skin (the stratum granulosum). Specifically, keratinocytes within the granulosum accumulate large amounts of lipids (mainly in the form of acylceramides) in complex multivesicular-like structures called lamellar bodies, which are subsequently secreted into the extracellular space; a process that may be analogous to the secretion of lipids from the tapetal cells onto the surface of pollen grains. Once secreted, the acylceramides are hydrolyzed to produce a complex mixture of ceramides, nonesterified fatty acids, and cholesterol.
Although TAG is not a major component of lipids within the lamellar body, TAG metabolism has recently been shown to be essential for the biosynthesis of the acylceramides that are enriched within these structures, again perhaps somewhat similar to the role that TAG metabolism plays in pollen grain formation. Specifically, the production of acylceramides involves the transfer of linoleic acid from TAGs or phospholipids onto the ω-hydroxyl group of ceramides (90). People harboring mutations in a gene called cgi-58, which encodes an LD-associated protein involved in TAG degradation and possibly phospholipid metabolism (91), exhibit substantially reduced acylceramide biosynthesis and, as a result, accumulate high amounts of free ω-hydroxy ceramides and TAG-containing LDs. These changes in lipid composition apparently disrupt lamellar body structure and function, resulting in aberrant formation of the hydrophobic skin barrier, which is lethal in mice due to severe dehydration after birth (89). In addition to showing an increase in LDs in keratinocytes, patients harboring mutations in cgi-58 can also show an increase in LD content in other non lipid-storing tissues such as muscle and blood cells, the disease associated with these mutations being known as Chanarin-Dorfman syndrome or neutral lipid storage disease (92). Recently, characterization of Arabidopsis plants harboring a mutation in the plant gene homolog of cgi-58 revealed a similar accumulation of LD droplets in nonlipid storing tissues such as leaves and stems (3), suggesting that, in addition to the apparent shared features underlying development in anthers and skin, other fundamental aspects of LD biogenesis and TAG regulation in non lipid-storing tissues of plants and animals may be conserved.
APPROACHES TO IDENTIFY NOVEL PROTEINS INVOLVED IN LD BIOGENESIS IN PLANTS
Whereas TAG accumulation in plants mostly has been characterized in seed tissues, all plant cells and tissues have the capacity to synthesize TAG. However, the ability of nonseed (i.e., vegetative) cells and tissues to accumulate TAG can vary substantially (93). For example, LDs are not particularly abundant in leaf tissues, but they are prevalent in some fruits (e.g., avocado, palm, olive), roots/tubers (e.g., cotton, nutsedge), floral tissues and even stems (e.g., Mongolian oil wood) of certain species (94). What regulates the abundance of LDs in these tissues is unknown, but it appears that it is not through the well-characterized transcriptional programs operating in maturing seeds (95).
Perhaps informative experimental results about the conservation of plant proteins involved in the formation of LDs will come from analysis of nonseed tissues, where it would be expected that general biogenesis machinery is operational (versus oleosin-containing LDs that are specialized for desiccation). For example, there are homologs in plant genomes for lipodystrophy genes identified in humans (26) (Table 1), several of which affect LD formation and tissue-specific distribution. One pertinent example is CGI-58 that, as described above, results in accumulation of LDs in tissues of both plants and animals that do not normally store lipids (3, 96).
TABLE 1.
Human lipodystrophy genes (26) with their apparent homologs in Arabidopsis
| Human gene | Protein function | Candidate Arabidopsis homolog(s)a |
| AGPAT2 | LPAT, synthesis of phosphatidic acid | At1g80950; At1g51260; At3g57650; At3g18850; At1g75020; At4g30580 |
| BSCL2 | Seipin, role in LD morphology, number, size | At5g16460; At1g29760; At2g34380 |
| CAV1 | Caveolin 1, formation of membrane microdomains | No compelling homologc |
| LMNA | Lamin A, nuclear lamina protein subunit | No compelling homologc |
| PPARG | PPAR γ- transcription factor regulates lipid synthesis | No compelling homologc |
| AKT2 | Protein kinase B | At3g08730; At3g08720; At5g04510b; At310540b |
| ZMPSTE24 | Zinc metalloprotease; processing of lamin subunits | At4g01320 |
| CGI-58 | ABHD5; coactivator of ATGL, also has LPAT activity | At4g24160 |
| LIPA | Lysosomal acid lipase; hydrolyzes cholesteryl esters and TAGs | At5g14180; At2g15230 |
Best match by WU-BLAST against the Arabidopsis genome at TAIR (www.arabidopsis.org).
Contains Pleckstrin homology domains and has phosphoinositide-3-dependent kinase activity.
e value > 5.0.
Another potential plant candidate for a “lipid compartmentation” gene with cross-kingdom sequence conservation is seipin. This gene in humans is responsible for Berardinelli-Seip congenital lipodystrophy (97) and it affects LD biogenesis in humans (98), mice (99), and yeast (24). In yeast, seipin was shown to control LD size and is localized to ER-lipid droplet junctions (23), where it functions as an oligomer to somehow facilitate lipid droplet formation (100). At least three apparent seipin homologs exist in Arabidopsis (Table 1), one of which is expressed almost exclusively in seeds (At5g16460), whereas the second is more ubiquitously expressed (At1g29760) and the third has no publicly-available expression data but for which a cDNA has been identified (At2g34380). It remains to be determined if any of these Arabidopsis seipin-like genes influence LD morphology in seeds or nonseed tissues.
A third gene responsible for a human lipodystrophy that also has putative homologs in plants is lysosomal acid lipase, whose mutation results in Wolman's disease in humans (101). Loss of acid lipase results in accumulation of LDs enriched in TAG and sterol esters in the liver (101), suggesting that the protein, or perhaps a related autophagic process, may be important in facilitating the degradation of LDs in non lipid-storing tissues (102). Arabidopsis contains two genes with significant sequence similarity to the lysosomal acid lipase (At5g14180 and At2g15230) (Table 1), and one of them (At5g14180) is annotated as having partial localization to the vacuole (the lysosome equivalent) in plant cells. Notably, disruption of other gene (At2g15230) revealed that it was not involved in storage oil breakdown in germinating seeds (103), but its counterpart (At5g14180) is required for production of an antibiotic-like activity present in petiole exudates that suppress plant pathogen infection (104). These latter results suggest that the acid lipases are somehow involved in generating lipid signals involved in the innate immune response in plants but a role, if any, in LD formation/turnover similar to that observed in humans, remains to be determined.
Another method for identifying genes potentially involved in LD biogenesis in plants would be the use of unbiased genetic screens to identify Arabidopsis mutants with alterations in LD size, tissue distribution, and/or number per cell, like the extremely revealing screens conducted in yeast cells (23, 24). Table 2 shows the overlapping set of genes identified in three screens for aberrant lipid droplet morphologies in yeast and their candidate Arabidopsis homologs where identifiable. Of course, other genes known to be important for LD biogenesis in yeast cells that were not identified in these screens might also prove to be important for LD biogenesis in plant cells. For example, as mentioned briefly above (LD biogenesis in plants section), phosphatidic acid phosphohydrolase 1 (PAH1) in yeast cells plays an important role in LD biogenesis, likely by producing a localized pool of DAG in the ER that stimulates LD formation in a process that is distinct from the role of DAG as a substrate for TAG biosynthesis (18). Additional support for a role of PAH1 enzymes in LD formation is that, in mammals, mutation of the PAH1 ortholog (referred to as lipin) results in an 80% decrease in adipose tissue mass (105). Furthermore, DAG is known to recruit the mammalian PAT protein, TIP47, to the surface of LDs (106). Although it is currently unknown whether the PAH1 homologs in plants are similarly involved in LD formation, the enzymes are known to have phosphatidate phosphohydrolase activity (107, 108), and ectopic expression of the gene can functionally complement yeast mutants lacking PAH1 activity (109). Collectively, these observations suggest that the localized production of DAG may be conserved feature of LD biogenesis in all eukaryotic cells, including plant cells.
TABLE 2.
TAG “compartmentation” genes in yeast and their apparent homologs in Arabidopsis.
| Yeast gene | Protein function | Candidate Arabidopsis homolog(s)a |
| ADE8 | Phosphoribosyl-glycinamide transformylase (de novo purine biosynthesis) | At1g31220 |
| ADE12 | Adenylosuccinate synthase (purine biosynthesis) | At3g57610 |
| ANP1 | Alpha-1,6 mannosyltransferase transmembrane subunit | No compelling homologb |
| CHC1 | Clathrin heavy chain subunit | At3g08530; At3g11130 |
| CNM67 | Spindle pole body component required for proper nuclear migration | No compelling homologb |
| ERD1 | Membrane protein; retention of lumenal ER proteins | At5g35730; At2g32295 |
| EST3 | Telomerase component | No compelling homologb |
| KEM1 | Component of cytoplasmic processing (P) bodies involved in mRNA decay | At1g75660; At5g42540; At5g42540; At1g54490 |
| MDM20 | Non-catalytic subunit of the NatB N-terminal acetyltransferase | No compelling homologb |
| NEM1 | Catalytic subunit of Nem1p-Spo7p phosphatase; regulates P-lipid synthesis | At5g46410; AT5G11860; AT4G18140 (SCP1-like P-ases) |
| OST4 | Subunit of the oligosaccharyltransferase complex of the ER lumen | No compelling homologb |
| PAF1 | Component of the Paf1p complex; modulates the activity of RNA polymerases | At1g79730 |
| ROX3 | Subunit of the RNA polymerase II mediator complex | No compelling homologb |
| SPO7 | Regulatory subunit of Nem1p-Spo7p phosphatase | No compelling homologb |
| SSD1 | Translational repressor with a role in cell wall polar growth and integrity | At1g77680; At2g17510 |
| TPD3 | Subunit A of protein phosphatase 2A (PP2A); required for cell morphogenesis | At3g25800; At1g13320; At1g25490 |
| VMA6 | Integral membrane subunit of vacuolar H -ATPase (V-ATPase) | At3g28715; At3g28710 |
| VPS16 | Vacuole protein sorting subunit; essential for membrane docking and fusion | At2g38020 |
| VPS66 | Cytoplasmic protein of unknown function involved in vacuolar protein sorting | No compelling homologb |
| YLR404W | Seipin, lipid droplet size, homology to human BSCL2 involved in lipodystrophy | At5g16460; At1g29760; At2g34380 |
The identification of genes involved in LD formation in plants might be also uncovered by analyzing oil production in oleaginous fruits. For example, oil palm mesocarp (fruit) contains abundant LDs with no oleosins, whereas oil palm seeds contain LDs with conventional oleosins. Recent genomic information from deep-sequencing of oil palm fruit at different developmental stages provided substantial genetic information about the regulatory and metabolic programs operating in this oil-storing tissue system (110, 111). Perhaps future comparisons of transcriptional profiles between oil palm fruit and seeds (two adjacent tissue systems rich in lipid droplets, one without and one with oleosins), with an eye toward lipid-droplet biogenesis, may provide insights into the generalized and/or unique machinery involved in TAG packaging in plants. These studies will likely be complemented by proteomics analysis of LDs as well as emerging techniques for analyzing the lipid composition (i.e., lipidome) of individual LDs (112, 113).
NOVEL ROLES FOR LDS IN BIOTECHNOLOGY APPLICATIONS IN PLANTS
Although fundamental knowledge of LD biogenesis in mammals will likely stimulate the development of better treatments for debilitating lipid disorders in humans, knowledge of LD biogenesis in plants will instead serve to underpin the development of creative biotechnology applications that exploit novel aspects of LD structure and function. For instance, LDs are low density, lipid-rich particles and as such, they can easily be isolated by flotation centrifugation. This property has been exploited for many “molecular farming” applications, whereby a protein of interest is fused (using standard recombinant DNA techniques) to oleosin and then the chimeric gene is expressed in seeds wherein the fusion protein targets to and associates with LDs (114). Subsequent purification of these LDs and cleavage of the fusion protein thereby provides a simple method for preparing large amounts of purified protein.
LDs are also lipid-rich reservoirs that can serve as depots for lipophilic vitamins and nutrients, thereby providing opportunities for “biofortification” of foods with enhanced nutritional value (115). The emerging role of LDs in stress response and BR metabolism may also provide novel opportunities for increasing stress resistance and/or enhancing yield of plants, a field of research that is already receiving considerable attention (116). Finally, knowledge of LD formation in nonseed tissues may allow for increasing the total TAG content in vegetative biomass of plants (117–119), which could serve as a useful source of food, feed, fuel, or feedstocks for a variety of industrial applications (94, 120–122).
Acknowledgments
The authors thank Dr. Charlene Case for assistance with manuscript preparation.
Footnotes
Abbreviations:
- BL
- brassinolide
- BR
- brassinosteroid
- DAG
- diacylglycerol
- DGAT
- diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- HSD
- hydroxysteroid dehydrogenase
- LD
- lipid droplet
- PAH1
- phosphatidic acid phosphohydrolase 1
- PC
- phosphatidylcholine
- RNAi
- RNA interference
- TAG
- triacylglycerol
The authors’ work on LD formation in plants is supported in part by a grant from the US Department of Energy, Office of Science (BER), DE-SC0000797.
References
- 1.Chapman K. D., Ohlrogge J. 2011. TAG accumulation in plants. J. Biol. Chem. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh A. K., Chauhan N., Rajakumari S., Daum G., Rajasekharan R. 2009. At4g24160, a soluble acyl-coenzyme A-dependent lysophosphatidic acid acyltransferase. Plant Physiol. 151: 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James C. N., Horn P. J., Case C. R., Gidda S. K., Zhang D., Mullen R. T., Dyer J. M., Anderson R. G., Chapman K. D. 2010. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. USA. 107: 17833–17838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy D. J. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. Epub ahead of print. October 15, 2011. [DOI] [PubMed] [Google Scholar]
- 5.Frey-Wyssling A., Grieshaber E., Mühlethaler K. 1963. Origin of spherosomes in plant cells. J. Ultrastruct. Res. 8: 506–516. [Google Scholar]
- 6.Wanner G., Theimer R. R. 1978. Membranous appendices of spherosomes (oleosomes). Possible role in fat utilization in germinating oil seeds. Planta. 140: 163–169. [DOI] [PubMed] [Google Scholar]
- 7.Wanner G., Formanek H., Theimer R. R. 1981. The ontogeny of lipid bodies (spherosomes) in plant cells. Planta. 151: 109–123. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzenbach A. M. 1971. Observations on spherosomal membranes. Cytobiologie. 4: 145–147. [Google Scholar]
- 9.Yatsu L. Y., Jacks T. J. 1972. Spherosome membranes: half unit-membranes. Plant Physiol. 49: 937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparkes I. A., Frigerio L., Tolley N., Hawes C. 2009. The plant endoplasmic reticulum: a cell-wide web. Biochem. J. 423: 145–155. [DOI] [PubMed] [Google Scholar]
- 11.Park S. H., Blackstone C. 2010. Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11: 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynes E. M., Simmen T. 2011. Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim. Biophys. Acta. 1813: 1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., Rothstein S. J., Mullen R. T., Dyer J. M. 2006. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 18: 2294–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gidda S. K., Shockey J. M., Rothstein S. J., Dyer J. M., Mullen R. T. 2009. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 47: 867–879. [DOI] [PubMed] [Google Scholar]
- 15.Lacey D. J., Hills M. J. 1996. Heterogeneity of the endoplasmic reticulum with respect to lipid synthesis in developing seeds of Brassica napus L. Planta. 199: 545–551. [Google Scholar]
- 16.Sarmiento C., Ross J. H., Herman E., Murphy D. J. 1997. Expression and subcellular targeting of a soybean oleosin in transgenic rapeseed. Implications for the mechanism of oil-body formation in seeds. Plant J. 11: 783–796. [DOI] [PubMed] [Google Scholar]
- 17.Lacey D. J., Beaudoin F., Dempsey C. E., Shewry P. R., Napier J. A. 1999. The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J. 17: 397–405. [Google Scholar]
- 18.Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., Goodman J. M. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFie P. J., Stone S. L., Banman S. L., Stone S. J. 2010. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. J. Biol. Chem. 285: 37377–37387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stymne S., Stobart A. K., Glad G. 1983. The role of the acyl-CoA pool in the synthesis of polyunsaturated 18-carbon fatty acids and triacylglycerol production in the microsomes of developing safflower seeds. Biochim. Biophys. Acta. 752: 198–208. [DOI] [PubMed] [Google Scholar]
- 21.Sperling P., Heinz E. 1993. Isomeric sn-1-octadecenyl and sn-2-octadecenyl analogues of lysophosphatidylcholine as substrates for acylation and desaturation by plant microsomal membranes. Eur. J. Biochem. 213: 965–971. [DOI] [PubMed] [Google Scholar]
- 22.Harwood J. L. 1996. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta. 1301: 7–56. [DOI] [PubMed] [Google Scholar]
- 23.Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W. P., Agarwal A. K., Garg A., Anderson R. G., Goodman J. M. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104: 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., Yang H. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180: 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., et al. 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7: e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg A., Agarwal A. K. 2009. Lipodystrophies: disorders of adipose tissue biology. Biochim. Biophys. Acta. 1791: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanghellini J., Wodlei F., von Grünberg H. H. 2010. Phospholipid demixing and the birth of a lipid droplet. J. Theor. Biol. 264: 952–961. [DOI] [PubMed] [Google Scholar]
- 28.Kalantari F., Bergeron J. J., Nilsson T. 2010. Biogenesis of lipid droplets–how cells get fatter. Mol. Membr. Biol. 27: 462–468. [DOI] [PubMed] [Google Scholar]
- 29.Goodman J. M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy D. J. 1993. Structure, function and biogenesis of storage lipid bodies and oleosins in plants. Prog. Lipid Res. 32: 247–280. [DOI] [PubMed] [Google Scholar]
- 31.Napier J. A., Stobart A. K., Shewry P. R. 1996. The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol. Biol. 31: 945–956. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt M. A., Herman E. M. 2008. Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant. 1: 910–924. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M., Shinohara Y., Ohsaki Y., Fujimoto T. 2011. Lipid droplets: size matters. J. Electron Microsc. (Tokyo). 60(Suppl 1): S101–S116. [DOI] [PubMed] [Google Scholar]
- 34.Huang A. H. C. 1992. Oil bodies and oleosins in seeds. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 177–200. [Google Scholar]
- 35.Hsieh K., Huang A. H. 2004. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136: 3427–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H. U., Hsieh K., Ratnayake C., Huang A. H. 2002. A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 277: 22677–22684. [DOI] [PubMed] [Google Scholar]
- 37.Huang C. Y., Chung C. I., Lin Y. C., Hsing Y. I., Huang A. H. 2009. Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol. 150: 1192–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh K., Huang A. H. 2007. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 19: 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beaudoin F., Napier J. A. 2000. The targeting and accumulation of ectopically expressed oleosin in non-seed tissues of Arabidopsis thaliana. Planta. 210: 439–445. [DOI] [PubMed] [Google Scholar]
- 40.Wahlroos T., Soukka J., Denesyuk A., Wahlroos R., Korpela T., Kilby N. J. 2003. Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis. 35: 125–132. [DOI] [PubMed] [Google Scholar]
- 41.Domenico S. D., Bonsegna S., Lenucci M. S., Poltronieri P., Sansebastiano G. P. D., Santino A. 2011. Localisation of seed oil body proteins in tobacco protoplasts reveals specific mechanisms of protein targeting to leaf lipid droplets. J. Int. Plant Biol. Epub ahead of print. September 26, 2011; doi: 10.1111/j.1744-7909.2011.01077.x. [DOI] [PubMed] [Google Scholar]
- 42.Hills M. J., Watson M. D., Murphy D. J. 1993. Targeting of oleosins to the oil bodies of oilseed rape (Brassica napus L.). Planta. 189: 24–29. [DOI] [PubMed] [Google Scholar]
- 43.Abell B. M., High S., Moloney M. M. 2002. Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J. Biol. Chem. 277: 8602–8610. [DOI] [PubMed] [Google Scholar]
- 44.Beaudoin F., Napier J. A. 2002. Targeting and membrane-insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: the central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta. 215: 293–303. [DOI] [PubMed] [Google Scholar]
- 45.Abell B. M., Holbrook L. A., Abenes M., Murphy D. J., Hills M. J., Moloney M. M. 1997. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 9: 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M., Murphy D. J., Lee K. H., Wilson R., Smith L. J., Clark D. C., Sung J. Y. 2002. Purification and structural characterization of the central hydrophobic domain of oleosin. J. Biol. Chem. 277: 37888–37895. [DOI] [PubMed] [Google Scholar]
- 47.Frandsen G. I., Mundy J., Tzen J. T. 2001. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 112: 301–307. [DOI] [PubMed] [Google Scholar]
- 48.Siloto R. M., Findlay K., Lopez-Villalobos A., Yeung E. C., Nykiforuk C. L., Moloney M. M. 2006. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell. 18: 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimada T. L., Shimada T., Takahashi H., Fukao Y., Hara-Nishimura I. 2008. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55: 798–809. [DOI] [PubMed] [Google Scholar]
- 50.Graham I. A. 2008. Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- 51.Kelly A. A., Quettier A. L., Shaw E., Eastmond P. J. 2011. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157: 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quettier A. L., Eastmond P. J. 2009. Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. 47: 485–490. [DOI] [PubMed] [Google Scholar]
- 53.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559. [DOI] [PubMed] [Google Scholar]
- 54.Beller M., Bulankina A. V., Hsiao H. H., Urlaub H., Jäckle H., Kühnlein R. P. 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12: 521–532. [DOI] [PubMed] [Google Scholar]
- 55.Hope R. G., Murphy D. J., McLauchlan J. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277: 4261–4270. [DOI] [PubMed] [Google Scholar]
- 56.Chen J. C., Tsai C. C., Tzen J. T. 1999. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 40: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 57.Naested H., Frandsen G. I., Jauh G. Y., Hernandez-Pinzon I., Nielsen H. B., Murphy D. J., Rogers J. C., Mundy J. 2000. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 44: 463–476. [DOI] [PubMed] [Google Scholar]
- 58.Katavic V., Agrawal G. K., Hajduch M., Harris S. L., Thelen J. J. 2006. Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics. 6: 4586–4598. [DOI] [PubMed] [Google Scholar]
- 59.Jiang P. L., Chen J. C., Chiu S. T., Tzen J. T. 2009. Stable oil bodies sheltered by a unique caleosin in cycad megagametophytes. Plant Physiol. Biochem. 47: 1009–1016. [DOI] [PubMed] [Google Scholar]
- 60.Frandsen G., Müller-Uri F., Nielsen M., Mundy J., Skriver K. 1996. Novel plant Ca(2+)-binding protein expressed in response to abscisic acid and osmotic stress. J. Biol. Chem. 271: 343–348. [DOI] [PubMed] [Google Scholar]
- 61.Hanano A., Burcklen M., Flenet M., Ivancich A., Louwagie M., Garin J., Blée E. 2006. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 281: 33140–33151. [DOI] [PubMed] [Google Scholar]
- 62.Jiang P. L., Tzen J. T. 2010. Caleosin serves as the major structural protein as efficient as oleosin on the surface of seed oil bodies. Plant Signal. Behav. 5: 447–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu T. H., Chyan C. L., Li F. Y., Tzen J. T. 2009. Stability of artificial oil bodies constituted with recombinant caleosins. J. Agric. Food Chem. 57: 2308–2313. [DOI] [PubMed] [Google Scholar]
- 64.Meesapyodsuk D., Qiu X. 2011. A peroxygenase pathway involved in the biosynthesis of epoxy fatty acids in oat. Plant Physiol. 157: 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mosblech A., Feussner I., Heilmann I. 2009. Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47: 511–517. [DOI] [PubMed] [Google Scholar]
- 66.Gerhardt B., Fischer K., Balkenhohl T. J., Pohnert G., Kühn H., Wasternack C., Feussner I. 2005. Lipoxygenase-mediated metabolism of storage lipids in germinating sunflower cotyledons and beta-oxidation of (9Z,11E,13S)-13-hydroxy-octadeca-9,11-dienoic acid by the cotyledonary glyoxysomes. Planta. 220: 919–930. [DOI] [PubMed] [Google Scholar]
- 67.Weichert H., Kolbe A., Kraus A., Wasternack C., Feussner I. 2002. Metabolic profiling of oxylipins in germinating cucumber seedlings–lipoxygenase-dependent degradation of triacylglycerols and biosynthesis of volatile aldehydes. Planta. 215: 612–619. [DOI] [PubMed] [Google Scholar]
- 68.Poxleitner M., Rogers S. W., Lacey Samuels A., Browse J., Rogers J. C. 2006. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 47: 917–933. [DOI] [PubMed] [Google Scholar]
- 69.Purkrtova Z., Le Bon C., Kralova B., Ropers M. H., Anton M., Chardot T. 2008. Caleosin of Arabidopsis thaliana: effect of calcium on functional and structural properties. J. Agric. Food Chem. 56: 11217–11224. [DOI] [PubMed] [Google Scholar]
- 70.Hsiao E. S., Tzen J. T. 2011. Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol. Biochem. 49: 77–81. [DOI] [PubMed] [Google Scholar]
- 71.Partridge M., Murphy D. J. 2009. Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol. Biochem. 47: 796–806. [DOI] [PubMed] [Google Scholar]
- 72.Aubert Y., Vile D., Pervent M., Aldon D., Ranty B., Simonneau T., Vavasseur A., Galaud J. P. 2010. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 51: 1975–1987. [DOI] [PubMed] [Google Scholar]
- 73.Kim Y. Y., Jung K. W., Yoo K. S., Jeung J. U., Shin J. S. 2011. A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 52: 874–884. [DOI] [PubMed] [Google Scholar]
- 74.Lin L. J., Tai S. S., Peng C. C., Tzen J. T. 2002. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 128: 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Penning T. M., Drury J. E. 2007. Human aldo-keto reductases: function, gene regulation, and single nucleotide polymorphisms. Arch. Biochem. Biophys. 464: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Penning T. M. 2003. Hydroxysteroid dehydrogenases and pre-receptor regulation of steroid hormone action. Hum. Reprod. Update. 9: 193–205. [DOI] [PubMed] [Google Scholar]
- 77.Penning T. M. 2011. Human hydroxysteroid dehydrogenases and pre-receptor regulation: insights into inhibitor design and evaluation. J. Steroid Biochem. Mol. Biol. 125: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.d'Andréa S., Canonge M., Beopoulos A., Jolivet P., Hartmann M. A., Miquel M., Lepiniec L., Chardot T. 2007. At5g50600 encodes a member of the short-chain dehydrogenase reductase superfamily with 11beta- and 17beta-hydroxysteroid dehydrogenase activities associated with Arabidopsis thaliana seed oil bodies. Biochimie. 89: 222–229. [DOI] [PubMed] [Google Scholar]
- 79.Clouse S. D., Sasse J. M. 1998. Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- 80.Kim T. W., Wang Z. Y. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- 81.Fujioka S., Yokota T. 2003. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164. [DOI] [PubMed] [Google Scholar]
- 82.Bajguz A. 2007. Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. 45: 95–107. [DOI] [PubMed] [Google Scholar]
- 83.Baud S., Dichow N. R., Kelemen Z., d'Andréa S., To A., Berger N., Canonge M., Kronenberger J., Viterbo D., Dubreucq B., et al. 2009. Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol. 50: 1463–1478. [DOI] [PubMed] [Google Scholar]
- 84.Li F., Asami T., Wu X., Tsang E. W., Cutler A. J. 2007. A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baud S., Mendoza M. S., To A., Harscoët E., Lepiniec L., Dubreucq B. 2007. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838. [DOI] [PubMed] [Google Scholar]
- 86.Rudolph M., Schlereth A., Körner M., Feussner K., Berndt E., Melzer M., Hornung E., Feussner I. 2011. The lipoxygenase-dependent oxygenation of lipid body membranes is promoted by a patatin-type phospholipase in cucumber cotyledons. J. Exp. Bot. 62: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tnani H., López I., Jouenne T., Vicient C. M. 2011. Protein composition analysis of oil bodies from maize embryos during germination. J. Plant Physiol. 168: 510–513. [DOI] [PubMed] [Google Scholar]
- 88.Coca M., San Segundo B. 2010. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63: 526–540. [DOI] [PubMed] [Google Scholar]
- 89.Radner F. P., Grond S., Haemmerle G., Lass A., Zechner R. 2011. Fat in the skin: triacylglycerol metabolism in keratinocytes and its role in the development of neutral lipid storage disease. Dermatoendocrinol. 3: 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wertz P. W., Downing D. T. 1990. Metabolism of linoleic acid in porcine epidermis. J. Lipid Res. 31: 1839–1844. [PubMed] [Google Scholar]
- 91.Oberer M., Boeszoermenyi A., Nagy H. M., Zechner R. 2011. Recent insights into the structure and function of comparative gene identification-58. Curr. Opin. Lipidol. 22: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamaguchi T., Osumi T. 2009. Chanarin-Dorfman syndrome: deficiency in CGI-58, a lipid droplet-bound coactivator of lipase. Biochim. Biophys. Acta. 1791: 519–523. [DOI] [PubMed] [Google Scholar]
- 93.Lin W., Oliver D. J. 2008. Role of triacylglycerols in leaves. Plant Sci. 175: 233–237. [Google Scholar]
- 94.Carlsson A. S., Yilmaz J. L., Green A. G., Stymne S., Hofvander P. 2011. Replacing fossil oil with fresh oil – with what and for what? Eur. J. Lipid Sci. Technol. 113: 812–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baud S., Lepiniec L. 2010. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49: 235–249. [DOI] [PubMed] [Google Scholar]
- 96.Agarwal A. K., Garg A. 2006. Genetic basis of lipodystrophies and management of metabolic complications. Annu. Rev. Med. 57: 297–311. [DOI] [PubMed] [Google Scholar]
- 97.Magré J., Delépine M., Khallouf E., Gedde-Dahl T., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. 2001. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28: 365–370. [DOI] [PubMed] [Google Scholar]
- 98.Agarwal A. K., Garg A. 2004. Seipin: a mysterious protein. Trends Mol. Med. 10: 440–444. [DOI] [PubMed] [Google Scholar]
- 99.Cui X., Wang Y., Tang Y., Liu Y., Zhao L., Deng J., Xu G., Peng X., Ju S., Liu G., et al. 2011. Seipin ablation in mice results in severe generalized lipodystrophy. Hum. Mol. Genet. 20: 3022–3030. [DOI] [PubMed] [Google Scholar]
- 100.Binns D., Lee S., Hilton C. L., Jiang Q. X., Goodman J. M. 2010. Seipin is a discrete homooligomer. Biochemistry. 49: 10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aslanidis C., Ries S., Fehringer P., Büchler C., Klima H., Schmitz G. 1996. Genetic and biochemical evidence that CESD and Wolman disease are distinguished by residual lysosomal acid lipase activity. Genomics. 33: 85–93. [DOI] [PubMed] [Google Scholar]
- 102.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. 2009. Autophagy regulates lipid metabolism. Nature. 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.El-Kouhen K., Blangy S., Ortiz E., Gardies A. M., Ferté N., Arondel V. 2005. Identification and characterization of a triacylglycerol lipase in Arabidopsis homologous to mammalian acid lipases. FEBS Lett. 579: 6067–6073. [DOI] [PubMed] [Google Scholar]
- 104.Louis J., Lorenc-Kukula K., Singh V., Reese J., Jander G., Shah J. 2010. Antibiosis against the green peach aphid requires the Arabidopsis thaliana MYZUS PERSICAE-INDUCED LIPASE1 gene. Plant J. 64: 800–811. [DOI] [PubMed] [Google Scholar]
- 105.Péterfy M., Phan J., Xu P., Reue K. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- 106.Skinner J. R., Shew T. M., Schwartz D. M., Tzekov A., Lepus C. M., Abumrad N. A., Wolins N. E. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284: 30941–30948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakamura Y., Koizumi R., Shui G., Shimojima M., Wenk M. R., Ito T., Ohta H. 2009. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA. 106: 20978–20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eastmond P. J., Quettier A. L., Kroon J. T., Craddock C., Adams N., Slabas A. R. 2010. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell. 22: 2796–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mietkiewska E., Siloto R. M., Dewald J., Shah S., Brindley D. N., Weselake R. J. 2011. Lipins from plants are phosphatidate phosphatases that restore lipid synthesis in a pah1Δ mutant strain of Saccharomyces cerevisiae. FEBS J. 278: 764–775. [DOI] [PubMed] [Google Scholar]
- 110.Tranbarger T. J., Dussert S., Joët T., Argout X., Summo M., Champion A., Cros D., Omore A., Nouy B., Morcillo F. 2011. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol. 156: 564–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bourgis F., Kilaru A., Cao X., Ngando-Ebongue G. F., Drira N., Ohlrogge J. B., Arondel V. 2011. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA. 108: 12527–12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Horn P. J., Ledbetter N. R., James C. N., Hoffman W. D., Case C. R., Verbeck G. F., Chapman K. D. 2011. Visualization of lipid droplet composition by direct organelle mass spectrometry. J. Biol. Chem. 286: 3298–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Horn P. J., Chapman K. D. 2011. Organellar lipidomics. Plant Signal. Behav. 6: 1594–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bhatla S. C., Kaushik V., Yadav M. K. 2010. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol. Adv. 28: 293–300. [DOI] [PubMed] [Google Scholar]
- 115.Mattoo A. K., Shukla V., Fatima T., Handa A. K., Yachha S. K. 2011. Genetic engineering to enhance crop-based phytonutrients (nutraceuticals) to alleviate diet-related diseases. Adv. Exp. Med. Biol. 698: 122–143. [DOI] [PubMed] [Google Scholar]
- 116.Divi U. K., Krishna P. 2009. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 26: 131–136. [DOI] [PubMed] [Google Scholar]
- 117.Slocombe S. P., Cornah J., Pinfield-Wells H., Soady K., Zhang Q., Gilday A., Dyer J. M., Graham I. A. 2009. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 7: 694–703. [DOI] [PubMed] [Google Scholar]
- 118.Andrianov V., Borisjuk N., Pogrebnyak N., Brinker A., Dixon J., Spitsin S., Flynn J., Matyszczuk P., Andryszak K., Laurelli M., et al. 2010. Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8: 277–287. [DOI] [PubMed] [Google Scholar]
- 119.Sanjaya, Durrett T. P., Weise S. E., Benning C. 2011. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9: 874–883. [DOI] [PubMed] [Google Scholar]
- 120.Durrett T. P., Benning C., Ohlrogge J. 2008. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54: 593–607. [DOI] [PubMed] [Google Scholar]
- 121.Dyer J. M., Stymne S., Green A. G., Carlsson A. S. 2008. High-value oils from plants. Plant J. 54: 640–655. [DOI] [PubMed] [Google Scholar]
- 122.Ohlrogge J., Chapman K. 2011. The seeds of green energy: expanding the contribution of plant oils as biofuels. The Biochemist. 33: 34–38. [Google Scholar]
- 123.Alexander L. G., Sessions R. B., Clarke A. R., Tatham A. S., Shewry P. R., Napier J. A. 2002. Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta. 214: 546–551. [DOI] [PubMed] [Google Scholar]
- 124.Fei W., Alfaro G., Muthusamy B. P., Klaassen Z., Graham T. R., Yang H., Beh C. T. 2008. Genome-wide analysis of sterol-lipid storage and trafficking in Saccharomyces cerevisiae. Eukaryot. Cell. 7: 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]