SUMMARY
Dendrites of the same neuron usually avoid each other. Some neurons also repel similar neurons through dendrite-dendrite interaction to tile the receptive field. Non-overlapping coverage based on such contact-dependent repulsion requires dendrites to compete for limited space. Here we show that Drosophila class IV dendritic arborization (da) neurons, which tile the larval body wall, grow their dendrites mainly in a two-dimensional (2D) space on the extracellular matrix (ECM) secreted by the epidermis. Removing neuronal integrins or blocking epidermal laminin production causes dendrites to grow into the epidermis, suggesting that integrin-laminin interaction attaches dendrites to the ECM. We further show that some of the previously identified tiling mutants fail to confine dendrites in a 2D plane. Expansion of these mutant dendrites in three dimensions results in overlap of dendritic fields. Moreover, overexpression of integrins in these mutant neurons effectively reduces dendritic crossing and restores tiling, revealing a novel mechanism for tiling.
INTRODUCTION
Precise sampling of sensory inputs from the environment is critical for the fitness and survival of animals. Biological systems utilize a variety of strategies to determine the location and strength of sensory inputs, including dendritic self-avoidance and tiling for organizing receptive fields of neurons (Grueber and Sagasti, 2010; Jan and Jan, 2010).
Self-avoidance, the phenomenon that dendrites of the same neuron avoid to fasciculate or overlap with one another, ensures maximal spreading of isoneuronal dendrites for better coverage of the receptive field. In both vertebrates and invertebrates, contact-mediated self-repulsion is likely a common mechanism underlying self-avoidance (Kramer and Stent, 1985; Sdrulla and Linden, 2006; Sugimura et al., 2003). In Drosophila, Down syndrome cell adhesion molecule (Dscam), a transmembrane immunoglobin (Ig) protein with 38,016 possible isoforms through alternative splicing, is required for self-avoidance in many neurons (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007; Wang et al., 2002; Zhu et al., 2006). Dscam mediates repulsion through homophilic interactions between identical isoforms on dendritic membranes of the same neuron. Vertebrate Dscam molecules, although lacking diverse alternative splicing, also mediate self-avoidance in subsets of retina neurons (Fuerst et al., 2009; Fuerst et al., 2008).
Dendritic tiling refers to partitioning of a receptive field by neurons of the same functional group without overlap, thereby ensuring complete but non-redundant coverage and unambiguous sampling of sensory inputs. Tiling has been observed in many neuronal types in both invertebrates and vertebrates (Grueber and Sagasti, 2010; Jan and Jan, 2010), and mutants with defective tiling have been found in Drosophila and C. elegans (Emoto et al., 2004; Emoto et al., 2006; Gallegos and Bargmann, 2004; Koike-Kumagai et al., 2009). Drosophila dendritic arborization (da) neurons, sensory neurons of the peripheral nervous system (PNS), spread dendritic arbors over the larval body wall (Grueber et al., 2002). Four classes of da neurons (I–IV) display increasing complexities of dendritic patterns (Grueber et al., 2002). Whereas all four classes of da neurons show self-avoidance, only class III and class IV display dendritic tiling (Grueber et al., 2003). Two types of experiments implicate homotypic repulsion between dendrites of the same class of neurons (heteroneuronal dendrites) in establishing tiling. First, when a class IV da neuron is ablated during embryonic stages, dendrites of neighboring class IV da neurons will grow into its territory (Grueber et al., 2003; Parrish et al., 2009; Sugimura et al., 2003). Second, duplication of class IV da neurons causes division of receptive fields with very little overlap between dendrites of the duplicated neurons (Grueber et al., 2003). Genetic analyses have revealed an intracellular signalling pathway important for tiling among class IV da neurons, which is composed of the Ste-20 family kinase Hippo (Hpo), a NDR family kinase Tricornered (Trc), and a Trc activator, Furry (Fry) (Emoto et al., 2004; Emoto et al., 2006). In trc and fry mutants, dendrites of adjacent class IV da neurons cross one another and thus have overlapping dendritic fields (Emoto et al., 2004). Interestingly, trc and fry mutants also display extensive isoneuronal dendritic crossing of class IV da neurons. Timelapse analyses in fry mutants showed that a much smaller percentage of dendrites display avoidance behaviors than in the wildtype, leading to the hypothesis that fry and trc directly regulate homotypic repulsion (Emoto et al., 2004). In addition, Target of rapamycin (Tor), SAPK-interacting protein 1 (Sin1), and rapamycin-insensitive companion of Tor (rictor), three components of TOR complex 2 (TORC2), act in the same pathway by phosphorylating and activating Trc (Koike-Kumagai et al., 2009).
Neuron-substrate interactions are important for neuronal development and function. Neuronal substrates typically derive from secreted molecules in the extracellular matrix (ECM). The ECM not only serves as an adhesive substrate permitting neurite outgrowth (Kapfhammer and Schwab, 1992), it also hosts guidance cues important for axon pathfinding (Nakamoto et al., 2004). A major class of cell membrane receptors for ECM are integrins, which anchor to the actin cytoskeleton through cytosolic signaling and adaptor proteins, providing a linkage between the local ECM milieu and intracellular signaling events (Cabodi et al., 2010). In vivo studies have shown that integrins play pivotal roles in many aspects of neural development, such as axon guidance, neuronal migration, neurite growth, arborization, and maintenance (Denda and Reichardt, 2007). In Drosophila, integrins are known to regulate axon guidance and synaptic plasticity (Grotewiel et al., 1998; Stevens and Jacobs, 2002), but their role in patterning dendritic fields is largely unexplored.
Class IV da neuron dendrites innervate the larval body wall by attaching to the epidermis. Self-avoidance and tiling together ensure even spreading of the dendrites. In theory, contact-dependent repulsion requires dendrites to be restricted within a two-dimensional (2D) space. Even though the receptive field of class IV da neurons, the larval epidermis, appears to be a 2D sheet, it is unclear whether the dendrites are indeed distributed within a 2D space. Specifically, how dendrites are positioned relative to epidermal cells is unknown. By using high-resolution confocal imaging, here we show that most dendrites of class IV da neurons are indeed distributed in a 2D space between the epidermal basal surface and the ECM. We further show that αPS1 and βPS integrins are cell-autonomously required in neurons for dendrites to attach to the ECM. Laminins secreted by the epidermis likely serve as ligands for integrins in the dendrite-ECM interaction. Unexpectedly, mutants of the TORC2/Trc pathway exhibit impaired dendrite attachment to the ECM: while dendrites attached to the ECM avoid one another, numerous dendrites detach from the ECM so as to cross other dendrites not confined to the same 2D space. Moreover, forced attachment of dendrites to the ECM by integrin overexpression in these tiling mutants rescued both isoneuronal and heteroneuronal dendritic crossing defects. Our results demonstrate the importance of 2D distribution in the tiling of class IV da neurons and reveal that the TORC2/Trc pathway plays a major role in ensuring tiling by confining dendrites to a 2D space rather than by mediating homotypic repulsion.
RESULTS
Class IV da dendrites are mainly distributed within a 2D plane
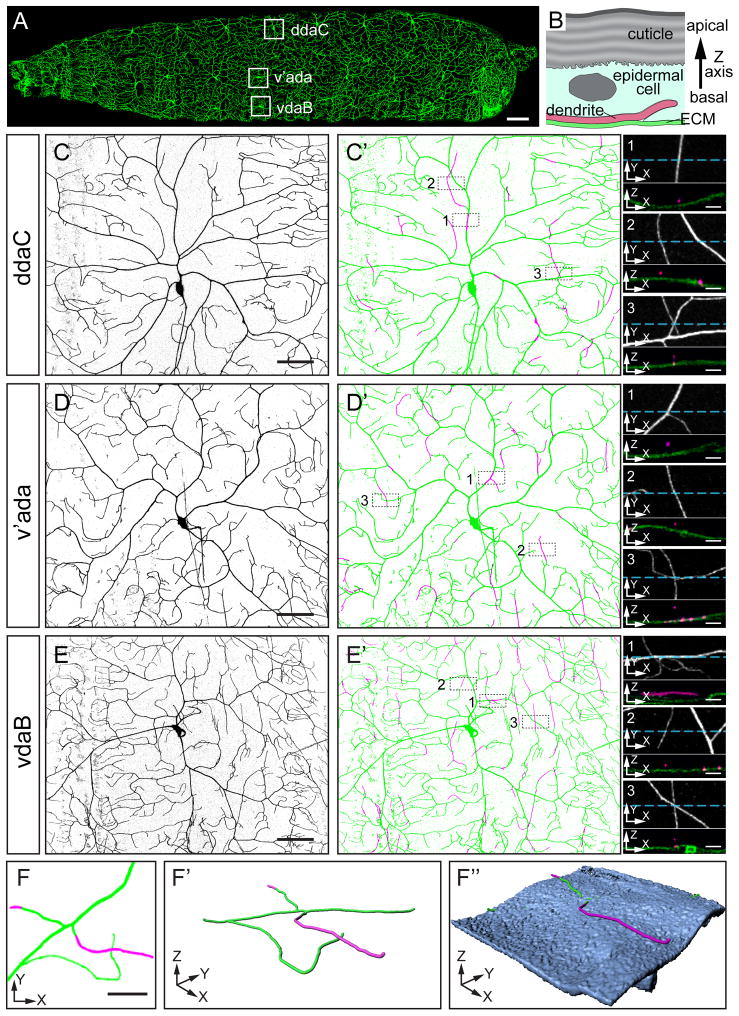
The Drosophila larval body wall is evenly covered by class IV da dendrites (Figure 1A). To understand how dendrites and epidermal cells are arranged relative to one another in a three-dimensional (3D) space, we simultaneously imaged class IV da dendrites and the ECM of the epidermis in live 3rd instar larvae by labelling dendrites with an improved class IV-specific membrane marker ppk-CD4-tdTom (Han et al., 2011, see Experimental Procedures) and the ECM with viking-GFP (vkg-GFP). Since the epidermis is comprised of a thin layer of epithelial cells, a detailed cross-section view of the epidermis requires 3D reconstruction of an image stack with a high resolution on the Z axis (i.e. the apical-basal axis of epidermal cells, Figure 1B). To maximize the resolving power on the Z axis, we adopted a high-resolution confocal imaging protocol combined with deconvolution (see Experimental Procedures), which greatly improves the visualization of the spatial relationship between dendrites and the ECM.
Figure 1. Positioning of class IV da dendrites relative to the ECM.
(A) A second instar larva showing class IV da neurons that completely and non-redundantly cover the entire body wall with dendritic arbors. The somas of the class IV da neurons in abdominal segment 3 (A3) are boxed. Scale bar, 100 μm. (B) Diagram of a cross section of the epidermis, showing relative positions of the cuticle, an epidermal cell, the ECM, and a terminal dendrite of a class IV da neuron. The tip of the terminal dendrite is depicted to extend into the epidermal cell. (C–E′) Dendritic fields of ddaC (C and C′), v’ada (B and B′), and vdaB (E and E′) labeled by ppk-CD4-tdTom. Live images of the neurons are inverted to show the dendritic patterns in (C), (D), and (E). The dendritic arbors attached to ECM (green) and those enclosed in the epidermal layer (magenta) are shown in (C′), (D′), and (E′). Three representative regions from each dendritic field are selected (boxed with broken outlines) to show enclosed dendrites that are within primary branches (1), are parts of terminal branches (2), or form crossings with other dendrites attached to the ECM (3). High magnification views of the selected regions are in small panels on the right, with both projections on the Z axis (upper panels) and cross sections along the blue lines (lower panels). Scale bars, 50 μm in (C), (D), and (E) or 5 μm in small panels. (F–F″) A 2D view (F) and 3D renderings (F′ and F″) of two crossing dendrites that are not in direct contact. The dendrites attached to the ECM are in green and the enclosed dendrites are in magenta. The surface rendering of the ECM (F″) is in blue. Part of the dendrites appear to be inside of the ECM due to the resolution limit of confocal microscopy. Scale bar, 10 μm. See also Movie S1.
Each abdominal hemisegment of the Drosophila larva contains three class IV da neurons, the dorsal ddaC, the ventro-lateral v’ada, and the ventral vdaB (Figure 1A). We examined the dendrite positioning of all three neurons along the Z axis. Most of the dendrites were found to directly contact the ECM (green dendrites in Figures 1C′, 1D′, and 1E′; also see Movie S1), while a small percentage of dendrites are detached from the ECM in the apical direction (magenta dendrites in Figures 1C′, 1D′, and 1E′). Since epidermal cells are located immediately apical to the ECM (Figure 1B), the dendrites detached from the ECM are enclosed within the epidermal layer (referred to as enclosed dendrites hereafter). We noticed several characteristics of these enclosed dendrites. First, they can be either segments in the middle of stabilized branches (small panel 1 in Figures 1C′, 1D′, and 1E′), or parts of terminal dendrites (small panel 2 in Figures 1C′, 1D′, and 1E′). Second, the enclosed dendrites may appear to cross other branches attached to the ECM in Z-axis projections, even though the overlapping dendrites are located at different depths in the epidermis and are not in direct contact (small panel 3 in Figures 1C′, 1D′, 1E′, and Figures 1F––1F″). We call this type of dendritic overlap non-contacting crossing. Third, the three class IV da neurons display different levels of dendrite enclosure in the epidermis, with ddaC showing the least and vdaB the most. Fourth, most of the enclosed dendrites are in the medial-lateral (or dorsal-ventral) orientation. With the exception of the enclosed dendrites, most class IV da dendrites are thus located in a 2D sheet at the interface of the epidermal basal surface and the ECM.
Distinct mechanisms contribute to epidermal enclosing of dendrites
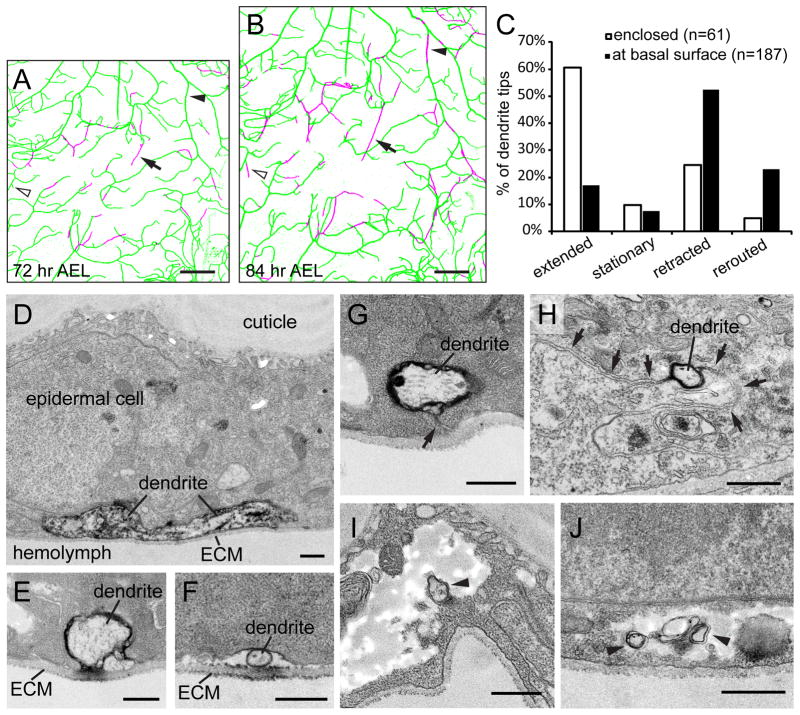
We then performed timelapse analyses of how epidermal cells enclose dendrites by imaging the ventral dendritic field of the same ddaC neurons at 72 hours after egg laying (AEL) (Figure 2A) and 84 hours AEL (Figure 2B) and comparing the distribution of enclosed dendrites. Newly enclosed dendrites were found to emerge in three different ways. First, stabilized branches initially attached to the ECM can subsequently become enclosed (arrowheads). Second, an enclosed dendrite tip can continue to grow within the epidermal layer (arrows). Third, a dendrite tip that was attached to the ECM can extend a new segment into the epidermal layer (open arrowheads).
Figure 2. Dynamics and TEM analysis of enclosed dendrites.
(A–B) A ventral ddaC dendritic field imaged at 72 hr AEL (A) and 84 hr AEL (B), with dendrites attached to the ECM in green and enclosed dendrites in magenta. The arrowheads point to a segment of primary dendrites that become enclosed during the 12 hr period. The open arrowheads point to a terminal branch that extended a new segment into the epidermal cell layer. The arrows point to an enclosed terminal branch that continued to grow in the epidermal cell layer. Scale bars, 30 μm. (C) Quantitative analysis of terminal dendrites that were enclosed (open bars) or at the basal surface of epidermal cells (filled bars) at both time points, showing the change of terminal dendrites over the 12 hr period. Rerouted denotes dendrites that changed direction after retraction (D–J) TEM images showing class IV da dendrites in cross sections of the larval body wall. Dendrites labeled by the dark DAB staining are either attached to the ECM (D–F) or enclosed in the epidermal cell layer (G–J). The arrow in (G) points to the channel formed by the basal cell membrane of the epidermal cell. The arrows in (H) point to the junction of two neighboring epidermal cells. The arrowheads in (I) and (J) point to the dendrites that are completely wrapped in the middle of epidermal cytoplasm. Scale bars, 500 nm.
How do existing and new dendrites grow into the epidermal layer? The first scenario can result from the basal plasma membrane of epidermal cells wrapping around an existing dendritic branch. The second and third scenarios indicate that dendrite tips can grow inside the epidermal layer either by “burrowing a tunnel” or by pushing through spaces between cells. To test these hypotheses, we further examined the spatial relationship of class IV da dendrites and the epidermis by transmission electron microscopy (TEM). In order to unequivocally identify the neuronal structures of interest, we used two pre-embedding staining strategies to specifically label the dendrites of class IV da neurons. The first strategy involved antibody staining against RFP in ppk-CD4-tdTom animals, with subsequent HRP-conjugated secondary antibody labelling. The second strategy was to express a membrane-tethered HRP transgene, UAS-HRP-DsRed-GPI, in class IV da neurons with an improved ppk-Gal4 that has higher and more specific expression (see Experimental Procedures). In both cases, the HRP reaction product Diaminobenzidine (DAB) can be detected by TEM (Larsen et al., 2003). Our TEM analysis revealed that, whereas most dendrites are located underneath the basal surface of epidermal cells and are in direct contact with the ECM (Figures 2D–2F), there are three types of dendrite enclosure in the epidermal layer. First, we observed thick enclosed dendrites connected to the ECM through a channel formed by opposing epidermal cell membranes (Figure 2H), suggesting the wrapping of existing dendrites by the epidermal basal surface. Second, some dendrites are located between cell junctions of neighboring epidermal cells (Figure 2G), confirming that dendrites can indeed grow between epidermal cells. Interestingly, in the third scenario, some dendrites are completely wrapped by the epidermal cell membrane inside the epidermis (Figures 2I and 2J), indicating that dendrites can also grow by burrowing into epidermal cells.
The observation of enclosed dendrite tips prompted us to ask whether epidermal cells pose a barrier for the growth of enclosed dendrite tips, which may explain the low frequency of dendrite enclosure. We therefore tracked the change of dendrite tips over 12 hours and compared the initially enclosed terminal dendrites to those attached to the ECM (Figure 2C), to see whether the enclosed dendrite tips are more likely to retract or remain stationary. Surprisingly, we found that more than half of the enclosed dendrite tips (61%, n=61) extended from the initial locations, while only 17% of the dendrite tips at the basal surface (n=187) extended along the original direction, suggesting that the preferred positioning of dendrites at the basal surface of epidermal cells cannot be accounted for by a growth disadvantage within the epidermis.
Integrin loss-of-function (LOF) leads to increased epidermal enclosing and dendritic crossings
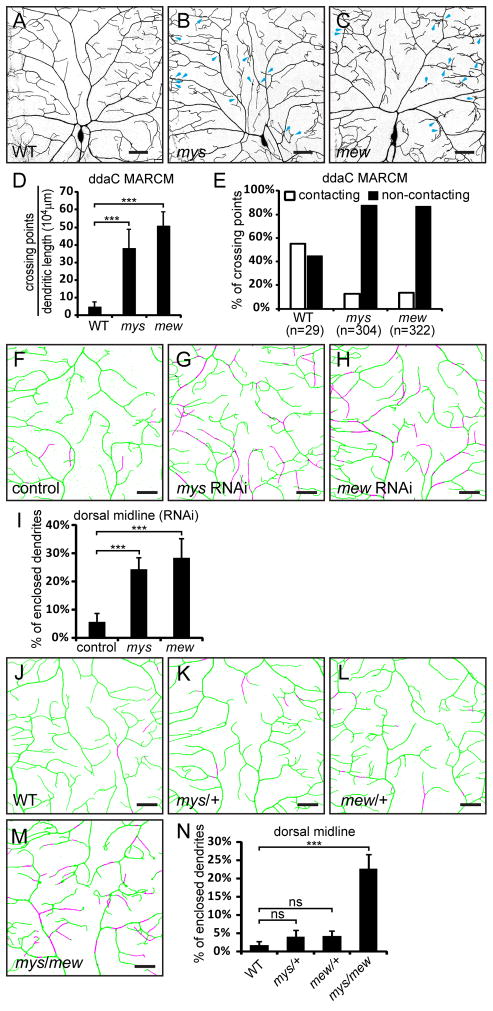
The restricted distribution of dendrites at the basal surface of the epidermis could be due to specific molecular mechanisms promoting the attachment of dendrites to the ECM. Integrins, the major class of cell surface receptors mediating cell-ECM interaction (Barczyk et al., 2010), are likely candidates. In Drosophila, there are five integrin α subunits (α1–5), encoded by multiple edematous wings (mew), inflated (if), scab (scb), αPS4, and αPS5, as well as two integrin β subunits encoded by myospheroid (mys) and βν integrin (βInt-ν) (Brower, 2003). To test if neuronal integrins are required for dendrite-ECM interaction, we first examined the cell-autonomous effects of null mutations of integrin genes mys, mew, if, and scb by mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999). Loss of integrin in class IV da neurons did not appear to affect the gross branching patterns or the total dendritic length (data not shown). However, clones of mys1 or mewM6 class IV da neurons showed a significant increase in dendritic crossings (Figures 3B–3C). We asked whether these crossings involve direct dendro-dendritic contacts by employing high-resolution imaging on the Z axis. Since the three class IV da neurons show different degrees of dendrite enclosure and isoneuronal dendritic crossing (Figure 1), we focused only on the dendritic crossing of ddaC. Most of the dendritic crossings in mys (88.14%) and mew (86.97%) mutant neurons turned out to be non-contacting (Figure 3E). As we could not distinguish contacting dendrites from non-contacting ones that are very close to each other (less than 550 nm apart on the Z axis) due to the diffraction-limited resolution of conventional confocal microscopy, the percentage of non-contacting crossing is likely higher than what we observed. These data suggest that loss of integrins contributes to non-contacting dendritic crossing, possibly by a loss of dendrite-ECM adhesion.
Figure 3. Integrin LOF leads to increased epidermal enclosing and dendritic crossings.
(A–C) Dendritic patterns of wildtype (A), mys1 (B), and mewM6 (C) ddaC neurons generated with MARCM. Some dendritic crossings in (B) and (C) are indicated by blue arrows. Scale bars, 100 μm. (D–E) Quantification of crossing points in wildtype, mys1 and mewM6 mutant neurons. (D) Number of crossing points normalized to total dendritic length. (E) Percentages of contacting (open bars) and non-contacting (filled bars) dendritic crossings. (F–H) Epidermal enclosing of dendrites at the dorsal midline of a control ddaC neuron (F), and neurons that express UAS-mys-RNAi (G) or UAS-mew-RNAi (H). See also Movie S2. (I) Quantification of enclosed dendrites in control and RNAi animals. (J–M) Epidermal enclosing of ddaC dendrites at the dorsal midline in wildtype (J), mys1 hemizygote (K), mewM6 hemizygote (L), and mys1/mewM6 transheterozygote animals (M). (N) Quantification of enclosed dendrites in wildtype, heterozygous and transheterozygous integrin mutant animals. The dendrites attached to the ECM are in green and enclosed dendrites are in magenta (F–H and J–M). Scale bars, 30 μm (F–H and J–M). Error bars represent standard deviations (D, I, and N). ***P<0.001; ns, not significant; one-way analysis of variance and Dunnett’s test.
To assess the role of integrins in dendrite-ECM interaction, we used UAS/Gal4-based RNA interference (RNAi) to knock down mys or mew. Expression of UAS-mys-RNAi or UAS-mew-RNAi in the wing causes severe wing blister (data not shown), a hallmark of defective integrin signaling (Brower, 2003). We knocked-down mys and mew in da neurons with Gal421–7 (Song et al., 2007) and examined the spatial relationship of class IV da dendrites and the ECM in 3rd instar larvae. The ddaC dendrites at the dorsal midline are usually attached to the ECM, with only 1.75% of dendritic length enclosed in the epidermis (Figure 3J). In RNAi control neurons with only UAS-Dicer-2 (UAS-Dcr-2) expression (Dietzl et al., 2007), the enclosed dendritic length is increased to 5.69% (Figure 3F). In contrast, with mys or mew knockdown, the enclosed dendritic length is increased to 24.33% or 28.32%, respectively (Figure 3G, 3H, 3I, and Movie S2), suggesting that defective positioning of dendrites is the underlying cause of the increased non-contacting dendritic crossings.
Integrins function by forming heterodimers of α and β subunits. If integrin α subunit Mew and β subunit Mys regulate dendrite positioning by forming a functional dimer, mutant alleles of mew and mys may genetically interact with each other. Indeed, in transheterozygotes of mys1 and mewM6, the percentage of enclosed dendrites is increased to 22.62%, compared to 4.09% in mys1/+ and 4.18% in mewM6/+ (Figures 3K–3N).
Collectively, these data show that integrin genes mew and mys are important for attaching the class IV da dendrites to the ECM and thus for non-overlapping coverage of dendritic fields.
Overexpression of integrins in class IV da neurons eliminates dendrite enclosure
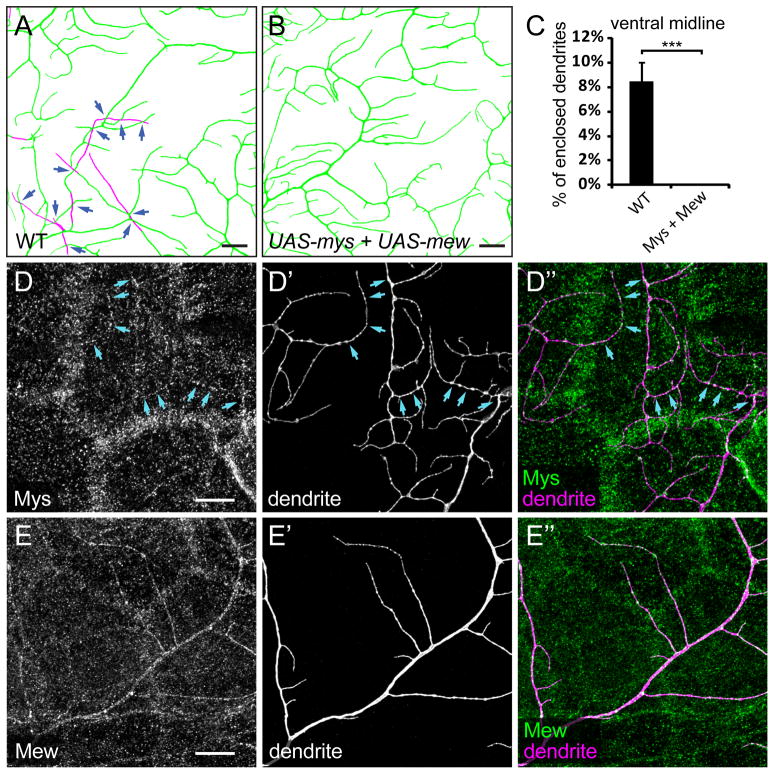
Since removal of mys and mew from class IV da neurons causes detachment of dendrites from the ECM, we tested if supplying more integrins in the dendrites promotes attachment to the ECM, by expressing UAS-mys and UAS-mew, individually or in combination, in class IV da neurons. Overexpression of Mys, but not Mew, in class IV da neurons causes significant dendritic reduction (Figures S1A and S1B). Expressing Mys and Mew simultaneously largely rescues the dendritic reduction associated with Mys overexpression (Figure S1C). Because Mys and Mew function as heterodimers and the balance of their dosages is likely important, we further analyzed the animals in which both Mys and Mew are overexpressed in class IV da neurons. At the ventral midline, the percentage of enclosed dendrites of vdaB is 8.43% (Figures 4A and 4C) in the wildtype control. In contrast, Mys and Mew coexpression in vdaB completely eliminated the dendrite enclosure and associated non-contacting dendritic crossing (Figures 4B and 4C), suggesting that Mys and Mew mediate the attachment of dendrites to the ECM.
Figure 4. Integrin overexpression forces dendrites to grow on the ECM.
(A–B) Epidermal enclosing of dendrites at the ventral midline, showing dendrites of wildtype vdaB neurons (A) and vdaB neurons expressing both UAS-mys and UAS-mew driven by ppk-Gal4 (B). The dendrites attached to ECM are in green and enclosed dendrites are in magenta. The dark blue arrows point to non-contacting dendritic crossings. Scale bars, 10 μm. (C) Quantification of enclosed dendrites in wildtype and integrin-overexpressing animals. Error bars represent standard deviations. ***P<0.001, Student’s t test. (D–E″) Immunostaining of Mys (D–D″) and Mew (E–E″) in animals expressing both UAS-mys and UAS-mew driven by ppk-Gal4. The light blue arrows (D–D″) point to Mys staining on the dendrites. Scale bars, 10 μm. See also Figure S1.
Since studies of loss or gain of integrin function implicate Mys and Mew activity in dendrites, including terminal branches, we asked whether Mys and Mew are localized on class IV da dendrites. Unfortunately, the high levels of epidermal expression of Mys and Mew at the basal surface (Figures S1D–S1E″) render it difficult to distinguish Mys and Mew on dendrites. In neurons overexpressing both Mys and Mew, however, Mys was localized in small patches along the dendrites (Figures 4D––4D″), while Mew was enriched in patches but also present at lower levels uniformly on the dendrites (Figures 4E––4E″). Thus, Mys and Mew can be efficiently delivered to dendrites.
Epidermis-derived laminins are likely the ligands mediating integrin-regulated dendrite-ECM interaction
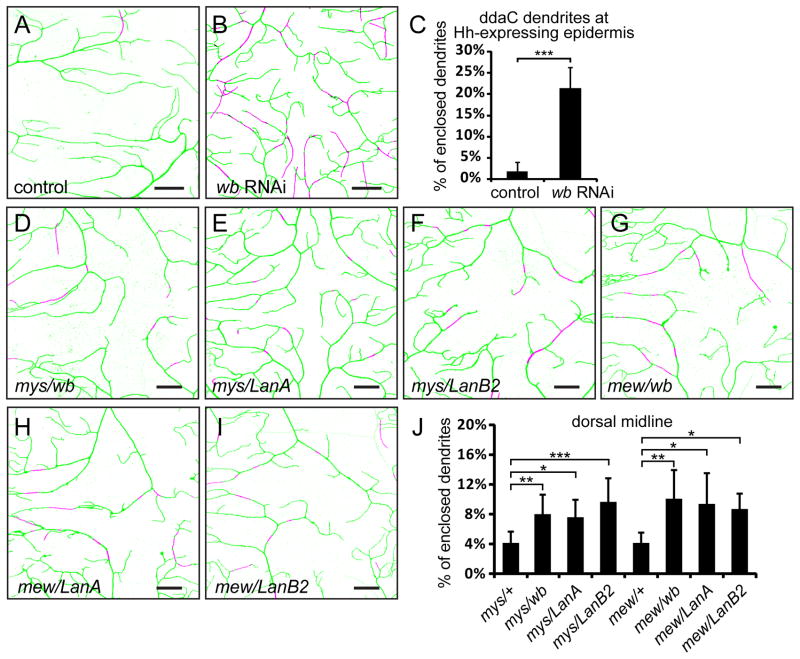
Integrins are cell-surface receptors of ligands residing in the ECM, including collagen, fibronectin, laminin, tenascin, and vitronectin (Humphries et al., 2006). The binding specificity of the integrin heterodimer is largely determined by the α subunit (Barczyk et al., 2010). As the α1 subunit Mew belongs to the laminin-binding type α family (Hynes and Zhao, 2000), we tested the role of laminins, which are protein complexes formed by three subunits (Durbeej, 2010). In Drosophila, wing blister (wb) and Laminin A (LanA) encode two α subunits, whereas LanB1 and LanB2 code for the β and γ subunits, respectively. Since null mutations in laminin genes cause lethality at embryonic or early larval stages, we first sought to use the RNAi strategy to knock down laminins. Expression of a UAS-wb-RNAi construct in the wing caused wing blisters, mimicking the wb LOF phenotype, whereas the available UAS-LanA-RNAi did not yield obvious wing phenotypes (data not shown). We therefore only tested wb RNAi, which did not generate obvious phenotype when expressed in class IV da neurons (data not shown). Since the basal surface of the larval epidermis contacts no other tissues except at the muscle attachment sites, the epidermis is likely the major source of laminins in the ECM. When we drove the expression of UAS-wb-RNAi in posterior epidermal cells of every segment with hh-Gal4, 21.36% of dendrites were found to be enclosed by the epidermis (Figures 5B and 5C), compared to just 1.92% in the control (Figures 5A and 5C). The defective positioning of dendrites is unlikely a result of altered epithelial polarity of epidermal cells, as the localization of several cell polarity markers are normal in cells where wb is knocked down (Figure S2), suggesting that wb is important for dendrite attachment to the ECM.
Figure 5. Epidermis-derived laminins mediate the attachment of dendrites to the ECM.
(A–B) Enclosure of ddaC dendrites in hh-expressing epidermal cells in a control animal (A) and an animal expressing hh-Gal4 UAS-wb-RNAi(B). Scale bars, 20 μm. See also Figure S2 and Movies S3 and S4. (C) Quantification of enclosed dendrites in wildtype and wb knock-down animals. Error bars represent standard deviations. ***P<0.001, Student’s t test. (D–I) Epidermal enclosing of ddaC dendrites at the dorsal midline in transheterozygous mutants of integrin and laminin genes. Scale bars, 30 μm. (J) Quantification of enclosed dendrites in hemizygous integrin mutants and transheterozygous mutants of integrin and laminin genes. Error bars represent standard deviations. *P<0.05, **P<0.01, ***P<0.001, one-way analysis of variance and Dunnett’s test. In all images of dendritic fields, dendrites attached to the ECM are in green and enclosed dendrites are in magenta.
We next asked if laminin mutants genetically interact with mys and mew. Transheterozygotes of mys1/wb09437 and mys1/LanA9–32 showed mild but statistically significant increase in dendrite enclosure at the dorsal midline (Figures 5D, 5E, and 5J) compared to mys1/+. Likewise, mewM6/wb09437 and mewM6/LanA9–32 showed similar enhancement (Figures 5G, 5H, and 5J) compared to mewM6/+. In addition, a hypomorphic allele of LanB2, LanB2MB04039, showed mild genetic interactions with mys1 and mewM6 (Figures 5F, 5I, and 5J). The weak genetic interactions between integrin mutants and laminin mutants may indicate that laminins are abundantly expressed in the epidermis even with only one functional copy.
Taken together, our data support the idea that laminins are the ECM ligands of neuronal integrins in the dendrite-ECM interaction.
The tiling mutants fry, trc, and Sin1 show increased dendrite enclosure in the epidermis
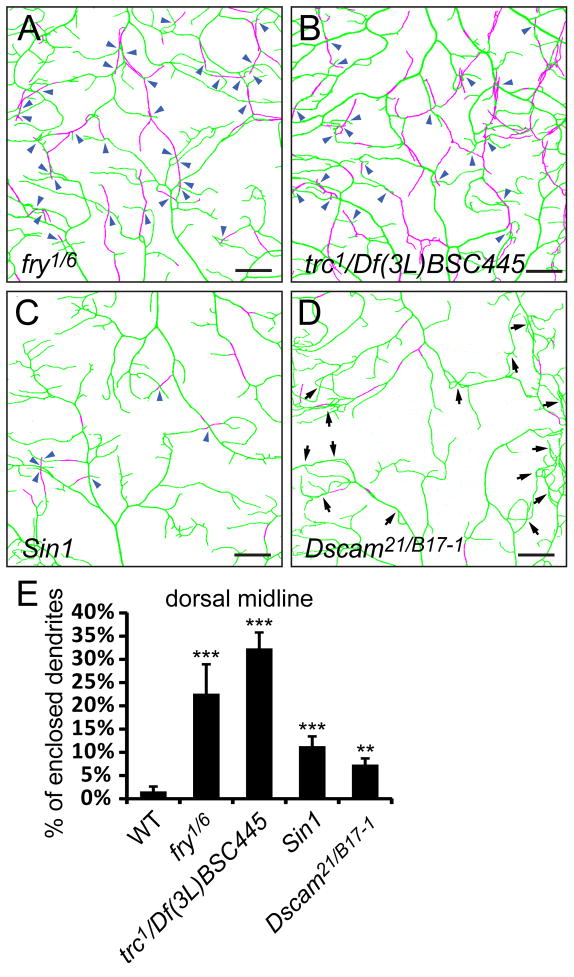
Enclosed dendrites have a higher frequency of non-contacting crossing with other dendrites, as exemplified by the integrin LOF phenotypes. Previous studies of tiling mutants that exhibit increased isoneuronal and heteroneuronal dendritic crossings employed methods with insufficient resolution on the Z axis and could not distinguish contacting and non-contacting dendritic crossings (Emoto et al., 2004; Koike-Kumagai et al., 2009). We therefore reexamined the nature of the dendritic crossings in those tiling mutants by first asking whether the dendrites are properly positioned on the body wall in LOF mutants of fry, trc, and Sin1. Drastic increases of enclosed dendrites were seen at the dorsal midline of fry1/fry6 and trc1/Df(3L)BSC445 mutants (Figures 6A, 6B and 6E). Sin1e03756 mutant larvae also showed a weak yet significant increase of enclosed dendrites (Figures 6C and 6E). Interestingly, most of the dendritic crossings in these mutants are between enclosed dendrites and dendrites attached to the ECM and thus are non-contacting crossings, a result likely caused by enclosure of dendrites in the epidermis. Consistent with the previous report that fry and trc act cell-autonomously in regulating tiling (Emoto et al., 2004), MARCM clones of class IV da neurons mutant for fry or trc show significant increases in enclosed dendrites and the number of non-contacting crossings (Figure S3).
Figure 6. Tiling mutants show increased dendrite enclosure in the epidermis.
(A–D) Enclosure of ddaC dendrites in the epidermis at the dorsal midline in fry1/fry6 (A), trc1/Df(3L)BSC445 (B), Sin1e03756 (C), and Dscam21/DscamB17-1 (D) mutant larvae. The dendrites attached to the ECM are in green and enclosed dendrites are in magenta. Scale bars, 30 μm. Blue arrowheads in (A)–(C) point to non-contacting crossings. Arrows in (D) point to crossings formed by contacting dendrites attached to the ECM. (E) Quantification of enclosed dendrites in fry, trc, Sin1, and Dscam mutants. Error bars represent standard deviations. **P<0.01, ***P<0.001, one-way analysis of variance and Dunnett’s test compared to the wildtype. See also Figure S3 and Movie S5.
Increased dendritic crossing caused by loss of Dscam is unrelated to epidermal enclosing of dendrites
Self-avoidance is required for preventing isoneuronal dendritic crossing. In Dscam mutants, dendrites of da neurons form bundles and cross one another (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). Although we observed a mild increase of enclosed dendrites in Dscam mutants at the dorsal midline (Figures 6D and 6E), most of the dendritic crossings in Dscam mutants (89.1%, n=303) were between contacting dendrites attached to the ECM (arrows in Figure 6D), indicating that lack of repulsion is the primary cause of dendritic crossings in Dscam mutants, as suggested by previous studies (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). We noticed that dendrites of Dscam mutant neurons are more convoluted and the enclosed dendrite segments are more often in the middle of stabilized dendritic branches, indicating that loss of Dscam function may change the stiffness or the tendency of dendrites to curve and indirectly cause more enclosure.
Dendrites of fry mutant animals exhibit normal homotypic repulsion
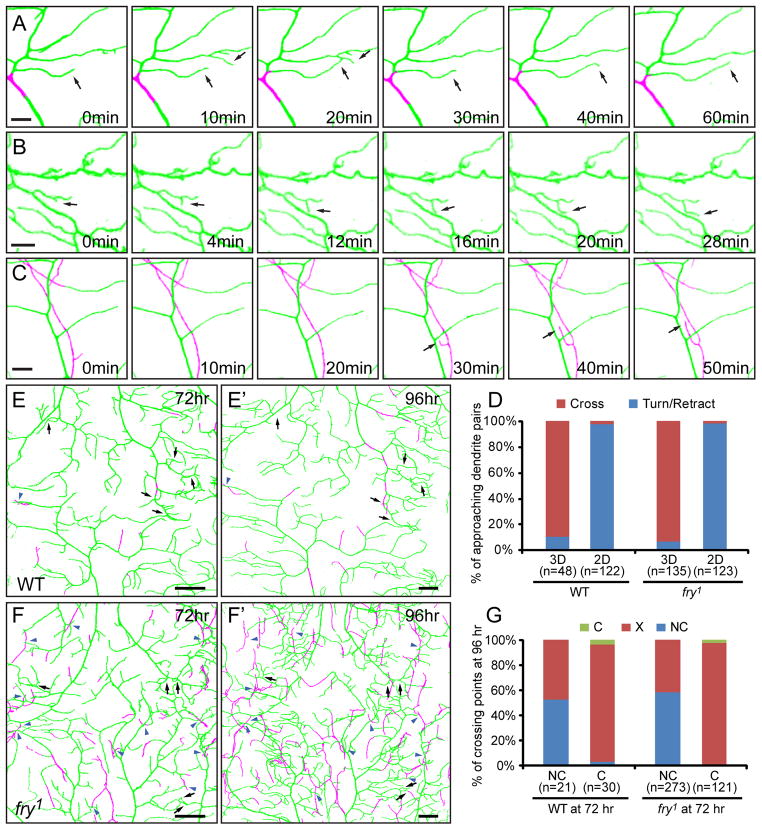
Previous timelapse analyses comparing dendrite distribution over a 16 hour period showed that a much higher percentage of dendrite branches can cross sister dendrites in fry mutants compared to the wildtype, even though turning of dendrites is also present in fry mutants at a lower frequency. This led to the hypothesis fry is required for homotypic repulsion of dendrites (Emoto et al., 2004). To further analyze dendrite interactions with dendrite enclosure taken into account, we conducted short-term timelapse imaging in 3D in fry1 homozygous mutant animals. Approaching dendrites were found to exhibit distinct behaviors depending on whether they are located on the same plane. When two approaching dendrites are both attached to the ECM, they either retract (Figure 7A) or alter the direction of extension (Figure 7B) to avoid crossing (98.4%, n=123). In some cases there was merging of fluorescent signals from two dendrites (Figure 7A as one example), suggesting that direct dendro-dendritic contacts may have occurred preceding retraction or turning. Conversely, when two dendrites are located on different planes, such as one attached to the ECM and the other enclosed, the extending dendrite usually passes the other to result in crossing (Figure 7C, 93.3%, n=135). Similar dendrite interactions were also observed in wildtype animals (Figure 7D). These data show that fry mutant neurons have normal dendritic repulsion and indicate that dendritic crossing in those tiling mutants is caused by impaired confinement of dendritic growth in a 2D space rather than defects in homotypic repulsion.
Figure 7. Class IV da dendrites show normal avoidance behaviors in fry mutants.
(A–C) Timelapse imaging of class IV da dendrites in fry1 mutant animals. These images show the retraction of two approaching dendrites that are both attached to the ECM (A), turning of an extending arbor when coming in close proximity to another branch, with both branches attached to the ECM (B), and passing of an enclosed terminal branch over another dendrite attached to the ECM (C). The time points relative to the first frame in each series are indicated. Scale bars, 5 μm. (D) Quantification of behaviors of approaching dendrites in wildtype and fry1 mutant animals. The dendrite pairs were classified as “both attached to the ECM” (2D) or “located on different focal planes” (3D). In each group, the dendrites either avoid each other (Turn/Retract) or continue extending to form crossing (Cross). (E–F′) Dorsal midline of a wildtype (E and E′) and a fry1 (F and F′) larvae imaged at 72 hr and 96 hr AEL. Contacting crossings (black arrows) and non-contacting crossings (blue arrowheads) are indicated for the dendritic fields at 72 hr AEL and the corresponding positions are also indicated for 96 hr AEL. Scale bars, 30 μm. (G) Quantification of changes of dendritic crossings in wildtype and fry1 mutant animals during 72–96 hr AEL. At 72 hr AEL, dendritic crossings were identified as either non-contacting (NC) or contacting (C). At 96 hr AEL, they either disappear (X) or remain as crossings (NC or C). In all images of dendritic fields, dendrites attached to the ECM are in green and enclosed dendrites are in magenta.
We next asked whether repulsions between contacting dendrites induce dendrite enclosure over time and contribute to the increase of non-contacting crossings in the fry mutant. To address this question, we compared dendrite enclosure and crossing of the same neurons at 72 hours and 96 hours AEL in fry mutant animals. Most contacting crossings (97.5%, n=121) disappeared during the intervening 24 hours due to retraction of dendrites (black arrows in Figures 7F and 7F′). Since retraction is a normal exploratory behavior of class IV da dendrites, which may occur in the absence of repulsion, we also analyzed the dynamics of non-contacting crossings as a control. In contrast to contacting crossings, more than half of non-contacting crossings in fry1 (58.6%, n=273) remained after 24 hours (blue arrowheads in Figure 7F and 7F′). Similar results were obtained from wildtype animals (Figures 7E and 7E′). These data indicate that the repulsive signal between contacting dendrites destabilizes them.
Integrin overexpression rescues the crossing phenotypes of tiling mutants
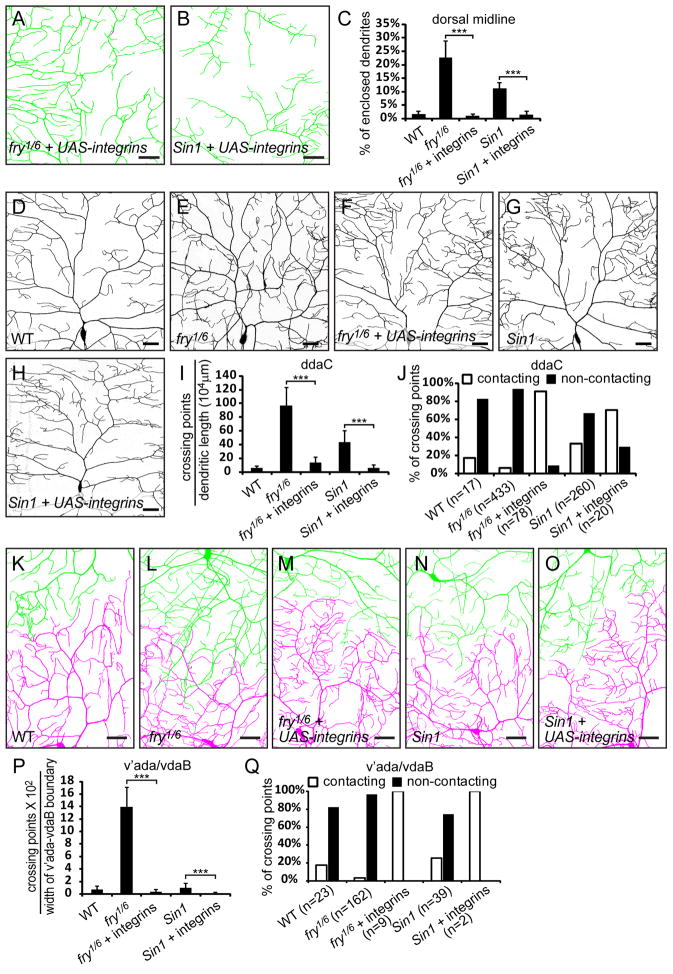
To ask whether forcing the dendrite growth onto the ECM would rescue the crossing phenotypes in mutants of the TORC2/Trc pathway, we first tested if overexpression of Mys and Mew can restore the attachment of ddaC dendrites to the ECM in fry and Sin1 mutants. Indeed, the enclosed dendrites at the dorsal midline were brought back to the wildtype level in fry1/fry6 (1.08%, Figures 8A and 8C) and Sin1e03756 (1.43%, Figures 8B and 8C) mutant animals when UAS-mys and UAS-mew were coexpressed in class IV da neurons.
Figure 8. Integrin overexpression rescues both isoneuronal and heteroneuronal crossing phenotypes in tiling mutants.
(A–B) Dendrite enclosure of ddaC neurons overexpressing both UAS-mys and UAS-mew (UAS-integrins) driven by ppk-Gal4 in fry1/fry6 (A) and Sin1e03756 (B) mutants at the dorsal midline. The dendrites attached to the ECM are in green and enclosed dendrites are in magenta. Scale bars, 30 μm. (C) Quantification of enclosed dendrites. (D–H) Representative dendritic fields of ddaC neurons in wildtype, fry, and Sin1 mutants without and with overexpression of UAS-mys and UAS-mew driven by ppk-Gal4. Scale bars, 30 μm. (I–J) Quantification of crossings of ddaC dendrites, showing the number of dendritic crossings normalized to total dendritic length (I) and percentages of contacting and non-contacting dendritic crossings (J). (K–O) Dendritic patterns at the interface between v’ada (green) and vdaB (magenta) dendritic fields in wildtype and fry, Sin1 mutants without and with overexpression of UAS-mys and UAS-mew driven by ppk-Gal4. Scale bars, 30 μm. (P–Q) Quantification of heteroneuronal crossings between v’ada and vdaB dendrites, showing the number of dendritic crossings normalized to the width of v’ada-vdaB interface (P) and percentages of contacting and non-contacting dendritic crossings (Q). The error bars represent standard deviations (C, I, and P). ***P<0.001, one-way analysis of variance and Bonferroni test for selected pairs of columns.
We next examined isoneuronal dendritic crossing in ddaC neurons. Consistent with previous reports (Emoto et al., 2004; Koike-Kumagai et al., 2009), both fry and Sin1 mutant larvae showed higher frequency of dendritic crossing within the ddaC dendritic field (Figures 8E, 8G, and 8I) than the wildtype (Figures 8D and 8I). The majority of these crossings do not involve direct dendritic contacts (Figure 8J). Overexpression of Mys and Mew in ddaC largely rescued the crossing defects (Figures 8F, 8H, and 8I). The crossing dendrites contact each other at the majority of the remaining few crossing points in the rescued animals (Figure 8J).
Lastly, we tested if integrin overexpression can rescue the tiling defects in fry and Sin1 mutants by examining the interface between v’ada and vdaB neurons. fry1/fry6 larvae show extensive overlap of v’ada and vdaB dendritic fields (Figure 8L), which is also caused by non-contacting dendritic crossings (Figures 8P and 8Q). Overexpression of Mys and Mew in class IV da neurons completely rescued this phenotype (Figures 8M and 8P). We did not observe a significant increase of heteroneuronal crossings in Sin1e03756 mutant larvae at the v’ada/vdaB interface (Figures 8N and 8P), but found a reduction of such crossings by overexpression of Mys and Mew (Figures 8O and 8P). It is worth noting that although integrins rescued both isoneuronal and heteroneuronal dendritic crossing in fry mutant animals, they did not appear to rescue the overbranching phenotype (Figures 8F and 8M), a defect associated with fry and trc that was shown to be independent of the crossing phenotype (Emoto et al., 2004).
Taken together, our results indicate that tiling mutants of the TORC2/Trc pathway cause dendritic crossings that result in overlapping dendritic fields primarily by releasing dendrites from their confinement to the 2D space specified by the ECM.
DISCUSSION
Self-avoidance and tiling are fundamental mechanisms governing the proper patterning of dendritic fields. Both mechanisms involve homotypic repulsion of dendrites to ensure non-redundant coverage of dendritic fields. In principle, such repulsion could arise from contact-dependent repulsion and/or short-range diffusible repulsive signals. For Drosophila class IV da neuron, there is substantial evidence for the involvement of contact-dependent dendritic repulsion (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007, this study). For the contact-dependent dendritic repulsion to work with high fidelity, it is essential that growing dendrites encounter each other reliably when they enter a shared territory, which is only possible if they grow on the same substrate in a restricted space such as a 2D sheet.
In this study we demonstrate the dendrites of class IV da neurons mostly grow between the basal surface of the epidermal cells and the ECM secreted by the epidermis, which effectively limits the dendrites to a 2D sheet. This restriction is imposed by the interaction between neuronal integrins and epidermal cell-derived laminins in the ECM. Loss of this interaction leads to dendrites’ detachment from the ECM and increased enclosure of dendrites by epidermal cells. As a result, the dendrites are no longer restricted in a 2D space and can cross other dendrites without direct dendro-dendritic contacts. Conversely, increasing the adhesive force between dendrites and the ECM by supplying more integrins to the dendrites eliminates enclosure of dendrites in the epidermis.
The role of TORC2/Trc pathway in limiting the class IV da dendrites to a 2D space
Previous studies revealed that class IV da neurons deficient for components of the TORC2/Trc pathway display many more “dendritic crossings” (as viewed as projections onto the x–y plane) between sister isoneuronal branches as well as between branches of adjacent class IV da neurons (Emoto et al., 2004; Koike-Kumagai et al., 2009), leading to the hypothesis that the “dendritic crossing” phenotype is caused by a defect in the like-repels-like mechanisms. However, the technical limitations of those earlier studies in the resolution along the Z-axis precluded the possibility to distinguish the “dendritic crossings” when two dendrites are separated by a small distance along the Z-axis from the cases in which the two dendrites actually make contact. With improved capacity of high resolution imaging, we take into consideration the 3D nature of the larval epidermis and demonstrate that the crossing defects in those tiling mutants arise from a substantial increase in non-contacting overlap of dendrites located at different depths of the epidermal layer. In fact, both the isoneuronal and heteroneuronal dendritic crossing in mutants of the TORC2/Trc pathway can be accounted for by growth of dendrites in a 3D space instead of defective dendritic repulsion. Importantly, forced dendrite attachment to the ECM in those mutants by integrin overexpression effectively restores the non-redundant coverage of dendritic fields.
How do fry and trc promote dendrite attachment to the ECM? One possibility is that fry and trc function upstream of integrins to regulate integrin interaction with ECM. However, the rescue of the fry phenotype by integrin overexpression suggests that integrin activation does not rely on Fry activity. Alternatively, fry and trc may regulate other adhesion molecules in a pathway parallel to integrin-mediated adhesion. A recent study has also implicated turtle (tutl), a gene encoding a transmembrane Ig protein, in preventing isoneuronal dendritic crossing of class IV da neurons (Long et al., 2009). In light of our 3D analysis of dendrite distribution, it remains to be determined whether tutl is required for dendro-dendritic repulsion or proper attachment of dendrites to the ECM.
Functional significance of integrin-mediated dendrite-ECM interaction
Integrin overexpression experiments suggest that the amount of integrins on dendrites may be a limiting factor determining whether a branch will be attached to the ECM or enclosed in the epidermis. Interestingly, wildtype neurons have a small percentage of dendrites enclosed in the epidermis. The degree of dendrite enclosure appears to roughly correlate with the location of the dendritic field along the dorsal/ventral axis of the body wall. This raises the question whether a certain level of dendrite enclosure is desirable for the function of class IV da neurons. These neurons may fine-tune the degree of dendrite enclosure through controlled integrin expression to achieve the most efficient sensing of certain sensory inputs such as mechanical stimuli. The physiological significance of dendrite enclosure awaits future studies.
Comparison with tiling in other nervous systems
The crossing defects of integrin and Sin1/fry/trc mutant neurons illustrate the importance of spatial restriction for a tiling system based on homotypic repulsion. For neurons normally tiling a 2D territory, without spatial restraints, repulsion would drive homologous dendrites to disperse into a 3D space with potentially considerable overlap of dendritic coverage in a given receptive field. In the case of Drosophila class IV da neurons, the restricted space is a 2D sheet between the basal surface of epidermal cells and the ECM. As highlighted by our study, tiling requires high precision in this spatial restriction: a slight deviation of dendrites from this 2D space is sufficient to circumvent homotypic repulsion and cause overlap of dendritic fields.
What about other tiling systems? One vertebrate tiling system that has some similarity with the Drosophila class IV da neurons is the fish somatosensory neurons, which innervate the skin with axon arbors (Sagasti et al., 2005). Like the dendrites of class IV da neurons, these peripheral axons exhibit contact-dependent repulsion and expansion after ablation of adjacent neurons. It will be interesting to determine whether the processes of these sensory neurons are restricted to a 2D layer within the skin and whether the interaction of those neurites with ECM or skin cells are important for their tiling.
The vertebrate retina is a classical system for studying neuronal tiling. Certain types of retinal ganglion cells (RGCs) and amacrine cells perfectly tile the retina in the x–y plane. Ablation studies suggest that dendro-dendritc repulsion exists between dendrites of neighboring RGCs (Perry and Linden, 1982). The dendrites of RGCs and amacrine cells innervate various layers of retinal inner plexiform in a neuronal type-specific manner (Dacey et al., 2003; Kolb et al., 1992; Mariani, 1990). It will be intriguing to find out whether the dendrites of a given type of ganglion cells or amacrine cells that tile are indeed restricted to a molecularly defined 2D layer(s) in the inner plexiform, and, if so, whether interactions between cell adhesion molecules and the ECM contribute to the restricted dendritic distribution and tiling.
Theoretically, spatial restraints should be a general prerequisite for homotypic repulsion, which may or may not be limited to 2D. Tiling of neuritic fields could conceivably be established by homotypic repulsion in restricted 3D spaces, such as columnar tiling of the transient neurites of murine retinal horizontal cells during postnatal development (Huckfeldt et al., 2009). However, besides spatial restraints, there could be additional mechanisms to enhance the effectiveness of homotypic repulsion in partitioning 3D neuritic fields. One potential mechanism is to have more homologous neurites within a given space, which would increase the possibility of neurite encounter and repulsion. Homotypic interaction could also be enhanced by dynamic behaviors of neurites, such as constant exploratory behaviors and rapid retraction upon contact. These properties of neurites may help to maintain dynamic boundaries between neuritic fields of like neurons. Lastly, if homotypic repulsion also involves short-range diffusible molecules, such signals secreted by the arbor of a neuron may create a 3D pocket inaccessible to the neurites of like neurons.
Mechanisms of spatial restriction
Integrin-ECM interaction plays a critical role in restricting class IV da dendrites to a 2D space. Similar neurite-ECM interactions may be at work to create spatial restraints in other neuronal systems that display homotypic repulsion. However, Drosophila class IV da neurons are sensory neurons that receive sensory rather than synaptic inputs, and thus may bear significant difference in the patterning of dendritic fields from neurons in the central nervous system (CNS). It is conceivable that CNS neurons may employ alternative or additional mechanisms than neurite-ECM interaction to create spatial restriction. One mechanism may be the interaction between pre- and postsynaptic partners. For example, homophilic interactions mediated by Ig domain-containing adhesion molecules between pre- and post-synaptic partners are critical for restricting dendrites of some RGCs and amacrine cells to specific sublaminae of the inner plexiform layer (Fuerst et al., 2010; Yamagata and Sanes, 2008, 2010; Yamagata et al., 2002). Another example is cerebellar Purkinje cells, which align complex dendritic arbors in sagittal planes and show minimal overlap between sister dendrites; this monoplanar arrangement of arborization depends on afferent inputs from climbing fiber axons (Kaneko et al., 2011). Neurite growth could also be constrained by the availability of growth promoting or inhibiting, or guidance factors, which may only be present on certain substrates or in limited spaces.
Concluding remarks
Together with previous studies demonstrating the existence of homotypic repulsion between class IV da dendrites, our study provides a more complete view of tiling by revealing the essential role of spatial constraints to ensure such dendritic interaction. On the one hand, tiling involves recognition and repulsion of homologous dendrites through as yet unidentified molecular pathway(s); on the other, it critically relies on spatial confinement of dendrites imposed by the cell adhesion machinery to facilitate interactions among dendrites encroaching on overlapping territories.
EXPERIMENTAL PROCEDURES
Fly Stocks
mys1 (Bunch et al., 1992), mewM6 (Brower et al., 1995), UAS-βPS (UAS-mys) (Beumer et al., 1999), UAS-αPS1(UAS-mew) (Martin-Bermudo et al., 1997), wb09437(Martin et al., 1999), LanA9–32 (Henchcliffe et al., 1993) trc1 (Geng et al., 2000), fry1(Cong et al., 2001), fry6 (Emoto et al., 2004), Dscam21 (Hummel et al., 2003), DscamB17-1 (Wang et al., 2004), Sin1e03756 (Hietakangas and Cohen, 2007), Gal421-7 (Song et al., 2007), UAS-EGFP (Halfon et al., 2002), and UAS-CD4-tdTom (Han et al., 2011) have been described previously. Df(3L)BSC445 and LanB2MB04039 were obtained from Bloomington Stock Center. UAS-mys-RNAi (v29619), UAS-mew-RNAi (v44890), UAS-wb-RNAi (v3141), and UAS-Dcr-2 (Dietzl et al., 2007) were obtained from Vienna Drosophila RNAi Center (VDRC). vkg-GFPG00205 (Morin et al., 2001) was obtained from FlyTrap (Kelso et al., 2004). For imaging class IV da dendrites, we used either CD4-tdTom (Han et al., 2011) or spGFP11-CD4-tdTom driven by a ppk enhancer (Grueber et al., 2003). spGFP11-CD4-tdTom is otherwise the same as CD4-tdTom except for the use of a synthetic signal peptide and the small split-GFP fragment (Feinberg et al., 2008) before CD4 and the lack of an ER exit signal from Kir2.1. Both ppk-CD4-tdTom and ppk-spGFP11-CD4-tdTom were constructed in pHemmar, a dual-platform transgenic vector that endows high expression in the Drosophila nervous system (Han et al., 2011). ppk-CD4-tdTom and ppk-spGFP11-CD4-tdTom behave similarly in labelling class IV da dendrites and both are referred to as ppk-CD4-tdTom in the text.
Molecular Cloning
To make the more specific and stronger ppk-Gal4 than one described previously (Grueber et al., 2007), pDEST-Hemmar (Han et al., 2011) was modified to make pDEST-APIGH, a Gal4-coding destination vector to be driven by any enhancer. The ppk enhancer was then cloned into pDEST-APIGH by Gateway cloning (Invitrogen). To make UAS-HRP-DsRed-GPI, a HRP-DsRed-GPI fusion gene was assembled in pCS2 vector by sequential restriction cloning to contain, from 5′ to 3′, the signal peptide sequence of wingless (AA1–AA37), HRP cDNA, DsRedT1 cDNA (Clontech), GPI sequence of dally-like (AA695–AA765). The HRP-DsRed-GPI fragment was then cloned into pUAST (Brand and Perrimon, 1993) between EcoRI and XbaI. Trangenic animals were obtained via P-element-mediated transformation with a standard protocol.
MARCM Analysis
MARCM analyses of mys and mew were performed as described previously (Grueber et al., 2002). mys1 FRT19A/FM7c or mewM6 FRT19A/FM7c female flies were crossed with tub-Gal80 FRT19A; hs-Flp Gal4109(2)80 UAS-mCD8-GFP to generate marked neurons mutant for mys or mew, respectively. Embryos were collected for 2 hr and allowed to develop for 3hr at 25°C, then heat-shocked for 1 hr at 38°C. Heat-shocked embryos were then reared on grape agar plates at 25°C until the time of living imaging at about 96 hr AEL.
RNAi
RNAi knock-down of mys and mew in da neurons were carried out with driver Gal421-7 UAS-Dcr-2. Knock-down of wb in the larval epidermis was carried out with driver UAS-Dcr-2; hh-Gal4 UAS-EGFP. UAS-EGFP was used to label epidermal cells that express the RNAi constructs. The effectiveness of UAS-mew-RNAi and UAS-wb-RNAi in the wing was tested with UAS-Dcr-2; hh-Gal4 UAS-EGFP. As cross between UAS-mys-RNAi and UAS-Dcr-2; hh-Gal4 UAS-EGFP produces progeny dying at early larval stages, knock-down by UAS-mys-RNAi was tested with a wing-specific Gal4, MS1096 (Lunde et al., 1998).
Live Imaging
Animals were reared at 25°C in density-controlled vials. The vials containing the first batch of embryos were discarded as the dendritic morphology of da neurons is less consistent in those animals. 3rd instar larvae at 96hr AEL (unless specified otherwise) were mounted in halocarbon oil and confocal images of class IV da dendrites were collected with a Leica SP5 laser scanning microscope. For high-resolution imaging on the Z axis, the larvae were lightly anesthetized with isoflurane before mounting. Image stacks with a Z step size between 0.16–0.2 μm were acquired with a 40X 1.25NA oil lens. For quantification of dendritic phenotypes, 8–10 image stacks were collected from class IV da neurons in A2–A3 segments for every genotype. For short-term time-lapse imaging of dendritic dynamics, the larvae were mounted in a imaging chamber constructed with a thin aluminum slide with a hole in the middle. The bottom of the hole was covered with an oxygen-permeable membrane (model 5793; YSI). The larvae were mounted on the membrane in halocarbon oil.
Image Analysis
Confocal image stacks were deconvoluted with Autoquant (MediaCybernetics) and analyzed in Imaris (Bitplane). Detailed methods for image analysis and quantification are described in Supplemental Experimental Procedures.
Immunohistochemistry
Antibodies used in this study are mouse anti-Mys (1:50, CF.6G11, DSHB), mouse anti-Mew (1:10, DK.1A4, DSHB), rabbit anti-DsRed (1:200, Clontech). Secondary antibodies conjugated to DyLight dyes (Jackson ImmunoResearch) were used at 1:400 dilution. Immunostaining of Drosophila larvae was performed as described (Grueber et al., 2002). Briefly, 3rd instar larvae were dissected in cold PBS, fixed in 4% formaldehyde/PBS for 20 min at room temperature (RT), and stained with the proper primary antibodies and subsequent secondary antibodies, each for 2 hr at room temperature.
Transmission Electron Microscopy
Detailed methods for TEM are described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Wes Gruber for communicating results prior to publication. We thank members of the Jan lab for discussion; Chung-hui Yang for help in cloning and making transgenic lines; Yang Xiang for testing of electrophysilogical experiments; Sandra Barbel for help in dendrite tracing; Mark Krasnow, Frieder Schoeck, Jian Wang, Bloomington Stock Center, VDRC, and FlyTrap for fly stocks; DSHB for antibodies. This work was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund to C.H., a California Institute for Regenerative Medicine (CIRM) grant to S.Z., NIH grant (2R01 GM063891), American Cancer Society (RSG-07-051), and the Knowledge Innovation Program of the Chinese Academy of Sciences KSCX2-YW-R-263 to X. L., and by NIH grant (2R37NS040929) to Y.N.J.. L.Y.J. and Y.N.J. are investigators of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Rohrbough J, Prokop A, Broadie K. A role for PS integrins in morphological growth and synaptic function at the postembryonic neuromuscular junction of Drosophila. Development. 1999;126:5833–5846. doi: 10.1242/dev.126.24.5833. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brower DL. Platelets with wings: the maturation of Drosophila integrin biology. Curr Opin Cell Biol. 2003;15:607–613. doi: 10.1016/s0955-0674(03)00102-9. [DOI] [PubMed] [Google Scholar]
- Brower DL, Bunch TA, Mukai L, Adamson TE, Wehrli M, Lam S, Friedlander E, Roote CE, Zusman S. Nonequivalent requirements for PS1 and PS2 integrin at cell attachments in Drosophila: genetic analysis of the alpha PS1 integrin subunit. Development. 1995;121:1311–1320. doi: 10.1242/dev.121.5.1311. [DOI] [PubMed] [Google Scholar]
- Bunch TA, Salatino R, Engelsgjerd MC, Mukai L, West RF, Brower DL. Characterization of mutant alleles of myospheroid, the gene encoding the beta subunit of the Drosophila PS integrins. Genetics. 1992;132:519–528. doi: 10.1093/genetics/132.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabodi S, Di Stefano P, Leal Mdel P, Tinnirello A, Bisaro B, Morello V, Damiano L, Aramu S, Repetto D, Tornillo G, Defilippi P. Integrins and signal transduction. Adv Exp Med Biol. 2010;674:43–54. doi: 10.1007/978-1-4419-6066-5_5. [DOI] [PubMed] [Google Scholar]
- Cong J, Geng W, He B, Liu J, Charlton J, Adler PN. The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development. 2001;128:2793–2802. doi: 10.1242/dev.128.14.2793. [DOI] [PubMed] [Google Scholar]
- Dacey DM, Peterson BB, Robinson FR, Gamlin PD. Fireworks in the primate retina: in vitro photodynamics reveals diverse LGN-projecting ganglion cell types. Neuron. 2003;37:15–27. doi: 10.1016/s0896-6273(02)01143-1. [DOI] [PubMed] [Google Scholar]
- Denda S, Reichardt LF. Studies on integrins in the nervous system. Methods Enzymol. 2007;426:203–221. doi: 10.1016/S0076-6879(07)26010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Durbeej M. Laminins. Cell Tissue Res. 2010;339:259–268. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Harris BS, Johnson KR, Burgess RW. A novel null allele of mouse DSCAM survives to adulthood on an inbred C3H background with reduced phenotypic variability. Genesis. 2010;48:578–584. doi: 10.1002/dvg.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewiel MS, Beck CD, Wu KH, Zhu XR, Davis RL. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Sagasti A. Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol. 2010;2:a001750. doi: 10.1101/cshperspect.a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Gisselbrecht S, Lu J, Estrada B, Keshishian H, Michelson AM. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis. 2002;34:135–138. doi: 10.1002/gene.10136. [DOI] [PubMed] [Google Scholar]
- Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci U S A. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchcliffe C, Garcia-Alonso L, Tang J, Goodman CS. Genetic analysis of laminin A reveals diverse functions during morphogenesis in Drosophila. Development. 1993;118:325–337. doi: 10.1242/dev.118.2.325. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev. 2007;21:632–637. doi: 10.1101/gad.416307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckfeldt RM, Schubert T, Morgan JL, Godinho L, Di Cristo G, Huang ZJ, Wong RO. Transient neurites of retinal horizontal cells exhibit columnar tiling via homotypic interactions. Nat Neurosci. 2009;12:35–43. doi: 10.1038/nn.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Yamaguchi K, Eiraku M, Sato M, Takata N, Kiyohara Y, Mishina M, Hirase H, Hashikawa T, Kengaku M. Remodeling of monoplanar Purkinje cell dendrites during cerebellar circuit formation. PLoS One. 2011;6:e20108. doi: 10.1371/journal.pone.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhammer JP, Schwab ME. Modulators of neuronal migration and neurite growth. Curr Opin Cell Biol. 1992;4:863–868. doi: 10.1016/0955-0674(92)90112-p. [DOI] [PubMed] [Google Scholar]
- Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32:D418–420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Kumagai M, Yasunaga K, Morikawa R, Kanamori T, Emoto K. The target of rapamycin complex 2 controls dendritic tiling of Drosophila sensory neurons through the Tricornered kinase signalling pathway. EMBO J. 2009;28:3879–3892. doi: 10.1038/emboj.2009.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H, Linberg KA, Fisher SK. Neurons of the human retina: a Golgi study. J Comp Neurol. 1992;318:147–187. doi: 10.1002/cne.903180204. [DOI] [PubMed] [Google Scholar]
- Kramer AP, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. II. Experimentally induced variations in the branching pattern. J Neurosci. 1985;5:768–775. doi: 10.1523/JNEUROSCI.05-03-00768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CW, Hirst E, Alexandre C, Vincent JP. Segment boundary formation in Drosophila embryos. Development. 2003;130:5625–5635. doi: 10.1242/dev.00867. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Long H, Ou Y, Rao Y, van Meyel DJ. Dendrite branching and self-avoidance are controlled by Turtle, a conserved IgSF protein in Drosophila. Development. 2009;136:3475–3484. doi: 10.1242/dev.040220. [DOI] [PubMed] [Google Scholar]
- Lunde K, Biehs B, Nauber U, Bier E. The knirps and knirps-related genes organize development of the second wing vein in Drosophila. Development. 1998;125:4145–4154. doi: 10.1242/dev.125.21.4145. [DOI] [PubMed] [Google Scholar]
- Mariani AP. Amacrine cells of the rhesus monkey retina. J Comp Neurol. 1990;301:382–400. doi: 10.1002/cne.903010305. [DOI] [PubMed] [Google Scholar]
- Martin-Bermudo MD, Dunin-Borkowski OM, Brown NH. Specificity of PS integrin function during embryogenesis resides in the alpha subunit extracellular domain. EMBO J. 1997;16:4184–4193. doi: 10.1093/emboj/16.14.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Zusman S, Li X, Williams EL, Khare N, DaRocha S, Chiquet-Ehrismann R, Baumgartner S. wing blister, a new Drosophila laminin alpha chain required for cell adhesion and migration during embryonic and imaginal development. J Cell Biol. 1999;145:191–201. doi: 10.1083/jcb.145.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto T, Kain KH, Ginsberg MH. Neurobiology: New connections between integrins and axon guidance. Curr Biol. 2004;14:R121–123. [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, Linden R. Evidence for dendritic competition in the developing retina. Nature. 1982;297:683–685. doi: 10.1038/297683a0. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr Biol. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Sdrulla AD, Linden DJ. Dynamic imaging of cerebellar Purkinje cells reveals a population of filopodia which cross-link dendrites during early postnatal development. Cerebellum. 2006;5:105–115. doi: 10.1080/14734220600620908. [DOI] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Jacobs JR. Integrins regulate responsiveness to slit repellent signals. J Neurosci. 2002;22:4448–4455. doi: 10.1523/JNEUROSCI.22-11-04448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci. 2010;30:3579–3588. doi: 10.1523/JNEUROSCI.6319-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.