Abstract
Cryptosporidium is a major cause of diarrhea in children in developing countries. However, there is no vaccine available and little is known about immune responses to protective antigens. We investigated antibody responses to p23, a putative vaccine candidate, in children in Bangladesh with cryptosporidiosis and diarrhea (cases) and uninfected children with diarrhea (controls), and p23 gene polymorphisms in infecting species. Serum IgM, IgG, and IgA responses to p23 were significantly greater in cases than controls after three weeks of follow-up. Cases with acute diarrhea had significantly greater serum IgA and IgM responses than those with persistent diarrhea, which suggested an association with protection from prolonged disease. The p23 sequences were relatively conserved among infecting species and subtype families. Although most children were infected with Cryptosporidium hominis, there was a cross-reactive antibody response to C. parvum antigen. These results support further development of p23 as a vaccine candidate.
Introduction
Cryptosporidium spp. of the phylum Apicomplexa are a significant cause of diarrheal illness worldwide,1,2 particularly in untreated patients with human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS) and children in resource-poor countries.3–5 In these countries, cryptosporidiosis in early childhood is associated with subsequent malnutrition, impairment in growth, physical fitness, and cognitive development.3,4 The immune system of the host is critical in mediating protection from and resolution of cryptosporidiosis.6 In immunocompetent hosts, the infection is generally asymptomatic or self-limited. However, in immunocompromised hosts including patients with HIV/AIDS, the disease can be chronic, severe, and possibly fatal.7
Therapeutic options for cryptosporidiosis are limited. Nitazoxanide is effective in immunocompetent hosts and is the only drug that has been approved by the Food and Drug Administration for treatment of cryptosporidiosis in the United States.8 However, this drug is not effective in the immunocompromised host9 and has not been widely tested in children in developing countries. Children in these countries are considered an important target group for vaccine development.10 However, there is no vaccine available for cryptosporidiosis. Thus, identification of putative protective antigens and characterization of human immune responses to them is essential for development of effective vaccines for this disease in vulnerable populations.
One of these antigens is p23 (also known as Cp23, p27 or 27-kDa antigen), which is a surface-associated, immunodominant antigen present on invasive stages of the parasite and shed from their surface during gliding motility.11–14 Monoclonal and bovine colostral antibodies to p23 protect against Cryptosporidium parvum challenge in mice and calves, respectively.11,12,15–18 The p23 antigen induces serum, mucosal, humoral, and cell-mediated immune responses in experimentally infected or immunized animals,19–26 and active immunization with DNA or peptide vaccines targeting p23 has been shown to confer varying degrees of protection in animal models.27–29
The p23 antigen is one of the most immunodominant Cryptosporidium antigens and is consistently recognized by serum from actively infected or previously exposed humans.30–36 The presence of pre-existing antibodies to this antigen was associated with reduced oocyst shedding and protection from diarrhea in infected human volunteers.37 In addition, serum antibody responses to p23 were associated with a reduced risk of diarrheal illness in immunosuppressed persons.38 The p23 antigen also induced cell-mediated and antibody responses in persons previously exposed to Cryptosporidium spp.39 Taken together, these findings identify p23 as a putative vaccine candidate. This antigen is considered one of the most promising candidates for vaccine development.40 However, there have been few clinical studies of immune responses to this antigen in well-defined cohorts, particularly in children in resource-poor countries.
Most Cryptosporidium infections are caused by two species: C. hominis almost exclusively infects humans, whereas C. parvum infects humans in addition to animals.41 Most human infections, particularly in resource-poor countries, are caused by C. hominis. Genes encoding other surface-associated Cryptosporidium proteins including gp4042,43 and Muc4 and 544 are highly polymorphic among clinical isolates of different species and subtypes. If p23 is to be considered as a vaccine candidate, it is essential to investigate polymorphisms in the gene that encodes it among clinical isolates in different geographic areas. A previous investigation of Cryptosporidium isolates found ten nucleotide polymorphisms between C. hominis and C. parvum, three of which result in amino acid changes.45 However, polymorphisms in this gene have not been extensively investigated in samples from clinical studies in different geographic areas.
The present study is part of a prospective case–control study to evaluate the clinical, epidemiologic, and immunologic features of cryptosporidiosis in children with diarrhea coming to the Dhaka Hospital of the International Center for Diarrheal Disease Research (ICDDR, B) in Dhaka, Bangladesh. We previously reported the clinical and epidemiologic features and immune responses to total antigens in oocyst lysate preparations from C. parvum46 and C. hominis43 and to gp15,43 another immunodominant Cryptosporidium antigen in the same cohort of children.
The aim of the current study was to investigate serum IgG, IgM, and IgA responses to p23 in serum samples from these children using recombinant C. parvum p23 (rp23) protein as antigen in enzyme-linked immunosorbent assays (ELISAs), to determine if there are any clinical or epidemiologic associations with antibody responses to p23 and to compare antibody responses to p23 with those to C. parvum lysate and gp15. An additional aim was to characterize polymorphisms in the p23 nucleotide and deduced amino acid sequences among the Cryptosporidium spp. infecting these children.
Methods
Patients
Children 15 days to 5 years of age with diarrhea who came to the Dhaka Hospital of the ICDDR, B were enrolled in the original study.46 Details of the recruitment procedure have been described.46 Informed consent was obtained from the parents or guardians of all children recruited to the study according to the guidelines of the Ethical Review Committee of the ICDDR, B, which approved the study.46 Diarrhea was defined as ≥ 3 watery stools within a 24-hour period. A diarrheal episode was defined as diarrhea for ≥ 72 hours. The end of a diarrheal episode was defined as absence of diarrhea for 48 hours. Acute diarrhea was defined as a diarrheal episode lasting < 14 days. Persistent diarrhea was defined as a diarrheal episode lasting ≥ 14 days.
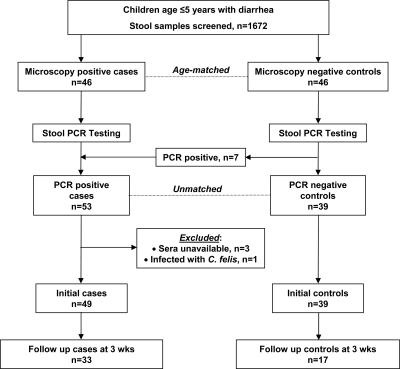
In the original study, 46 children with diarrhea and stools positive for Cryptosporidium spp. by microscopy were enrolled as cases and an equal number of age-matched children with diarrhea but whose stools were negative for Cryptosporidium spp. by microscopy were enrolled as controls.46 Subsequently, Cryptosporidium was detected by polymerase chain reaction (PCR) at the 18s ribosomal RNA locus in all 46 case-patients and in 7 control subjects.47 Thus, for subsequent studies, the study design was unmatched to include 53 cases and 39 controls. For this study, serum was available for 49 cases and 39 controls at hospitalization and for 32 cases and 17 controls at follow-up (Figure 1).
Figure 1.
Schematic showing numbers of cases and controls at initial and follow-up time points enrolled in the current study of children with cryptosporidiosis in Bangladesh.
Recombinant p23
The plasmid containing the p23 insert cloned into the pGEX 4T-2 vector was obtained from Dr. Jeffrey Priest (Centers for Disease Control and Prevention, Atlanta, GA) under a Materials Transfer Agreement with North Carolina State University (Raleigh, NC) and over-expressed in Escherichia coli BL21 cells as described.30 The recombinant p23 (rp23)–glutathione-S-transferase fusion protein was purified by affinity chromatography using a glutathione Sepharose 4B column (Amersham Biosciences, Piscataway, NJ). The glutathione-S-transferase tag was cleaved off with thrombin as described.30 Protein concentration was measured using the Micro BCA protein assay kit (Pierce, Rockford, IL).
Gene sequencing of p23
DNA was extracted from two stool samples representative of each of five C. hominis and one C. parvum subtype families identified from children in the study47 and from three children infected with the subtype IIc family in Vellore, India48 by using a QIAmp Stool Extraction Kit (QIAGEN, Valencia, CA). A 549-basepair fragment of the p23 gene was amplified by using primers 5′-ATTATTTTTACGTTCCTTCCACTTG-3′ and 5′-AACCTTAATAAAAAACACTCTATTG-3′ and the proofreading polymerases ProofStart (QIAGEN) or PFX-4 (Invitrogen, Carlsbad, CA).
When using the ProofStart enzyme, PCR conditions were 35 amplification cycles at 95°C for 5 minutes, 94°C for 1 minute, 50°C for 1 minute, followed by 72°C for 1 minute. For the PFX-4 enzyme, PCR conditions were 94°C for 5 minutes (initial activation), followed by 35 cycles at 94°C for 15 seconds, 50°C for 30 seconds, and 68°C for 1 minute. The PCR products were sequenced at Mclab (Molecular Cloning Laboratories, South San Francisco, CA) using an ABI 3730XL sequencer (Applied Biosystems, Foster City, CA).
In addition, p23 PCR products from two samples belonging to the C. parvum IIm subtype family from this study and two samples from the C. parvum subtype IIc family from India, which were identified by PCR restriction fragment length polymorphism analysis or sequence analysis of the gp40/15 gene as described,47,49 were cloned into the pCR II-TOPO vector using the TOPO TA cloning Dual promoter kit (Invitrogen). Plasmid DNA was isolated from the bacterial cultures by using the QIAprep Spin Miniprep Kit (QIAGEN) and sequenced.
Nucleotide and deduced amino acid sequences were aligned by using the Clustal W algorithm in the Align X program of Vector NTI Advance v 11.5 software (Invitrogen). The consensus p23 sequences from ≥ 2 clones from the IIm and IIc subtype families were identified by using the same program. Phylogenetic analysis was performed by using the maximum-likelihood method and the Phylogeny.fr server (http://www.phylogeny.fr/).50 Predicted sites of N and O-glycosylation were identified by using the NetNGlyc 1.0 and NetOGlyc 3.1 servers respectively at the ExPASy proteomics server (http://expasy.org/). Nucleotide and deduced amino acid sequences from this study were deposited in GenBank (accession nos. JF927201-12).
ELISA
Serum samples were shipped to Boston on dry ice and stored at –80°C. Approval for use of de-identified serum samples from children in the study was obtained from the Tufts Medical Center Institutional Review Board. Serum IgG, IgA, and IgM responses to p23 were assessed by ELISA using rp23 as antigen as described.43 Briefly, 96-well microtiter plates (Nunc, Rochester, NY) were coated with 0.1 μg of rp23 per well, overnight at 4°C. The plates were washed three times with 0.05% Tween 20 in 20 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl (phosphate-buffered saline [PBS]) by using an automated plate washer (BioTek-Elx50, Winooski, VT). Non-specific binding was blocked with 0.25% bovine serum albumin (BSA) in PBS for 2 hours at 37°C. Serum diluted 1:100 in 0.25% PBS-BSA was added and plates were incubated for 1 hour at 37°C.
After three washes with 0.05% Tween 20-PBS, alkaline phosphatase–conjugated IgG (γ chain specific), IgA (α chain specific), or IgM (μ chain specific) (Southern Biotech, Birmingham, AL) diluted in 0.25% PBS-BSA were added and the plates were incubated at 37°C for 1 hour. After three washes, substrate solution containing 1 mg/mL of p-nitrophenyl phosphate (Sigma, St. Louis, MO) in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2 was added, and the plates were incubated at room temperature for 30 minutes in the dark.
Absorbance at 405 nm (A405 nm) was measured with a Bio-Rad Microplate Reader (Model 550; Bio-Rad Laboratories, Hercules, CA). The same Cryptosporidium-positive and -negative serum samples (positive or negative for reactivity with p23 by Western blotting of a C. parvum lysate) were run on each plate. All samples were run in triplicate, and the mean was determined.
To control for plate-to-plate variability, values were normalized by dividing the A405 nm of each sample on a plate by the A405nm of the positive control for that plate and multiplying by 100. The effect of such normalization on inter-plate and intra-plate variability in ELISAs using the same serum samples was validated previously by using mixed effects regression models that indicated that the normalized values represent the original measures and are invariant to plate performance.43 Therefore, results presented are expressed in normalized ELISA units.
Statistical analysis
Statistical analysis was performed using S-plus version 8 (Insightful, Inc., Seattle, WA) statistical software and data plotted with Prism software version 5 (GraphPad Software Inc., San Diego, CA). Dichotomous demographic and clinical characteristics were compared by using Fisher's exact test. Continuous variables were compared using the unpaired t-test with Welch's correction for normally distributed variables, as defined by a standard test of normality, or the Mann-Whitney U test for non-normally distributed variables. To accommodate for the right skew in the distribution of antibody levels, the summary statistics are presented as the median and interquartile range (25th to 75th percentiles). Serum antibody levels to p23 at the initial and follow-up time points between cases and controls and between cases with acute or persistent diarrhea were compared by using the Mann-Whitney U test. Serum antibody levels at the initial and follow-up time points within cases and controls were compared by using the Wilcoxon matched pairs test. Changes in serum antibody levels from the initial to the follow-up time points in cases and controls were compared by using the unpaired t-test with Welche's correction. Associations between antibody levels to p23 and gp15 or C. parvum lysate were measured by using Spearman's correlation.
The weight-for-age, height-for-age, and weight-for-height Z-scores were calculated by using World Health Organization Anthro software (http://www.who.int/childgrowth/software/en). Multivariate analysis of antibody responses was performed by using a generalized linear model to control for potential contributing clinical and epidemiologic factors such as age, sex, duration of diarrhea, nutritional status (expressed by the weight-for-age scores), water source, and exposure to family members with diarrhea.
Results
Clinical and epidemiologic features
The demographic and clinical features of age-matched case and control children in the original age-matched study have been described.46 In the current unmatched case–control study, there were no significant differences in demographic and clinical characteristics such as age, sex, and nutritional status between cases or controls (Table 1). However, as in the original study, the duration of diarrhea at hospitalization was significantly greater in cases than in controls. In addition, 18 cases had persistent diarrhea (≥ 14 days) compared with none in the controls (Table 1). Of note, there was no significant difference in the duration of diarrhea or occurrence of persistent diarrhea in the patients that returned for follow-up compared than in those who did not return for follow-up. Frequencies of reported family members with diarrhea or animal contact or type of water supply were similar in cases and controls.
Table 1.
Demographic, clinical, and epidemiologic characteristics of cases of cryptosporidiosis in children and controls at presentation, Bangladesh
| Characteristic | Cases (n = 49) | Controls (n = 39) | P* |
|---|---|---|---|
| Age in months, median (25th, 75th percentile) | 12 (9, 15) | 11 (7, 15) | 0.68† |
| Male sex, no. (%) | 29 (59) | 27 (69) | 0.38‡ |
| Duration of diarrhea in days, median (25th, 75th percentile) | 8 (4, 13.5) | 3 (1, 4) | < 0.0001† |
| Persistent diarrhea (> 14 days), no. (%) | 18 (36.7) | 0 (0) | < 0.0001† |
| Nutritional status | |||
| Weight-for-age Z score, mean (SD) | −2.73 (1.13) | −2.46 (1.18) | 0.29§ |
| Height-for-age Z score, mean (SD) | −1.94 (1.28) | −1.66 (1.75) | 0.41§ |
| Weight-for-height Z score, mean (SD) | −2.37 (1.37) | −2.09 (1.51) | 0.38§ |
| Exposure to family member with diarrhea, no. (%) | 6 (12) | 5 (13) | 1.00‡ |
| Contact with animals, no. (%) | 7 (14) | 7 (18) | 0.77‡ |
| Water source | |||
| Tube well, no. (%) | 30 (61) | 23 (59) | 1.00‡ |
| Municipal water supply, no. (%) | 19 (39) | 16 (41) | 1.00‡ |
P values < 0.05 are indicated in bold.
By Mann-Whitney test.
By Fisher's exact test.
By unpaired t-test with Welch's correction.
Serum antibody responses to C. parvum p23
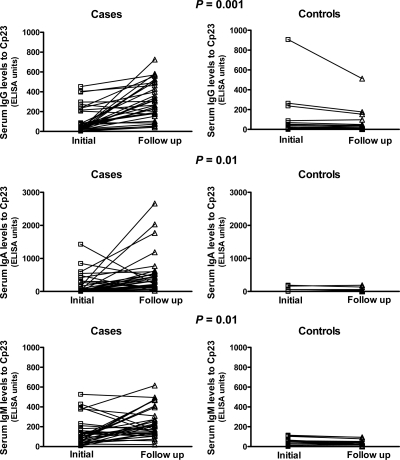
Serum IgA and IgM (but not IgG) levels to p23 were significantly greater in cases than in controls at hospitalization (Table 2). However, when antibody levels were compared by multivariate analysis using a generalized linear regression model to control for co-variates such as age, sex, nutritional status, water source, and exposure to family members with diarrhea, there were no significant differences in these levels between cases and controls. In addition, there were no significant differences in serum anti-p23 IgG, IgM, or IgA levels between the initial and follow-up time points in controls. Of note, there were no significant differences between the initial antibody levels of cases or controls that returned for follow-up versus those who did not return for follow-up. Serum p23 IgG, IgA, and IgM levels to p23 at the three-week follow-up time point were significantly higher in cases than in controls (Table 2). These differences remained significant after multivariate analysis. In addition, there was a significantly greater increase in serum anti-p23 IgG, IgA, and IgM levels from the initial to the follow-up time points in cases compared with controls (Table 2 and Figure 2). These differences also remained significant in multivariate analysis.
Table 2.
Antibody levels to Cryptosporidium (C) p23 at the initial and follow-up time points and their change from initial to follow-up time points in children with cryptosporidiosis in Bangladesh*
| Characteristic | Cases† | Controls‡ | P§ |
|---|---|---|---|
| IgG | |||
| Initial | 62.4 (23.91, 190.3) | 53.14 (23.71, 154.5) | 0.98 |
| Follow-up | 307 (185.4, 478.7) | 32.79 (10.5, 73.75) | < 0.0001 |
| Change¶ | 207.4 ± 187.6 | −35.59 ± 97.05 | < 0.0001 |
| IgA | |||
| Initial | 47.18 (0, 304.2) | 0 (0, 55) | 0.03 |
| Follow-up | 305.7 (140.9, 541.5) | 20 (20, 38.82) | < 0.0001 |
| Change¶ | 315 ± 671.7 | 6.45 ± 21.43 | 0.01 |
| IgM | |||
| Initial | 113.5 (62.92, 221) | 53.2 (31.3, 100.5) | < 0.0001 |
| Follow-up | 170.6 (133.9, 291.2) | 36.92 (21.45, 50.62) | < 0.0001 |
| Change¶ | 84.57 ± 169.7 | −4.24 ± 15.48 | 0.01 |
Values are medians (25th, 75th percentiles) and comparisons were made by using the Mann-Whitney test unless otherwise indicated.
Cases: initial, n = 49; follow-up, n = 32.
Controls: initial, n = 39; follow-up, n = 17.
P values < 0.05 are indicated in bold.
Change in antibody levels from initial to follow-up time points are represented as mean ± SD and compared by using the unpaired t-test with Welch's correction.
Figure 2.
Median serum IgG, IgM, and IgA levels to Cryptosporidium (C) p23 in cases and controls at the initial and follow-up time points determined by enzyme-linked immunosorbent assay with recombinant p23 as antigen in children with cryptosporidiosis in Bangladesh. Changes in antibody levels over the three-week follow-up period were compared by using the unpaired t-test with Welch's correction.
Serum antibody responses to p23 in cases with acute versus persistent diarrhea
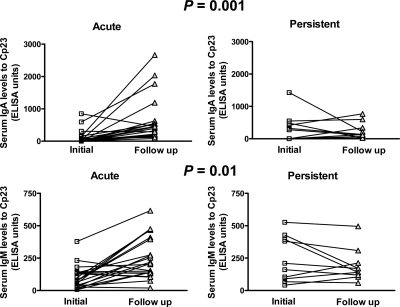
The only significant differences in clinical and epidemiologic features between cases and controls were the duration of diarrhea and occurrence of persistent diarrhea. We compared the antibody responses to p23 in 22 cases with acute (< 14 days) diarrhea with 10 cases with persistent (≥ 14 days) diarrhea. There were no significant differences in IgG levels (at the initial or follow-up time points or in the change from initial to follow-up time points) among cases with acute or persistent diarrhea. However, the initial IgA and IgM levels were significantly lower in those with acute diarrhea than in those with persistent diarrhea (P = 0.05 and P = 0.01 for IgA and IgM, respectively). In addition, the change in IgA and IgM levels from the initial to follow-up time points was significantly greater in those with acute diarrhea than in those with persistent diarrhea (Figure 3).
Figure 3.
Median serum IgM and IgA levels to Cryptosporidium (C) p23 in cases with acute or persistent diarrhea at the initial and follow-up time points determined by enzyme-linked immunosorbent assay with recombinant p23 as antigen in children with cryptosporidiosis in Bangladesh. Changes in antibody levels over the three-week follow-up period were compared by using the unpaired t-test with Welch's correction.
Correlation of serum antibody responses to p23 with those to C. parvum gp15 and C. parvum lysate
To determine if there was any association between serum antibody levels to p23 and those to the immunodominant gp15 antigen, or to total antigens in C. parvum lysate, we compared antibody levels to p23 with each of these antigen preparations from previous studies in the same children.43,46 There were significant correlations in the initial, follow-up, and change between the IgG, IgA, and IgM (except for IgM follow-up) antibody levels to p23 with those to gp15 in cases over the three-week follow-up period (Table 3). In addition, there were significant correlations in antibody levels (except for IgG follow-up and change) to p23 with those to C. parvum lysate at the initial, follow-up, and change between them (Table 3).
Table 3.
Correlation of antibody levels to Cryptosporidium (C) p23 with antibody levels to gp15 and C. parvum lysate at initial and follow-up points and their change from initial to follow-up time points in children with cryptosporidiosis, Bangladesh*
| Characteristic | Spearman correlation values |
|---|---|
| Cp23 vs. gp15 | |
| IgG | |
| Initial | 0.53† |
| Follow-up | 0.39‡ |
| Change | 0.51† |
| IgA | |
| Initial | 0.36‡ |
| Follow-up | 0.47† |
| Change | 0.64† |
| IgM | |
| Iintial | 0.70† |
| Follow-up | 0.12 |
| Change | 0.57† |
| Cp23 vs. C. parvum lysate | |
| IgG | |
| Initial | 0.50† |
| Follow-up | 0.14 |
| Change | 0.10 |
| IgA | |
| Initial | 0.68† |
| Follow-up | 0.48† |
| Change | 0.71† |
| IgM | |
| Initial | 0.46† |
| Follow-up | 0.53† |
| Change | 0.57† |
Cases: initial, n = 49; follow-up, n = 32; change, n = 32.
P < 0.01.
P < 0.05.
Analysis of p23 polymorphisms among infecting Cryptosporidium spp. subtypes
Molecular characterization of the Cryptosporidium spp. infecting case children in the study identified seven subtype families from C. parvum (IIm), C. hominis (Ia, Ib, Id, Ie and If), and C. felis (IIa).47 The C. parvum IIm subtype family has only recently been identified in Bangladesh,47 India,49 and Nigeria,51 and is considered an anthroponotic subtype because it has not been identified in animals. The other anthroponotic subtype family, IIc,41 was not identified in this study. We therefore amplified the p23 sequence from DNA from stool samples of three children infected with this subtype family in a previous study on cryptosporidiosis in children with diarrhea in Vellore, India.48
To determine if there were differences in the p23 sequences from subtype families of C. parvum and C. hominis spp. infecting children in this study with those in the published C. parvum52 and C. hominis53 genomes, as well as other p23 sequences deposited in GenBank, we performed phylogenetic analysis. This analysis showed that the p23 nucleotide and deduced amino acid sequences from the major species clustered separately for the most part (Figure 4).
Figure 4.
Multiple alignment of deduced amino acid p23 sequences. Samples from the study of cryptosporidiosis in children in Bangladesh are indicated by the letter B, followed by the sample number. Samples from India are indicated by the letter I, followed by the sample number. Sequences from GenBank are indicated by the accession number. The species and subtype family for each sequence, if known, are indicated after the sample number or GenBank accession number. Differences are in bold and underlined. The predicted N glycosylation site is in bold. The predicted O-glycosylation sites are in bold and italics. The QDKPAD peptide is double underlined.
The most divergent p23 sequence was from a C. parvum cervine genotype human isolate (GenBank accession no. DQ389174), which was reported to have low identity to C. parvum and C. hominis sequences and a multiple repeat region54 (Figure 4). Three other sequences from the mouse, rabbit, and pig II genotypes55 were more similar to each other than to the other sequences (Figure 4). All C. parvum sequences (except IIc sequences and one of the IIm sequences [B 7] from this study) clustered together as did the IIc and the second IIm (B 7) sequences (Figure 4). Finally all C. hominis p23 sequences including those from this study (Ia, Ib, Id, Ie, and If) and that of Sturbaum and others45 (Ia, Ib, Id, and Ie) clustered together. The deduced amino acid sequences of all C. parvum p23 sequences (except the IIc and the IIm B 7 sequence) were identical with each other and with that of the p23 sequence (which belongs to the IIa subtype family) from the C. parvum genome52 (Figure 4).
Similarly, all C. hominis sequences were identical with each other and with that of the published sequence (which belongs to the Ia subtype family) from the C. hominis genome53 (Figure 4). As reported,17,45 there were 10 nucleotide differences, which translated into three amino acid changes, P to S, A to S, and D to E (as indicated in Figure 4), between most C. parvum and C. hominis sequences. However, all three C. parvum IIc sequences and one C. parvum IIm (B 21) sequence (Figure 4) were identical with each other, but differed from other C. parvum and C. hominis sequences in that they shared the same P, A, and D residues as the other C. parvum sequences but had an A to S change in the C-terminal most residues compared with the rest of the C. parvum and all the C. hominis sequences.
The predicted N-linked glycosylation site NKS (indicated in bold in Figure 4) is conserved among all p23 sequences as are 4 predicted O-linked glycosylation sites (indicated in bold and italics in Figure 4). An additional predicted O-glycosylated S residue is conserved among all C. parvum and C. hominis sequences. All C. hominis sequences share another putative O-glycosylated S residue, and the C-terminal-most S residue in all IIc and B 7 IIm sequences is predicted to be O-glycosylated (Figure 4). The C-terminal QDKPAD peptide against which the neutralizing 7A10 monoclonal antibody is directed45 is conserved among all (except the C. parvum cervine genotype) sequences, and the second QDKPAD peptide is conserved among all C. parvum sequences analyzed in this study (Figure 4). However, the C terminal D residue is replaced with an E in all C. hominis sequences (Figure 4).
Discussion
Although p23 is considered one of the most promising vaccine candidates for cryptosporidiosis,40 there have been few clinical studies in well-defined cohorts that have characterized immune responses to this antigen and none that have analyzed polymorphisms in the gene encoding it from Cryptosporidium spp. and subtype families infecting patients in the study. In this case–control study of children less than five years of age with diarrhea in Bangladesh, we found that Cryptosporidium-infected case children, but not uninfected controls, showed development of statistically significant serum IgG, IgA, and IgM responses to this antigen over a three-week follow-up period. Serum IgA and IgM responses were significantly lower in children with persistent diarrhea than in those with acute diarrhea over the three-week follow-up period, which suggested that these responses may be associated with protection from prolonged diarrhea.
The p23 nucleotide and deduced amino acid sequences from Cryptosporidium spp. infecting these children were relatively conserved among different C. parvum and C. hominis subtype families. Although most children were infected with different subtype families of C. hominis, overall, there were significant antibody responses to the C. parvum antigen in cases compared with controls, which suggested that these cross-reactive responses are directed at conserved epitopes. These results support further development of p23 as a component of a subunit vaccine for cryptosporidiosis.
A number of previous studies have reported serum antibody responses to p2330–36 in immunocompetent and immunocompromised hosts. However, most studies were serosurveys to estimate prevalence and investigate outbreaks32,33,56–60 or to demonstrate the utility of p23 as an antigen for ELISAs.30,35 Priest and others investigated serum antibody responses to a 27-kDa antigen (same as p23) in a birth cohort of children in Peru by using the same recombinant C. parvum protein we used as antigen for the ELISA. As in our study, most children in their study were infected with C. hominis. However, serum antibody responses to the C. parvum p23 antigen occurred in children infected with a number of different species and subtype families. Serum IgG responses to p23 increased with age and with repeated infections.31 In a study of HIV-infected persons from Australia, Frost and others reported that a strong serologic response to p23 was associated with a reduced risk of diarrhea without weight loss, but not in those who had weight loss in addition to diarrhea.38
In this study, there were no significant differences in serum IgG, IgM, or IgA levels to p23 between cases and controls after controlling for covariates in multivariate analysis. However, after three weeks of follow-up, levels of all three isotypes were significantly higher in cases than controls, as was the change in antibody levels from the initial to the follow-up time points. These differences remained significant after controlling for covariates. This finding is in contrast to antibody responses to gp15 in the same children in whom only IgG levels at follow-up and in the change from the initial to follow-up time points were significantly greater in cases than in controls by multivariate analysis.43 This finding suggests that p23 induces IgA and IgM responses (possibly reflecting transfer from mucosal surfaces), which persist for a longer time.
Persistent diarrhea is a common consequence of cryptosporidiosis in children in developing countries, particularly in those who are malnourished.3,4 Approximately one-third of the cases, but none of the controls in this study, had persistent diarrhea. In these children, IgA and IgM levels were significantly higher in those with persistent diarrhea than in those with acute diarrhea. The higher IgA and IgM levels in those with persistent diarrhea may be related to the longer duration of diarrhea in these children. However, there was a significantly greater increase in IgA and IgM levels from the initial to follow-up time points in those with acute diarrhea than in those with persistent diarrhea. This finding suggests that IgA and IgM responses (possibly mucosal) that persist for a longer time may protect against development of persistent diarrhea. These results are similar to those obtained in our studies of antibody responses to total Cryptosporidium antigens in oocyst lysates46 and to the immunodominant gp15 antigen43 in the same children.
Although there were significant correlations in antibody levels to p23 with those to C. parvum lysate, serum IgG levels to antigens in the C. parvum lysate did not correlate well with the p23 response at the follow-up time point. The reason for this finding is not clear, but may reflect differences in the dynamics of the IgG response to different antigens present in the C. parvum lysate compared with that to p23.
In this study, PCR was used to re-classify seven microscopy-negative controls as cases. Because PCR is currently the most sensitive method for detection of Cryptosporidium spp., it is unlikely although possible that additional controls may have been misclassified. Three of the controls had relatively high levels of serum IgG (but not IgM) to p23, which decreased at the follow-up time point. This finding may represent previous symptomatic or asymptomatic infection with Cryptosporidium spp.
Cell-mediated immunity is critical for protection from and resolution of cryptosporidiosis. Although antibody responses to specific antigens such as gp15 and p23 have been associated with protection from symptoms of cryptosporidiosis37,38,61 it is not known whether these responses are themselves protective or whether they merely reflect protective cellular responses.62 Smith and others examined serum antibody and T cell proliferative responses to p2339 in residents of Haiti previously exposed to Cryptosporidium spp., as suggested by high levels of seropositivity. They found that antibody responses to p23 were greater in persons who displayed proliferative T cell responses to this antigen, suggesting that antibody responses may correlate with cellular responses. In our study, it was not possible to assess cell-mediated responses for logistical reasons.
The finding that there were relatively few single amino acid polymorphisms in the p23 deduced amino acid sequences from different C. parvum and C. hominis subtype families in this study and in others from different geographic locations indicates that this antigen is relatively conserved and supports its consideration as a vaccine candidate. This is the first report of p23 sequences from the anthroponotic C. parvum IIc and IIm subtype families. The finding that these sequences clustered separately from the other C. parvum sequences is interesting and consistent with those of previous studies, which showed that they are divergent at other loci including that of the gp40/15 (also called gp60)41 and Muc4 genes.44
Although most children in our study were infected with different C. hominis subtype families,47 the finding that they developed significant antibody responses to the C. parvum antigen is consistent with that reported by Priest and others.31 The results from both studies suggest that these responses are directed at epitopes conserved among species and different subtype families. The neutralizing monoclonal antibodies 7D10 and C6B6, which were first used to identify p23, are directed at linear (QDKPAD) and conformational epitopes, respectively, in the C-terminal region of p23.11 Sturbaum and others reported that 9 of 10 monoclonal antibodies generated against C. parvum p23 were reactive with the p23 antigen from three C. hominis isolates, confirming that for the most part p23 is antigenically conserved among isolates from both species.17,45 However, the epitopes recognized by human serum antibodies to p23 remain to be determined.
Because E. coli-expressed recombinant p23 was used as antigen for ELISAs, the findings of this study indicate that antibody responses were directed at peptide epitopes. However, as shown in the present study, p23, like many other surface-associated Cryptosporidium spp. antigens,42 is predicted to contain conserved N and O-glycosylation sites, and we have shown that this antigen binds to Helix pomatia agglutinin, suggesting that it displays mucin-type O glycosylation (Ward H. D., unpublished data). However, it is not known whether immune responses to this antigen are directed against glycosylated as well as peptide epitopes.
As noted, there were a number of limitations with the parent study, including a small sample size and significant loss to follow-up in an urban hospital-based setting.43,46 In addition, we were not able to determine whether cases or controls experienced symptomatic or asymptomatic cryptosporidiosis previously. Furthermore, case children came to the hospital at various times after the onset of diarrhea. In the current study, assessment of immune responses was limited to measurement of systemic serum antibody levels to p23. For logistical reasons, we were not able to assess cell-mediated or mucosal responses to this antigen. Nonetheless, the findings of this study provide useful data that support further development of p23 as a vaccine candidate and will inform larger, community-based, longitudinal, studies on systemic and mucosal cell-mediated and humoral responses to protective peptide and glycosylated epitopes on this and other putative vaccine candidates.
ACKNOWLEDGMENTS
We thank study participants and field and laboratory staff for participating in the study and Dr. Jeffery Priest (Centers for Disease Control, Atlanta, GA) for providing the p23 plasmid and helpful discussions.
Footnotes
Financial support: This study was supported by an opportunity pool grant and in part by grants UO1 AI45508, U01 AI058935, and RO1 AI52786 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) and K24 AT003683 from the National Center for Complementary and Alternative Medicine, NIH. Anoli J. Borad was supported by T32 AI007438, and Geneve M. Allison was supported by T32 AI07389 from NIAID, NIH.
Authors' addresses: Anoli J. Borad, Division of Infectious Diseases, Department of Medicine, Yale School of Medicine, New Haven, CT, E-mail: anoli.borad@yale.edu. Geneve M. Allison, Anne V. Kane, Joy Moy, and Honorine D. Ward, Division of Geographic Medicine and Infectious Diseases, Department of Medicine, Tufts Medical Center, Boston, MA, E-mails: gallison@tuftsmedicalcenter.org, akane@tuftsmedicalcenter.org, joy.moy@gmail.com, and hward@tuftsmedicalcenter.org. David Wang, Department of Biomedical Engineering, Tufts University School of Engineering, Medford, MA, E-mail: david.e.wang@gmail.com. Sabeena Ahmed, Mohammad M. Karim, and Wasif A. Khan, Clinical Sciences Division, Centre for Health and Population Research, International Center for Diarrheal Disease Research, Bangladesh, Dhaka, Bangladesh, E-mails: mahbubul@icddrb.org, sabeena@hotmail.com, and wakhan@icddrb.org. Patricia L. Hibberd, Division of Global Health, Department of Pediatrics, Massachusetts General Hospital, Boston, MA, E-mail: phibberd@partners.org. Sitara Swarna Rao Ajjampur and Gagandeep Kang, Department of Gastrointestinal Sciences, Christian Medical College, Vellore, India, E-mails: sitararao@cmcvellore.ac.in and gkang@cmcvellore.ac.in. Stephen B. Calderwood and Edward T. Ryan, Division of Infectious Diseases, Massachusetts General Hospital, Boston, MA, E-mails: scalderwood@partners.org and etryan@partners.org. Elena Naumova, Tufts Initiative for the Forecasting and Modeling of Infectious Diseases, Tufts University School of Engineering, Medford, MA, E-mail: elena.naumova@tufts.edu.
References
- 1.Collinet-Adler S, Ward HD. Cryptosporidiosis: environmental, therapeutic, and preventive challenges. Eur J Clin Microbiol Infect Dis. 2010;29:927–935. doi: 10.1007/s10096-010-0960-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leav BA, Mackay M, Ward HD. Cryptosporidium species: new insights and old challenges. Clin Infect Dis. 2003;36:903–908. doi: 10.1086/368194. [DOI] [PubMed] [Google Scholar]
- 3.Dillingham RA, Lima AA, Guerrant RL. Cryptosporidiosis: epidemiology and impact. Microbes Infect. 2002;4:1059–1066. doi: 10.1016/s1286-4579(02)01630-1. [DOI] [PubMed] [Google Scholar]
- 4.Huang DB, White AC. An updated review on Cryptosporidium and Giardia. Gastroenterol Clin North Am. 2006;35:291–314. doi: 10.1016/j.gtc.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Snelling WJ, Xiao L, Ortega-Pierres G, Lowery CJ, Moore JE, Rao JR, Smyth S, Millar BC, Rooney PJ, Matsuda M, Kenny F, Xu J, Dooley JS. Cryptosporidiosis in developing countries. J Infect Dev Ctries. 2007;1:242–256. [PubMed] [Google Scholar]
- 6.Borad A, Ward H. Human immune responses in cryptosporidiosis. Future Microbiol. 2010;5:507–519. doi: 10.2217/fmb.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connor RM, Shaffie R, Kang G, Ward HD. Cryptosporidiosis in patients with HIV/AIDS. AIDS. 2011;25:549–560. doi: 10.1097/QAD.0b013e3283437e88. [DOI] [PubMed] [Google Scholar]
- 8.Cabada MM, White AC., Jr Treatment of cryptosporidiosis: do we know what we think we know? Curr Opin Infect Dis. 2010;23:494–499. doi: 10.1097/QCO.0b013e32833de052. [DOI] [PubMed] [Google Scholar]
- 9.Abubakar I, Aliyu SH, Arumugam C, Usman NK, Hunter PR. Treatment of cryptosporidiosis in immunocompromised individuals: systematic review and meta-analysis. Br J Clin Pharmacol. 2007;63:387–393. doi: 10.1111/j.1365-2125.2007.02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Graaf DC, Spano F, Petry F, Sagodira S, Bonnin A. Speculation on whether a vaccine against cryptosporidiosis is a reality or fantasy. Int J Parasitol. 1999;29:1289–1306. doi: 10.1016/S0020-7519(99)00082-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perryman LE, Jasmer DP, Riggs MW, Bohnet SG, McGuire TC, Arrowood MJ. A cloned gene of Cryptosporidium parvum encodes neutralization-sensitive epitopes. Mol Biochem Parasitol. 1996;80:137–147. doi: 10.1016/0166-6851(96)02681-3. [DOI] [PubMed] [Google Scholar]
- 12.Enriquez FJ, Riggs MW. Role of immunoglobulin A monoclonal antibodies against P23 in controlling murine Cryptosporidium parvum infection. Infect Immun. 1998;66:4469–4473. doi: 10.1128/iai.66.9.4469-4473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrowood MJ, Sterling CR, Healey MC. Immunofluorescent microscopical visualization of trails left by gliding Cryptosporidium parvum sporozoites. J Parasitol. 1991;77:315–317. [PubMed] [Google Scholar]
- 14.Arrowood MJ, Mead JR, Mahrt JL, Sterling CR. Effects of immune colostrum and orally administered antisporozoite monoclonal antibodies on the outcome of Cryptosporidium parvum infections in neonatal mice. Infect Immun. 1989;57:2283–2288. doi: 10.1128/iai.57.8.2283-2288.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riggs MW, Schaefer DA, Kapil SJ, Barley-Maloney L, Perryman LE. Efficacy of monoclonal antibodies against defined antigens for passive immunotherapy of chronic gastrointestinal cryptosporidiosis. Antimicrob Agents Chemother. 2002;46:275–282. doi: 10.1128/AAC.46.2.275-282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perryman LE, Kapil SJ, Jones ML, Hunt EL. Protection of calves against cryptosporidiosis with immune bovine colostrum induced by a Cryptosporidium parvum recombinant protein. Vaccine. 1999;17:2142–2149. doi: 10.1016/s0264-410x(98)00477-0. [DOI] [PubMed] [Google Scholar]
- 17.Sturbaum GD, Schaefer DA, Jost BH, Sterling CR, Riggs MW. Antigenic differences within the Cryptosporidium hominis and Cryptosporidium parvum surface proteins P23 and GP900 defined by monoclonal antibody reactivity. Mol Biochem Parasitol. 2008;159:138–141. doi: 10.1016/j.molbiopara.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer DA, Auerbach-Dixon BA, Riggs MW. Characterization and formulation of multiple epitope-specific neutralizing monoclonal antibodies for passive immunization against cryptosporidiosis. Infect Immun. 2000;68:2608–2616. doi: 10.1128/iai.68.5.2608-2616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyatt CR, Perryman LE. Detection of mucosally delivered antibody to Cryptosporidium parvum p23 in infected calves. Ann N Y Acad Sci. 2000;916:378–387. doi: 10.1111/j.1749-6632.2000.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 20.Wyatt CR, Brackett EJ, Savidge J. Evidence for the emergence of a type-1-like immune response in intestinal mucosa of calves recovering from cryptosporidiosis. J Parasitol. 2001;87:90–95. doi: 10.1645/0022-3395(2001)087[0090:EFTEOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 21.Bonafonte MT, Smith LM, Mead JR. A 23-kDa recombinant antigen of Cryptosporidium parvum induces a cellular immune response on in vitro stimulated spleen and mesenteric lymph node cells from infected mice. Exp Parasitol. 2000;96:32–41. doi: 10.1006/expr.2000.4545. [DOI] [PubMed] [Google Scholar]
- 22.de Graaf DC, De Coninck H, De Clercq C, Peeters JE. Screening of the T- and B-cell antigenicity in neonatal calves of the His-tagged Cryptosporidium parvum antigens CP15, CP15/60, P23 and TRAP-C1. Folia Parasitol (Praha) 2002;49:319–322. doi: 10.14411/fp.2002.059. [DOI] [PubMed] [Google Scholar]
- 23.Ehigiator HN, Romagnoli P, Priest JW, Secor WE, Mead JR. Induction of murine immune responses by DNA encoding a 23-kDa antigen of Cryptosporidium parvum. Parasitol Res. 2007;101:943–950. doi: 10.1007/s00436-007-0565-0. [DOI] [PubMed] [Google Scholar]
- 24.Geriletu Xu R, Jia H, Terkawi MA, Xuan X, Zhang H. Immunogenicity of orally administrated recombinant Lactobacillus casei Zhang expressing Cryptosporidium parvum surface adhesion protein P23 in mice. Curr Microbiol. 2011;62:1573–1580. doi: 10.1007/s00284-011-9894-4. [DOI] [PubMed] [Google Scholar]
- 25.Shirafuji H, Xuan X, Kimata I, Takashima Y, Fukumoto S, Otsuka H, Nagasawa H, Suzuki H. Expression of P23 of Cryptosporidium parvum in Toxoplasma gondii and evaluation of its protective effects. J Parasitol. 2005;91:476–479. doi: 10.1645/GE-364R1. [DOI] [PubMed] [Google Scholar]
- 26.Takashima Y, Xuan X, Kimata I, Iseki M, Kodama Y, Nagane N, Nagasawa H, Matsumoto Y, Mikami T, Otsuka H. Recombinant bovine herpes virus-1 expressing p23 protein of Cryptosporidium parvum induces neutralizing antibodies in rabbits. J Parasitol. 2003;89:276–282. doi: 10.1645/0022-3395(2003)089[0276:RBHEPP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 27.Hong-Xuan H, Lei C, Cheng-Min W, Kai Z, Yi T, Xi-Ming Q, Ming-Xing D. Expression of the recombinant fusion protein CP15-23 of Cryptosporidium parvum and its protective test. J Nanosci Nanotechnol. 2005;5:1292–1296. doi: 10.1166/jnn.2005.210. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, Zai D, Zhang D, Wei Q, Han G, Gao H, Huang B. Divalent Cp15-23 vaccine enhances immune responses and protection against Cryptosporidium parvum infection. Parasite Immunol. 2010;32:335–344. doi: 10.1111/j.1365-3024.2009.01191.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Luo J, Amer S, Guo Y, Hu Y, Lu Y, Wang H, Duan M, He H. Multivalent DNA vaccine induces protective immune responses and enhanced resistance against Cryptosporidium parvum infection. Vaccine. 2010;29:323–328. doi: 10.1016/j.vaccine.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 30.Priest JW, Kwon JP, Moss DM, Roberts JM, Arrowood MJ, Dworkin MS, Juranek DD, Lammie PJ. Detection by enzyme immunoassay of serum immunoglobulin G antibodies that recognize specific Cryptosporidium parvum antigens. J Clin Microbiol. 1999;37:1385–1392. doi: 10.1128/jcm.37.5.1385-1392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priest JW, Bern C, Xiao L, Roberts JM, Kwon JP, Lescano AG, Checkley W, Cabrera L, Moss DM, Arrowood MJ, Sterling CR, Gilman RH, Lammie PJ. Longitudinal analysis of Cryptosporidium species-specific immunoglobulin G antibody responses in Peruvian children. Clin Vaccine Immunol. 2006;13:123–131. doi: 10.1128/CVI.13.1.123-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss DM, Bennett SN, Arrowood MJ, Wahlquist SP, Lammie PJ. Enzyme-linked immunoelectrotransfer blot analysis of a cryptosporidiosis outbreak on a United States Coast Guard cutter. Am J Trop Med Hyg. 1998;58:110–118. doi: 10.4269/ajtmh.1998.58.110. [DOI] [PubMed] [Google Scholar]
- 33.Frost FJ, Calderon RL, Muller TB, Curry M, Rodman JS, Moss DM, de la Cruz AA. A two-year follow-up survey of antibody to Cryptosporidium in Jackson County, Oregon following an outbreak of waterborne disease. Epidemiol Infect. 1998;121:213–217. doi: 10.1017/s095026889800898x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priest JW, Bern C, Roberts JM, Kwon JP, Lescano AG, Checkley W, Cabrera L, Moss DM, Arrowood MJ, Sterling CR, Gilman RH, Lammie PJ. Changes in serum immunoglobulin G levels as a marker for Cryptosporidium sp. infection in Peruvian children. J Clin Microbiol. 2005;43:5298–5300. doi: 10.1128/JCM.43.10.5298-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priest JW, Li A, Khan M, Arrowood MJ, Lammie PJ, Ong CS, Roberts JM, Isaac-Renton J. Enzyme immunoassay detection of antigen-specific immunoglobulin g antibodies in longitudinal serum samples from patients with cryptosporidiosis. Clin Diagn Lab Immunol. 2001;8:415–423. doi: 10.1128/CDLI.8.2.415-423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandhu SK, Priest JW, Lammie PJ, Hubbard A, Colford JM, Jr, Eisenberg JN. The natural history of antibody responses to Cryptosporidium parasites in men at high risk of HIV infection. J Infect Dis. 2006;194:1428–1437. doi: 10.1086/508194. [DOI] [PubMed] [Google Scholar]
- 37.Moss DM, Chappell CL, Okhuysen PC, DuPont HL, Arrowood MJ, Hightower AW, Lammie PJ. The antibody response to 27-, 17-, and 15-kDa Cryptosporidium antigens following experimental infection in humans. J Infect Dis. 1998;178:827–833. doi: 10.1086/515377. [DOI] [PubMed] [Google Scholar]
- 38.Frost FJ, Tollestrup K, Craun GF, Fairley CK, Sinclair MI, Kunde TR. Protective immunity associated with a strong serological response to a Cryptosporidium-specific antigen group, in HIV-infected individuals. J Infect Dis. 2005;192:618–621. doi: 10.1086/431681. [DOI] [PubMed] [Google Scholar]
- 39.Smith LM, Priest JW, Lammie PJ, Mead JR. Human T and B cell immunoreactivity to a recombinant 23-kDa Cryptosporidium parvum antigen. J Parasitol. 2001;87:704–707. doi: 10.1645/0022-3395(2001)087[0704:HTABCI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins MC. Present and future control of cryptosporidiosis in humans and animals. Expert Rev Vaccines. 2004;3:669–671. doi: 10.1586/14760584.3.6.669. [DOI] [PubMed] [Google Scholar]
- 41.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Wanyiri J, Ward H. Molecular basis of Cryptosporidium-host cell interactions: recent advances and future prospects. Future Microbiol. 2006;1:201–208. doi: 10.2217/17460913.1.2.201. [DOI] [PubMed] [Google Scholar]
- 43.Allison G, Rogers K, Borad A, Karim M, Ahmed S, Kane A, Hibberd P, Naumova E, Calderwood S, Ryan E, Khan W, Ward H. Antibody responses to the immunodominant Cryptosporidium gp15 antigen and gp15 polymorphisms in a case-control study of cryptosporidiosis in children in Bangladesh. Am J Trop Med Hyg. 2011;85:97–104. doi: 10.4269/ajtmh.2011.11-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor RM, Burns PB, Ha-Ngoc T, Scarpato K, Khan W, Kang G, Ward H. Polymorphic mucin antigens CpMuc4 and CpMuc5 are integral to Cryptosporidium parvum infection in vitro. Eukaryot Cell. 2009;8:461–469. doi: 10.1128/EC.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturbaum GD, Jost BH, Sterling CR. Nucleotide changes within three Cryptosporidium parvum surface protein encoding genes differentiate genotype I from genotype II isolates. Mol Biochem Parasitol. 2003;128:87–90. doi: 10.1016/s0166-6851(03)00017-3. [DOI] [PubMed] [Google Scholar]
- 46.Khan WA, Rogers KA, Karim MM, Ahmed S, Hibberd PL, Calderwood SB, Ryan ET, Ward HD. Cryptosporidiosis among Bangladeshi children with diarrhea: a prospective, matched, case-control study of clinical features, epidemiology and systemic antibody responses. Am J Trop Med Hyg. 2004;71:412–419. [PubMed] [Google Scholar]
- 47.Hira KG, Mackay MR, Hempstead AD, Ahmed S, Karim MM, O'Connor RM, Hibberd PL, Calderwood SB, Ryan ET, Khan WA, Ward HD. Genetic diversity of Cryptosporidium spp. from Bangladeshi children. J Clin Microbiol. 2011;49:2307–2310. doi: 10.1128/JCM.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ajjampur SS, Gladstone BP, Selvapandian D, Muliyil JP, Ward H, Kang G. Molecular and spatial epidemiology of cryptosporidiosis in children in a semiurban community in South India. J Clin Microbiol. 2007;45:915–920. doi: 10.1128/JCM.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ajjampur SS, Liakath FB, Kannan A, Rajendran P, Sarkar R, Moses PD, Simon A, Agarwal I, Mathew A, O'Connor R, Ward H, Kang G. Multisite study of cryptosporidiosis in children with diarrhea in India. J Clin Microbiol. 2010;48:2075–2081. doi: 10.1128/JCM.02509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molloy SF, Smith HV, Kirwan P, Nichols RA, Asaolu SO, Connelly L, Holland CV. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg. 2010;82:608–613. doi: 10.4269/ajtmh.2010.09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 53.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 54.Wong PH, Ong CS. Molecular characterization of the Cryptosporidium cervine genotype. Parasitology. 2006;133:693–700. doi: 10.1017/S0031182006000990. [DOI] [PubMed] [Google Scholar]
- 55.Hu Y, Mi R, Chen Z, Yu H, Yue C. Cloning and sequence comparison of P23 gene of different Cryptosporidium genotypes. Dongwu Yixue Jinzhan (Chinese) 2010;31:14–18. [Google Scholar]
- 56.Frost FJ, Muller TB, Calderon RL, Craun GF. Analysis of serological responses to Cryptosporidium antigen among NHANES III participants. Ann Epidemiol. 2004;14:473–478. doi: 10.1016/j.annepidem.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Egorov A, Frost F, Muller T, Naumova E, Tereschenko A, Ford T. Serological evidence of Cryptosporidium infections in a Russian city and evaluation of risk factors for infections. Ann Epidemiol. 2004;14:129–136. doi: 10.1016/S1047-2797(03)00122-4. [DOI] [PubMed] [Google Scholar]
- 58.Guk SM, Chai JY, Shin YO, Seo M. Antibody responses to Cryptosporidium antigen in HIV-positive patients in the Republic of Korea. Korean J Parasitol. 2008;46:71–75. doi: 10.3347/kjp.2008.46.2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozisek F, Craun GF, Cerovska L, Pumann P, Frost F, Muller T. Serological responses to Cryptosporidium-specific antigens in Czech populations with different water sources. Epidemiol Infect. 2008;136:279–286. doi: 10.1017/S0950268807008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frost F, Craun G, Mihaly K, Gyorgy B, Calderon R, Muller T. Serological responses to Cryptosporidium antigens among women using riverbank-filtered water, conventionally filtered surface water and groundwater in Hungary. J Water Health. 2005;3:77–82. [PubMed] [Google Scholar]
- 61.Frost FJ, Roberts M, Kunde TR, Craun G, Tollestrup K, Harter L, Muller T. How clean must our drinking water be: the importance of protective immunity. J Infect Dis. 2005;191:809–814. doi: 10.1086/427561. [DOI] [PubMed] [Google Scholar]
- 62.Riggs MW. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 2002;4:1067–1080. doi: 10.1016/s1286-4579(02)01631-3. [DOI] [PubMed] [Google Scholar]