Abstract
The nucleotide sequence of 16S rDNA from Euglena gracilis chloroplasts has been determined representing the first complete sequence of an algal chloroplast rRNA gene. The structural part of the 16S rRNA gene has 1491 nucleotides according to a comparative analysis of our sequencing results with the published 5'- and 3'-terminal "T1-oligonucleotides" from 16S rRNA from E. gracilis. Alignment with 16S rDNA from Zea mays chloroplasts and E. coli reveals 80 to 72% sequence homology, respectively. Two deletions of 9 and 23 nucleotides are found which are identical in size and position with deletions observed in 16S rDNA of maize and tobacco chloroplasts and which seem to be characteristic for all chloroplast rRNA species. We also find insertions and deletions in E. gracilis not seen in 16S rDNA of higher plant chloroplasts. The 16S rRNA sequence of E. gracilis chloroplasts can be folded by base pairing according to the general 16S rRNA secondary structure model.
Full text
PDF

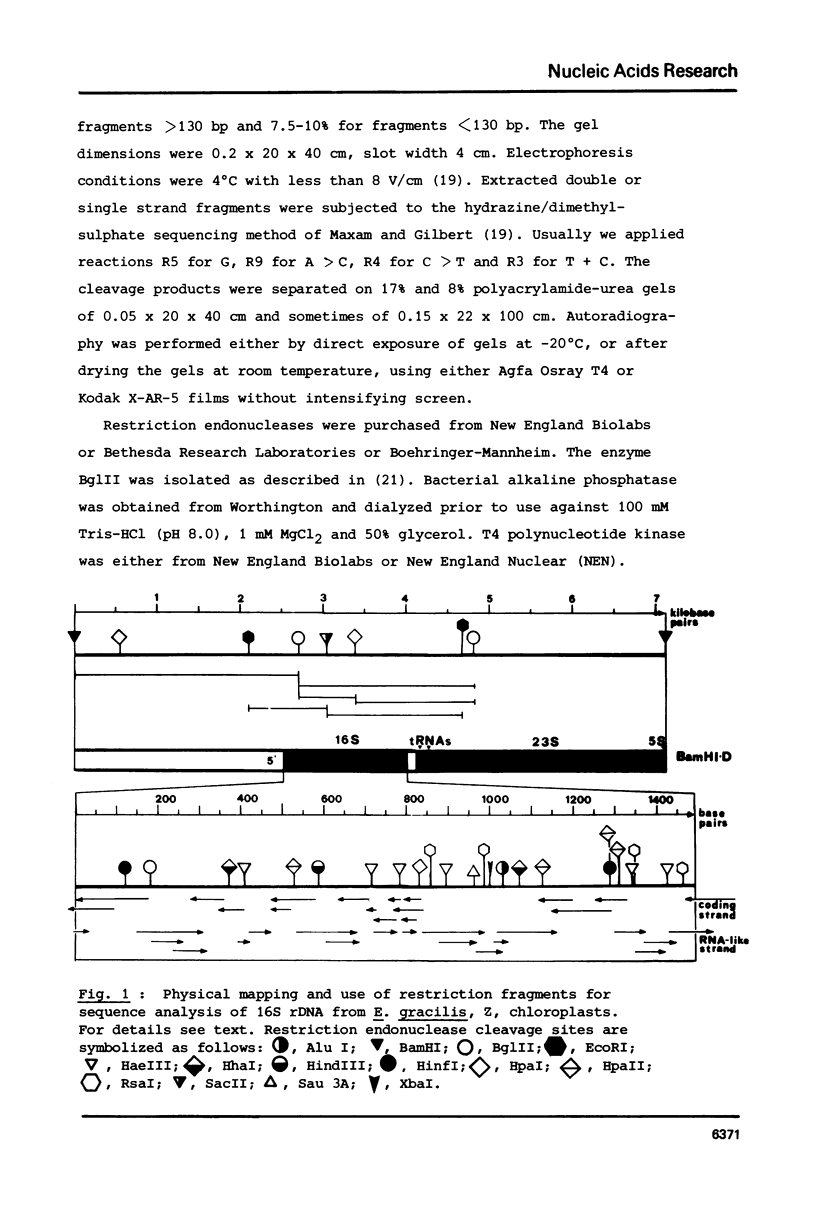



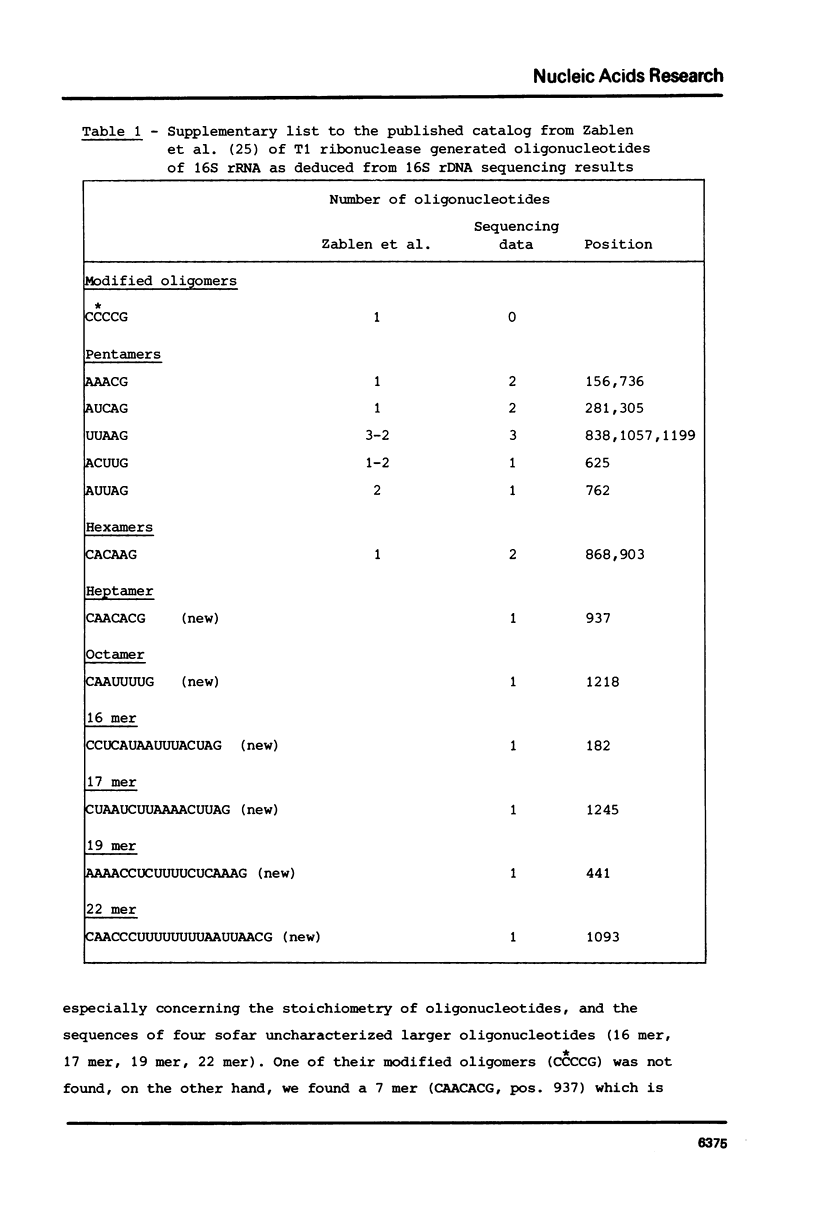
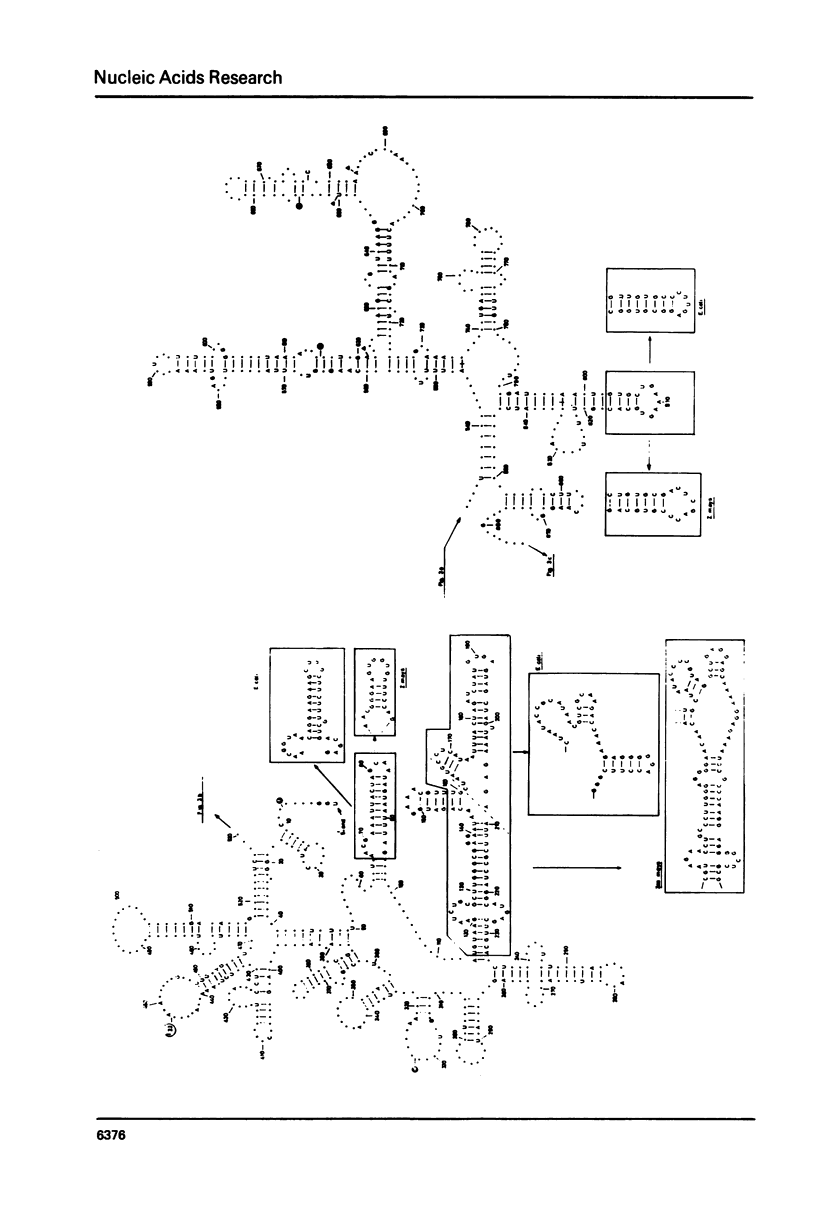
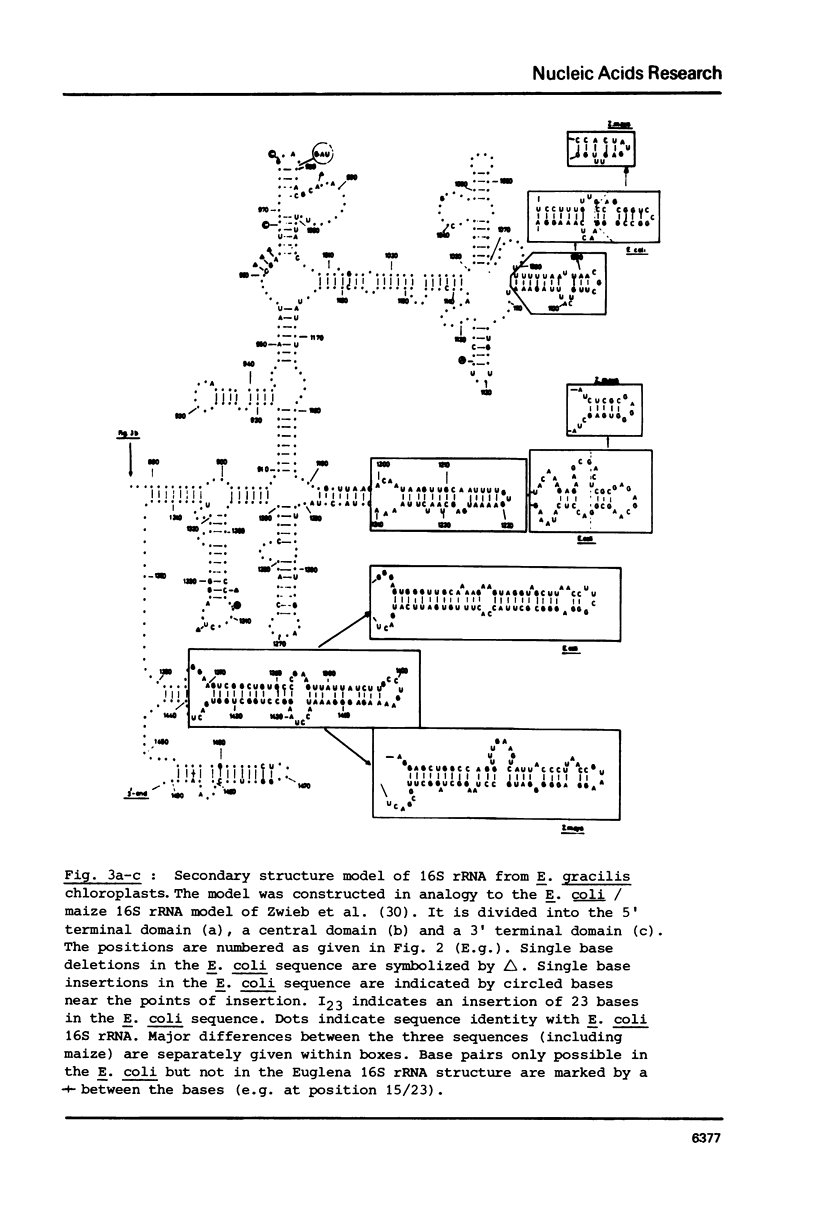




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bickle T. A., Pirrotta V., Imber R. A simple, general procedure for purifying restriction endonucleases. Nucleic Acids Res. 1977 Aug;4(8):2561–2572. doi: 10.1093/nar/4.8.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L., Doolittle W. F., Fox G. E. Cyanobacterial evolution: results of 16S ribosomal ribonucleic acid sequence analyses. Can J Biochem. 1979 Jun;57(6):879–888. doi: 10.1139/o79-108. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The sequence of Escherichia coli ribosomal 16 S RNA determined by new rapid gel methods. FEBS Lett. 1978 Oct 1;94(1):152–156. doi: 10.1016/0014-5793(78)80926-0. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf L., Kössel H., Stutz E. Sequencing of 16S--23S spacer in a ribosomal RNA operon of Euglena gracilis chloroplast DNA reveals two tRNA genes. Nature. 1980 Aug 28;286(5776):908–910. doi: 10.1038/286908a0. [DOI] [PubMed] [Google Scholar]
- Gray P. W., Hallick R. B. Physical mapping of the Euglena gracilis chloroplast DNA and ribosomal RNA gene region. Biochemistry. 1978 Jan 24;17(2):284–289. doi: 10.1021/bi00595a015. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jenni B., Stutz E. Analysis of Euglena gracilis chloroplast DNA: mapping of a DNA sequence complementary to 16 s rRNA outside of the three rRNA gene sets. FEBS Lett. 1979 Jun 1;102(1):95–99. doi: 10.1016/0014-5793(79)80936-9. [DOI] [PubMed] [Google Scholar]
- Jenni B., Stutz E. Physical mapping of the ribosomal DNA region of Euglena gracilis chloroplast DNA. Eur J Biochem. 1978 Jul 17;88(1):127–134. doi: 10.1111/j.1432-1033.1978.tb12429.x. [DOI] [PubMed] [Google Scholar]
- Knopf U. C., Stutz E. Molecular cloning of the gene region coding for the chloroplast rRNA of Euglena gracilis. Mol Gen Genet. 1978 Jul 6;163(1):1–6. doi: 10.1007/BF00268957. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Gray P. W., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. I. The location of transfer RNA, 5 S, 16 S, and 23 S ribosomal RNA genes. J Biol Chem. 1980 Nov 25;255(22):10991–10996. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Tohdoh N., Sugiura M. The complete nucleotide sequence of 16S ribosomal RNA gene from tobacco chloroplasts. Gene. 1982 Feb;17(2):213–218. doi: 10.1016/0378-1119(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Wienand U., Schwarz Z., Feix G. Electrophoretic elution of nucleic acids from gels adapted for subsequent biological tests. Application for analysis of mRNAs from maize endosperm. FEBS Lett. 1979 Feb 15;98(2):319–323. doi: 10.1016/0014-5793(79)80208-2. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]
- Zablen L. B., Kissil M. S., Woese C. R., Buetow D. E. Phylogenetic origin of the chloroplast and prokaryotic nature of its ribosomal RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2418–2422. doi: 10.1073/pnas.72.6.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]


