Abstract
The agents of human febrile illness can vary by region and country suggesting that diagnosis, treatment, and control programs need to be based on a methodical evaluation of area-specific etiologies. From December 2006 to December 2009, 9,997 individuals presenting with acute febrile illness at nine health care clinics in south-central Cambodia were enrolled in a study to elucidate the etiologies. Upon enrollment, respiratory specimens, whole blood, and serum were collected. Testing was performed for viral, bacterial, and parasitic pathogens. Etiologies were identified in 38.0% of patients. Influenza was the most frequent pathogen, followed by dengue, malaria, and bacterial pathogens isolated from blood culture. In addition, 3.5% of enrolled patients were infected with more than one pathogen. Our data provide the first systematic assessment of the etiologies of acute febrile illness in south-central Cambodia. Data from syndromic-based surveillance studies can help guide public health responses in developing nations.
Introduction
Limited resources and the great diversity of acute febrile illness (AFI) etiologies in tropical regions challenge diagnosis, treatment, and public health responses to endemic and epidemic diseases. Further confounding this is the fact that a majority of the patients present with non-descript symptoms (e.g., low-grade fever, general malaise, headache, and muscle ache) and usually no focal point of infection. Health care providers lacking proper diagnostic tools are usually unable to determine specific etiologies, often diagnosing patients presumptively based on clinical features and assumptions regarding circulating pathogens.
Syndromic-based disease surveillance provides a useful methodology to systematically identify and document causes of acute fever. This approach has been used by TropNetEuropä1,2 to diagnose fevers of unknown origin in Turkey,3 China,4 and India and a project designed by the U.S. Centers for Disease Control and Prevention (CDC, Atlanta, GA) for the same purpose along the United States-Mexico border.
In Southeast (SE) Asia, a prospective study of Thai patients presenting with acute undifferentiated febrile illness determined the etiologies of over 40% of cases, with the most prevalent being influenza, dengue, and rickettsial infections.5 In Latin America, surveillance identified the etiology of acute undifferentiated febrile illness in Ecuador in 40% of enrolled patients, identifying a number of pathogens that have not previously been isolated in the country.6
Emerging and re-emerging diseases are a concern in SE Asia during a dynamic time of population growth, urbanization, and global migration. Infections from dengue virus, malaria parasites, influenza viruses, hepatitis viruses, rickettsiae, and Leptospira have previously been shown to impact populations across SE Asia. However, among the SE Asian nations, limited data exists on the etiologies of AFI in the Kingdom of Cambodia. Earlier studies reported illnesses among Cambodian refugee populations that had either immigrated to the United States or occupied refugee camps in Thailand.7,8 These works documented infections by agents such as malaria, hepatitis, cryptosporidium, and tuberculosis that presented in an often stressed and undernourished patient cohort. More recent studies conducted in Cambodia used both prospective and/or syndromic approaches. Infectious etiologies of encephalitic syndromes among patients admitted to a provincial hospital near Phnom Penh9 were identified in 35 of 88 (40%) human immunodeficiency virus (HIV)-seronegative patients; 11 bacterial, 1 fungal, and 23 viral infections (Japanese encephalitis virus [JEV] the principal causative agent of disease). A hospital-based study among children in Phnom Penh was conducted to diagnose hemorrhagic fever, encephalitis, or hepatitis in children,10 reporting that hepatitis A and JEV contributed greatly to the burden of disease in this population. In 2006, we initiated clinic-based surveillance to ascertain the etiologies of AFI among patients seeking treatment at peri-urban and rural health care clinics and hospital outpatient departments in Cambodia. Herein, we describe the demographics, symptoms, and identified etiologies of the patients seeking health care for AFI from December 2006 to December 2009.
Materials and Methods
Ethical considerations
Eligible subjects voluntarily enrolled in accordance with an Institutional Review Board protocol approved by U.S. Naval Medical Research Unit Two (NAMRU2) in compliance with all applicable Federal regulations governing the protection of human subjects and the National Ethics Committee of the Royal Kingdom of Cambodia, Ministry of Health. Written consent was obtained from patients 18 years of age and older. For patients younger than 18 years of age, written consent was obtained from a parent or legal guardian. Additionally, written assent was obtained from patients between 8 and 17 years of age. For all individuals, the informed consent was verbally explained and questions solicited.
Site selection and study enrollment
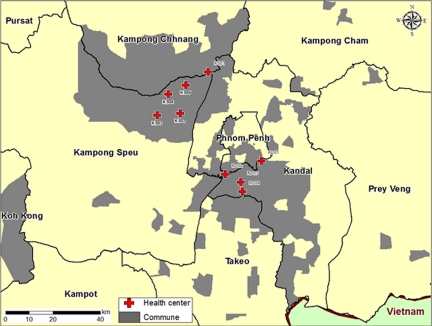
Clinic-based acute fever surveillance was initiated in December 2006. Patients were initially recruited from outpatient clinics of two referral hospitals, but during the course of the study, additional clinics were added: two in August 2007, one in October 2007, one in December 2007, one in February 2007, one in March 2008, and one in April 2008, for a total of nine. Five of the health care sites were located in Operational District A (peri-urban); four health care sites were located in Operational District B (rural). All health care sites were located within 50 km of Phnom Penh in south-central Cambodia (Figure 2).
Figure 2.
Catchment area (gray) of febrile surveillance study.
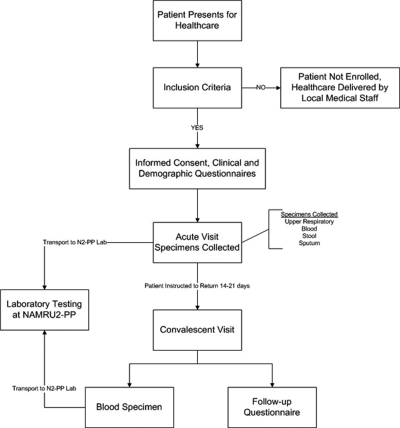
Patients were asked to enroll if they meet the inclusion criteria, which included: a measured tympanic membrane temperature > 38.0°C when at the study site and, as reported by the enrollee, a fever duration of at least 24 hours and < 10 days, were 2 years of age or older, and after medical examination, their illness was not able to be diagnosed during the patient’s physical examination or medical history.5 Patients were excluded if they declined to participate in the study or did not meet any of the inclusion criteria. A physician or medical assistant in each clinic obtained written informed consent, administered a pretested enrollment questionnaire, and performed a medical examination. Laboratory technicians collected blood and throat and nasal swabs. All study participants were directed to return 14 to 21 days for a follow-up appointment, in which a convalescent blood specimen was collected (Figure 1). All clinical specimens were transported to a central laboratory where testing for several infectious agents was performed.
Figure 1.
Flow chart of patient enrollment, information collected, and testing algorithms.
Definitions
Influenza-like illness was defined by fever (> 38.0°C), plus cough or sore throat.11–13 Diarrhea was defined as two or more loose liquid stools in any 24-hour period as all enrolled patients had the symptom of fever. Probable dengue was defined as only having a positive immunoglobulin M (IgM) result and confirmed dengue was defined as having a positive polymerase chain reaction (PCR) result. Recent travel was defined as the individual leaving their respective home for at least 5 consecutive days within 2 months preceding enrollment. Operational district is a designation used by the Cambodian government to denote a continuous geographic region under the purview of a single government office.
Specimen collection
For each enrollee, a blood specimen was obtained by venipuncture and one throat and one nasal swab were collected. For nasal swabs, a dry polyester swab was inserted into the nostril, parallel to the palate, slowly withdrawn, and placed in a vial containing 2–3 mL of virus transport medium. For throat swabs, both tonsils and the posterior pharynx were swabbed vigorously, and the swab placed in 2–3 mL of virus transport medium. All inoculated vials were kept at 4°C until transported to NAMRU2/NIPH’s laboratory. All specimens were accessioned at the laboratory between 24 and 72 hours post collection.
Malaria smears
At the time of enrollment a blood smear is prepared and transported (daily) to the laboratory for staining by Giemsa stain and interpretation by microscopy.
Blood cultures
A single blood culture was obtained from all enrolled patients in an aerobic BacT/Alert 3D (bioMerieux, Hazelwood, MO) bottle. Daily, inoculated bottles were transported to the NAMRU2 laboratory in Phnom Penh and then incubated for up to 7 days in the BacT/Alert blood culture system. A sample of blood from positive blood cultures was inoculated onto chocolate, MacConkey’s, and blood agar plates and incubated at 37°C, 5% CO2, for up to 48 hours. Bacterial identification was done using the API 20E biochemical identification system (bioMerieux).
Serological assays
All commercial enzyme-linked immunosorbent assay (ELISA) test assays were performed and interpreted according to the manufacturer’s instruction as Positive, Equivocal, and Negative. Dengue IgM serological assays were performed on all acute serum samples from all study participants using the Dengue IgM Capture ELISA Dx Select (Focus Diagnostics, Cypress, CA). Hepatitis A IgM was performed with the Hepalisa Anti-HAV IgM (PT INDEC DIAGNOSTICS, Jakarta, Indonesia). Hepatitis B surface antigen was detected using the Hepalisa HBsAg (PT INDEC DIAGNOSTICS). An IgM serological response to hepatitis E was detected using the HEV IgM ELISA 3.0 (MP Biomedicals, Singapore). An acute immunological response was detected to Leptospira using the Leptospira IgM ELISA (PANBIO, Brisbane, Australia).
The IgG serology was performed for dengue, Hantavirus, chikungunya, and Rickettsia on all convalescent serum samples. All commercial ELISA test assays were performed and interpreted according to the manufacturer’s instruction as Positive, Equivocal, and Negative. Convalescent specimens that were found to be IgG positive were paired with the acute serum and retested. A 4-fold increase in titer from acute to convalescent sample, or a result change going from negative to positive, was considered indicative of seroconversions. Detection of dengue IgG and hantavirus IgG were accomplished by using the Dengue IgG capture ELISA (PANBIO) and Hantavirus IgG ELISA Dx Select (Focus Diagnostics), respectively. Chikungunya virus-specific IgG was detected by ELISA as previously published.14 The IgG ELISA specific for typhus group and scrub typhus rickettsiae and spotted fever group ricketssiae were performed as previously described.15,16 A 4-fold increase in titer from acute to convalescent sample, or a result change going from negative to positive, was considered indicative of seroconversions.
PCR amplification
Influenza
The RNA was extracted from nasal and throat swabs using QIAamp viral RNA mini kits (QIAGEN, Hilden, Germany) following manufacturer’s instruction and stored at −80°C. Influenza virus genome was detected using a standardized real-time reverse transcription (rRT)-PCR assay developed by the U.S. CDC to detect influenza A and B viruses as well as influenza A viruses of H1, H3, and H5 subtypes. Real time assays were developed at the CDC and assays and primer sequences are available upon request under a Material Transfer Agreement. One-step rRT-PCR was performed in a final volume of 25 μL containing 5 μL of extracted RNA, 12.5 μL of buffer mix, and 0.5 μL Superscript III/Platinum Taq-Enzyme mix, 20 U of RNAse-out (Invitrogen, Carlsbad, CA), 0.8 μM for each primer, and 0.2 μM of probe. The Rotor-Gene 6000 real time thermocyler (Corbett Life Science, Sydney, Australia) was used for all PCR reactions. The thermocycling parameters for all targets consisted of 50°C for 30 minutes, 95°C for 2 minutes, and 45 cycles with 95°C for 15 seconds, 55°C for 30 seconds (U.S. CDC). In August 2009, a real-time PCR assay distributed by the U.S. CDC was introduced for the detection of pandemic H1N1 influenza virus. The Applied Biosystems 7500 real time thermocycler (Applied Biosystems, Foster City, CA) was used for all pH1N1 PCR reactions.
Nested conventional PCR for dengue
Dengue virus genome and serotype detection was based on previously published procedures with modifications to the Dengue-1 primer.17,18
Nested reverse real-time PCR for chikungunya virus
The RNA was extracted from serum using QIAamp viral RNA mini kits (QIAGEN) following manufacturer’s instruction and stored at −80°C. Chikungunya virus genome was detected using a nested RT-PCR assay, which comprises two assay steps; RT-PCR and nested PCR (nPCR). The RT reactions were performed in 25 μL containing 5 μL of Access AMV-Tfl buffer (Promega, Madison WI), 1.5 μL of 25 mM MgSO4, 1 μL of 10 mM deoxynucleoside triphosphate (dNTP; final concentration, 200 μM), 20 pico moles each of all outer primers sets, 0.5 μL of AMV-reverse transcriptase, and 0.5 μL of 5 U Tfl polymerase (Promega), and 5 μL of RNA template. The PCR thermal cycling was performed as follows: 48°C for 45 minutes, 94°C for 2 minutes, and 40 cycles with 94°C for 1 minute, 60°C for 1 minute, 68°C for 1 minute, and then 68°C for 7 minutes. All thermal cycling was performed with Applied Biosystems type 9700 machine. Inner primer pairs are then used in separate nPCR reactions using GeneAmp PCR Core reagent (Applied Biosystems). The nPCR reactions were performed in 25 μL containing 2.5 μL of ABI PCR buffer, 1.5 μL of 25 mM MgCl2, 2.0 μL of 10 mM dNTP (final concentration 200 μM), 1.0 μL of 10 μM forward primer, and 1.0 μL of reverse primer, 0.25 μL of 5 units of AmpliTaq DNA Polymerase, and 5 μL of diluted (1:50) RT-PCR product. The cycle conditions of the nPCR reaction were as follows; 94°C for 2 minutes, and 40 cycles with 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and then 72°C for 10 minutes. Outer and inner primer pairs are designed to amplify three structural gene regions, Capsid (C), Envelope-2 (E2), and an area overlapping the 6K and E1 genes (6K/E1).14 The PCR products were visualized on 2% agarose gels stained with ethidium bromide under UV light. A positive result was indicated upon observation of a 200 base pair PCR product, representing the capsid, and at least one of the envelope targets is present.
Quantitative real-time PCR (qPCR) for rickettsiae and Orientia tsutsugamushi (scrub typhus)
Deoxyribonucleic acid (DNA) was extracted from serum using QIAamp DNA mini kits (QIAGEN) following manufacturer’s instruction and stored at −80°C. The qPCR assays developed to detect Rickettsia spp. (Rick17), tick-borne rickettsiae (Trick), Rickettsia typhi (Rtyph), Rickettsia felis (Rfelis), and O. tsutsugamushi (Otsu47) were performed as previously described.19,20 Briefly, primer pairs were used in separate qPCR assays in a 25 μL final volume: Rick17, Trick, and Rtyph assays contained 2 μL of DNA, 12.5 μL of 2×TaqMan Universal PCR Master Mix (Cat. no. ABI 4304437), 0.4 μM for each primer, and 0.3 μM of probe. Orientia tsutsugamushi qPCR assay contained 5 μL of DNA, 12.5 μL of 2×TaqMan Universal PCR Master mix (Cat. no. ABI 4304437), 0.4 μM for each primer and 0.3 μM of probe; and the R. felis PCR assay contained 2 μL of DNA, 12.5 μL of 2×TaqMan Universal PCR Master mix (Cat. no. ABI 4304437), 0.4 μM for each primer and 0.4 μM of probe. The Applied Biosystems 7500 real time thermocycler (Applied Biosystems) was used for all real-time PCR assays. The thermocycling program used consisted of the following parameters; 50°C for 2 minutes, 95°C for 10 minutes, and 60 cycles with 95°C for 15 seconds, 60°C for 30 seconds.
rRT-PCR for hantaviruses
Hantavirus RNA was extracted from serum using QIAamp viral RNA mini kits (QIAGEN) following manufacturer’s instruction and stored at −80°C. An rRT-PCR assay was used to detect Hantaviruses serotypes Hantaan (HTN), Puumala (PUU), Seoul (SEO), and Dobrava-Belgrade (DOB).21 One-step rRT-PCR was performed in a final volume of 25 μL containing 5 μL of extracted RNA, 12.5 μL of buffer mix, 0.5 μL of MgSO4, and 0.625 μL Superscript III/Platinum Taq-Enzyme mix, 0.5 μL of RNAse-out (Invitrogen), 0.4 μM for each primer, and 0.25 μM of probe. The Rotor-Gene 6000 real time thermocyler (Corbett Life Science, Sydney, Australia) was used for all PCR reactions. The thermocycling parameters for all targets consisted of 50°C for 15 minutes, 95°C for 2 minutes, and 45 cycles with 95°C for 15 seconds, 55°C for 30 seconds, and 72°C for 1 minute.
Nested RT-PCR for detection of hepatitis E viruses
The RNA was extracted from serum using QIAamp viral RNA mini kits (QIAGEN) following manufacturer’s instruction and stored at −80°C. The nonstructural (NS) gene of Hepatitis E virus was detected using a nested reverse real-time PCR assay.22 The RT reactions were performed in 25 μL containing 5 μL of Qiagen buffer mix (Qiagen, Cat. no. 210212) and 1 μL of enzyme mix, 1 μL of 10 mM dNTP, 0.63 μM of each primer, 0.15 μL of 40 U RNase inhibitor (Promega, Cat. no. N2511), and 5 μL of RNA template. The PCR thermal cycling was performed as follows: 45°C for 30 minutes, 94°C for 15 minutes, and 40 cycles with 94°C for 1 minute, 45°C for 1 minute, 72°C for 1 minute, and then 72°C for 7 minutes. The PCR reactions were performed using the Applied Biosystems 9700 (Applied Biosystems). An nPCR reaction was carried out at a total volume of 25 μL. The reaction contained 2.5 μL of ABI PCR buffer (Applied Biosystems, Cat. no. N808-0009), 1.5 mM MgCl2, 2 μL of 10 mM dNTP, 0.75 μM forward primer and reverse primer, 0.125 μL of 5 U of AmpliTaq DNA Polymerase, and 5 μL of diluted (1:50) RT-PCR product. The cycle conditions of nPCR reaction are as follows: 94°C for 2 minutes, and 40 cycles with 94°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute, and then 72°C for 7 minutes. Products were visualized on 2% agarose gels stained with ethidium bromide under UV light.
Statistical analysis
All data was double-data entered into MS Access (Microsoft Inc., Redmond, WA). Data was imported into SAS v 9.1 (SAS, Cary, NC), which was used for all statistical analyses.
Results
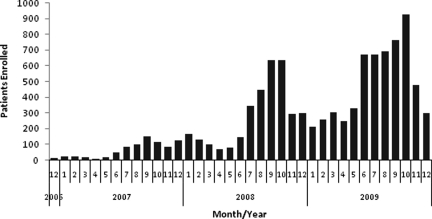
From December 2006 through December 2009, 9,997 patients were enrolled (Figure 3). All patients enrolled met the criteria of fever (> 38.0°C). Among the 9,997 patients, 89.3% had a complete questionnaire. A serum sample, nose and/or throat swab, and blood culture sample was available from 99.8%, 99.7%, and 98.2%, respectively.
Figure 3.
Enrollment by month Dec. 2006–Dec. 2009
The median age of enrolled patients was 13 years of age (interquartile range [IQR] 6–28) years of age (Table 1) and 53.9% were male. Patients reported both recent travel to other provinces or countries (7.9%) and travel into forested areas (7.1%). Study subject occupation was recorded, however students and children accounted for 63.5% of enrolled study subjects. Among study subjects reporting a specific occupation, grain farmers were most common (31.3%). Contact with cattle (8.3%), chickens (7.7%), and dogs (7.0%) were the most common reported animal exposures. Prior medication usage was reported by 36.6% of enrolled subjects. The most common symptoms reported by enrolled patients included headache (69.5%), cough (57.5%), sore throat (46.6%), chills (48.1%), and malaise (46.8%) (Table 1). Among enrolled participants, 65.7% presented with signs and symptoms characteristic of influenza-like illness, 17.1% with gastroenteritis, and 1.4% with jaundice. Close contact with an individual who had similar symptoms was reported among 9.0% of participants. Of the enrolled patients, 10.1% were hospitalized or referred for hospitalization.
Table 1.
Demographics of enrolled patients from Dec. 2006 to Dec. 2009
| Demographic and clinical information | n | % | Demographic and clinical information | n | % |
|---|---|---|---|---|---|
| Patients enrolled | 9,997 | Temperature, mean (SD) | 39 (0.6) | ||
| Age (years), mean (SD) | 19.6(16.9) | Malaise | 4,675 | 46.8 | |
| Age (years), median (SD) | 13.0(16.9) | Chills | 4,807 | 48.1 | |
| Gender | Muscle aches | 2,319 | 23.2 | ||
| Male | 5,392 | 54.0 | Rash | 197 | 2.0 |
| Female | 4,601 | 46.0 | Lesion | 96 | 1.0 |
| Medication | 3,627 | 36.3 | Joint pain | 1,894 | 19.0 |
| Travel history | Headache | 6,947 | 69.5 | ||
| Province/country | 793 | 7.9 | Seizure | 83 | 0.8 |
| Jungle | 708 | 7.1 | Sore throat | 4,655 | 46.6 |
| Occupation | Cough | 5,748 | 57.5 | ||
| Cattle/sheep farmer | 9 | 0.1 | Shortness of breath | 964 | 9.7 |
| Grain farmer | 3,133 | 31.3 | Nausea | 876 | 8.8 |
| Poultry farmer | 18 | 0.2 | Vomiting | 1,332 | 13.4 |
| Driver | 16 | 0.2 | Abdominal cramp | 2,412 | 24.2 |
| Factory worker | 387 | 3.9 | Diarrhea | 580 | 5.8 |
| Fisherman | 17 | 0.2 | Bloody stool | 66 | 0.7 |
| Office worker | 50 | 0.5 | Bloody urine | 20 | 0.2 |
| Other | 6,347 | 63.5 | Bleeding | 25 | 0.3 |
| Exposure history | Jaundice | 132 | 1.4 | ||
| Cats | 278 | 2.8 | |||
| Dogs | 700 | 7.0 | Person same symptom | 893 | 9.0 |
| Cattle | 824 | 8.3 | Patient disposition | ||
| Horses | 5 | 0.1 | Admitted at hospital | 990 | 9.9 |
| Goat/sheep | 5 | 0.1 | Outpatient follow-up | 8,975 | 89.9 |
| Pigs | 81 | 0.8 | Referred to hospital | 15 | 0.2 |
| Monkeys | 7 | 0.1 | |||
| Rodents | 8 | 0.1 | |||
| Chickens | 765 | 7.7 | |||
| Ducks | 186 | 1.9 |
Laboratory testing was performed for agents believed to be endemic to the region in addition to a variety of emerging pathogens (Table 2). Of enrolled patients, 19.9% (N = 1,983) were PCR positive for influenza. Dengue virus was considered the probable cause of illness (as defined by only a positive IgM result) in 842 enrollees (8.4%) and confirmed (as defined by a positive PCR result) in 883 cases (8.9%). All four serotypes of dengue were detected during the study period. Malaria was identified among 7.2% (N = 716) of patients, with Plasmodium falciparum identified in 30.2% of malaria cases. In addition, a total of 19 mixed infections of Plasmodiuom vivax and P. falciparum were identified. Acute leptospirosis was serologically identified in 20.8% of study subjects. Hepatitis A IgM testing was performed on the first 3,243 patients enrolled, 56 (1.7%) of the patients were positive. Hepatitis B surface antigen testing was done on the first 4,596 enrollees, and 261 (5.8%) were positive. Follow-up on testing of HBsAG-positive samples identified eight samples that were also positive for IgM. A hepatitis E IgM response was detected among 11.1% of participants. The PCR testing of 517 serum samples that were positive or equivocal for Hepatitis E IgM identified five samples positive for hepatitis E nucleic acid.
Table 2.
Distribution of pathogens identified among enrolled patients from Dec. 2006 to Dec. 2009
| Pathogens | Tested cases | Positive (N) | Positive (%) |
|---|---|---|---|
| Influenza viruses | 9,968 | 1,983 | 19.9 |
| H1N1 | 224 | 11.3 | |
| H3N2 | 827 | 41.7 | |
| H5N1 | 1 | < 0.1 | |
| Pandemic H1N1 | 286 | 14.4 | |
| Influenza B | 627 | 31.6 | |
| Influenza A, untyped | 18 | 0.9 | |
| H1N1 and influenza B | 1 | < 0.1 | |
| H3N2 and influenza B | 3 | 0.2 | |
| Pandemic H1N1 and H3N2 | 4 | 0.2 | |
| Malaria | 9,954 | 716 | 7.2 |
| P. falciparum | 216 | 30.2 | |
| P. vivax | 481 | 67.2 | |
| P. falciparum and P. vivax | 19 | 2.6 | |
| Dengue | 9,975 | ||
| IgM positive only (probable dengue) | 842 | ||
| PCR positive (confirmed dengue) | 883 | ||
| Serotype-1 | 83 | 9.4 | |
| Serotype-2 | 398 | 45.1 | |
| Serotype-3 | 108 | 12.2 | |
| Serotype-4 | 294 | 33.3 | |
| Blood-borne pathogen | 9,821 | 629 | 6.3 |
| Leptospirosis IgM | 9,975 | 2,076 | 20.8 |
| Hepatitis A IgM | 3,243 | 58 | 1.7 |
| Hepatitis B sAG | 4,596 | 267 | 5.8 |
| Hepatitis B IgM | 267 | 8 | 3.0 |
| Hepatitis E IgM | 9,353 | 1038 | 11.1 |
| Hepatitis E PCR | 517 | 5 | 1.0 |
Serological testing of paired samples was conducted for antibodies against chikungunya, hantavirus, and rickettsiae (scrub typhus group, typhus group, and spotted fever group) (Table 3). Testing of chikungunya IgG among the first 753 patients enrolled in 2007 identified a previous exposure in 16.3%. Chikungunya IgM was identified among 0.1% (2 of 1,438) of enrolled patients and PCR of patients presenting with fever and joint pain identified three cases out of 571 samples tested.
Table 3.
Pathogens identified from testing of acute and convalescent serum samples
| Pathogens | Tested cases | Positive (N) | Positive (%) |
|---|---|---|---|
| Chikungunya IgG (convalescent) | 753 | 123 | 16.3 |
| Chikungunya IgM | 1438 | 2 | 0. 1 |
| Chikungunya PCR | 571 | 3 | 0.5 |
| Hantavirus IgG (convalescent) | 5675 | 459 | 8.1 |
| Hantavirus 4×IgG rise and/or seroconverter | 459 | 71 | 15.5 |
| Hantavirus PCR | 79 | 0 | 0.0 |
| Scrub typhus group IgG (convalescent) | 1906 | 133 | 7.0 |
| Scrub typhus group-specific IgG 4-fold rise in titer or seroconversion | 133 | 35 | 26.3 |
| Orientia tsutsugamushi-specific qPCR | 67 | 2 | 3.0 |
| Typhus group IgG (convalescent) | 1946 | 261 | 13.4 |
| Typhus group-specific IgG 4-fold rise in titer or seroconversion | 261 | 56 | 21.5 |
| R. typhi-specific qPCR | 83 | 1 | 1.2 |
| Spotted fever group IgG (convalescent) | 1263 | 146 | 11.6 |
| Spotted fever group-specific IgG 4-fold rise in titer or seroconversion | 146 | 21 | 14.4 |
| Tick-borne rickettsiae-specific qPCR | 37 | 0 | 0.0 |
Hantavirus IgG was detected in convalescent specimens of 8.1% of patients tested. Paired testing of acute and convalescent samples identified 71 patients that had evidence of an acute hantavirus infection. No IgM-positive or PCR-positive cases of hantaviral infection were identified by IgM or molecular analysis.
Testing for scrub typhus group, typhus group, and spotted fever group rickettsiae-specific IgG in convalescent specimens identified seroprevalences of 7.0%, 13.4%, and 11.6%, respectively (Table 3). From positive convalescent samples, testing of paired samples to identify acute cases identified 35, 56, and 21 cases of scrub typhus, typhus, and spotted fever, respectively. Testing acute serum samples (seroconversion-positive samples) by qPCR assays confirmed two cases of scrub typhus and one case of murine typhus. No species of the spotted fever group were identified by qPCR.
Blood culture testing yielded a bacterial isolate in 6.3% (642 of 9,821) of study subjects (Table 4). The most common isolates were non-pathogenic skin flora (e.g., coagulase-negative Staphylococcus, micrococcus sp., bacillus sp. (non-anthracis); however, a number of known pathogens were also identified including Salmonella typhi (57), Staphylococcus aureus (4), Burkholderia pseudomallei (2), group C Streptococcus (1), and Moraxella catarrhalis (6) (Table 4).
Table 4.
Organisms isolated from 9821 patient blood cultures
| Organism | TOTAL | Organism | TOTAL |
|---|---|---|---|
| Pathogens: Achromobacter xylosoxidans | 1 | Potential pathogens: Acinetobacter baumannii/calcoaceticus | 3 |
| Actinomyces radingae | 3 | Acinetobacter lwoffii | 3 |
| Beta hemolytic streptococci group C | 1 | Bacillus sp. | 2 |
| Brevundimonas vesicularis | 7 | Chryseobacterium indologenes | 1 |
| Burkholderia cepacia | 1 | Mannheimia haemolytica/P. trehalosi | 3 |
| Burkholderia pseudomallei | 2 | Ochrobactrum anthropi | 1 |
| Edwardsiella tarda | 1 | Pseudomonas fluorescens | 1 |
| Escherichia coli | 12 | Pseudomonas oryzihabitans | 1 |
| Group D Salmonella sp. non-typhi | 1 | Pseudomonas putida | 1 |
| Gemella morbillorum | 2 | Pseudomonas stutzeri | 1 |
| Moraxella catarrhalis | 6 | ||
| Moraxella sp. non-catarrhalis | 3 | Likely contaminants: | |
| Neisseria sp. non-meningitidis | 2 | Aerococcus viridans | 1 |
| Pasteurella sp. | 1 | Arthrobacter sp. | 37 |
| Pseudomonas aeruginosa | 1 | Brevibacterium sp. | 1 |
| Ralstonia pickettii | 1 | Cellulomonas sp. | 1 |
| Rhodococcus sp. | 15 | Cellulosimicrobium cellulans | 1 |
| Salmonella choleraesuis | 10 | Coagulase negative Staphylococcus | 203 |
| Salmonella Group D sp. non-typhi | 1 | Corynebacterium sp. | 16 |
| Salmonella ser. Paratyphi A | 6 | Leifsonia aquatica | 1 |
| Salmonella typhi | 57 | Microbacterium sp. | 35 |
| Sphingomonas paucimobilis | 6 | Micrococcus sp. | 20 |
| Staphylococcus aureus | 4 | ||
| Stenotrophomonas maltophilia | 2 | ||
| Streptococcus constellatus | 1 | ||
| Streptococcus mitis | 1 | ||
| Streptococcus pneumonia | 1 | ||
| Aspergillus sp. | 2 | ||
| Candida sp. other than albicans | 14 |
To identify active co-infections, we limited analysis to patients who were positive by a combination of PCR, blood culture, and/or malaria microscopy. There were 94 (1.0%) patients whose specimens were positive by two or more of these tests (Table 5). Influenza was identified with confirmed dengue in 55 cases, with malaria in 8 cases, and an isolate from blood culture in 10 cases. Dengue was identified with malaria in 15 cases and a blood culture isolate in 4 cases (1 B. pseudomallei/dengue-2, 2 S. typhi/dengue-4, and 1 S. typhi/dengue-2). In one patient, influenza was identified with Salmonella cholerasuis and dengue-4.
Table 5.
List of unique or uncommon co-infections identified from polymerase chain reaction (PCR) positive
| Co-infection (n) | n | Notes |
|---|---|---|
| Influenza/dengue | 55 | 55 are PCR positive for dengue |
| Influenza/malaria | 8 | |
| Influenza/blood culture | 10 | |
| Dengue/malaria | 15 | 15 are PCR positive for dengue |
| Dengue/blood culture | 1 | B. pseudomallei and dengue-2 |
| Dengue/blood culture | 2 | S. typhi and dengue-4 |
| Dengue/blood culture | 1 | S. typhi and dengue-2 |
| Malaria/blood culture | 1 | |
| Influenza/dengue/blood culture | 1 | S. cholerasuis and dengue-4 |
Discussion
The purpose of this study was to identify the infectious etiologies of AFI among patients 2 years of age and older presenting for treatment at clinics in south-central Cambodia. This is the first comprehensive study to identify etiologies of AFI in the general population of Cambodia.
Our study enrolled nearly 10,000 patients over a 3-year period. During this time period, there was a seasonality to enrollment that corresponded to the rainy season, which roughly corresponds with the influenza season in Cambodia23 peaking in August–October of each year.
In this study, we identified influenza, Leptospira, dengue, hepatitis E, and malaria as the most frequently identified pathogens associated with AFI among Cambodians. Previous regional studies had identified similar etiologies among patients with AFI,5,6 with some variation of etiologies and distribution that could be based on the sampled population or the pathogen testing schemes. However, we identified that many of the acute serological responses to Leptospira and hepatitis E were associated with co-detection of other organisms (data not shown).
A large number of study subjects were classified chronic hepatitis B cases; additional testing revealed that only a small number of those individuals presented with an AFI caused by hepatitis B. A recent study of hepatitis B among blood donors found a similar percentage of HBsAG-positive volunteers24 in Cambodia.
Despite a lack of jaundice reported among the study population, a high percentage of patients showed an acute serological response to hepatitis E. Of the 175 patients presenting with clinical jaundice, only 29 (16.6%) were identified to have a positive hepatitis E IgM response. This represents a diagnostic challenge that may be caused by a long-term IgM response or continuous exposure in the environment to hepatitis E virus.
A large number of patients were positive for an acute serological response to Leptospira. A recent study focusing on hospitalized patients in Cambodia found a similar prevalence of acute or prior Leptospira infection, with approximately half of the patients presenting with an acute infection.25 Although leptospirosis has been documented to be endemic in the region,26 the more definitive microscopic agglutination testing (MAT) was not performed on these samples. In addition, a recent study showed poor diagnostic accuracy of the Leptospira IgM PanBio ELISA compared with MAT testing.27 Further laboratory studies and analysis is needed to understand the contribution of Leptospira to AFI in Cambodia.
Almost 1% of enrollees tested had results suggesting an acute hantavirus infection. However, PCR testing failed to identify Hantaan, Seoul, Puumala, or Dobrava strains among any study subjects. This may reflect genetic differences of Hantavirus strains in Cambodia that need to be further defined. Alternatively, the serology results may be from a cross-reaction from a similar yet previously unidentified pathogen. Previous studies conducted in Cambodia have identified the presence of Hantaviruses in rodent populations.28,29 In addition, because of biosafety and biosecurity limitations, we were unable to perform antibody neutralization studies on these samples.
The pathogen yield from blood culture was considerably low, likely caused by the single sampling used. However, we were able to detect a number of known pathogens (e.g., B. pseudomallei, S. typhi, S. paratyphi) as well as common skin contaminants (e.g., coagulase negative Staphylococcus, microbacterium). In addition, there were a number of pathogens that have limited reports of causing disease such as Sphingomonas paucimobilus,30 that require further investigation. Planned studies at these sites will expand molecular and serological techniques to determine the prevalence of bacterial pathogens among patients with AFI in Cambodia.
Co-detection was limited to PCR-positive results or culture/smear results where organisms were visualized. This work identified a number of unique co-infections such as Dengue and B. pseudomallei and S. typhi. The co-infections with malaria may represent asymptomatic infections among our study population. Currently, our group has several malaria-specific studies underway, accessing prevalence, incidence, and drug resistance. Our study identified over 50 co-infected cases of influenza and dengue, previous reports have also identified cases and concurrent outbreaks.5,31,32 Further studies are required to better understand the clinical outcome of patients who have tested positive for multiple pathogens.
The use of laboratory-based syndromic surveillance can alert regional officials and describe common infectious etiologies circulating in the region during a specific time period. Furthermore, laboratory-based syndromic surveillance can be used to help define symptoms and information that can be used for electronic-based surveillance programs and help to guide outbreak response and laboratory testing.
Testing for a number of pathogens in all samples as opposed to using a staged screening strategy allowed us to identify co-infections that may have otherwise been missed. The diversity of etiological agents, an inability to identify an etiology in the majority of enrolled patients, and the prevalence of multiple infections, demonstrate the complexity of diagnosis and treatment of medical care providers in this region of Cambodia.
ACKNOWLEDGEMENTS
We are grateful to the clinicians and medical staff at the field sites in Cambodia for their assistance in enrolling and sampling patients. We thank laboratory personnel and field staff of NAMRU2-Phnom Penh for the conduct of work performed in support of this study.
Disclaimer: The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the U.S. Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Financial support: This work was funded by the U.S. Department of Defense Global Emerging Infectious Systems (DoD-GEIS), the Biosecurity Engagement Program, U.S. Department of State, and the U.S. Centers for Disease Control and Prevention.
Disclosure: This work was prepared as part of the authors' official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Authors’ addresses: Matthew R. Kasper, Department of Bacteriology, U.S. Naval Medical Research Unit 6, Lima, Peru, E-mail: matthew.kasper@med.navy.mil. Patrick Blair, Naval Health Research Center, San Diego, CA, E-mail: Patrick.blair@med.navy.mil. Sok Touch, Communicable Disease Control Department, Phnom Penh, Kingdom of Cambodia, E-mail: touch358@moh.gov.kh. Buth Sokhal, National Institute of Public Health, Phnom Penh, Kingdom of Cambodia, E-mail: buthsokhal@yahoo.com. Chadwick Yasuda, U.S. Naval Medical Research Unit Two, Phnom Penh, Kingdom of Cambodia, E-mail: chad@namru2.org.kh. Maya Williams, Department of Virology, U.S. Naval Medical Research Unit 6, Lima, Peru, E-mail: Maya.williams@med.navy.mil. Allen Richards, Naval Medical Research Center, Silver Spring, MD, E-mail: Allen.Richards@med.navy.mil. Timothy Burgess, Infectious Disease Clinical Research Program, Uniformed Services University of Health Sciences, Bethesda, MD, E-mail: tburgess@idcrp.org. Thomas Wierzba, International Vaccine Institute, Seoul, Korea, E-mail: twierzba@ivi.int. Shannon Putnam, Naval Health Research Center, San Diego, CA, E-mail: Shan.putnam@med.navy.mil.
References
- 1.Jelinek T, Muhlberger N, Harms G, Corachan M, Grobusch MP, Knobloch J, Bronner U, Laferl H, Kapaun A, Bisoffi Z, Clerinx J, Puente S, Fry G, Schulze M, Hellgren U, Gjorup I, Chalupa P, Hatz C, Matteelli A, Schmid M, Nielsen LN, da Cunha S, Atouguia J, Myrvang B, Fleischer K. Epidemiology and clinical features of imported dengue fever in Europe: sentinel surveillance data from TropNetEurop. Clin Infect Dis. 2002;35:1047–1052. doi: 10.1086/342906. [DOI] [PubMed] [Google Scholar]
- 2.Jelinek T, Peyerl-Hoffmann G, Muhlberger N, Wichmann O, Wilhelm M, Schmider N, Grobusch MP, von Sonnenburg F, Gascon J, Laferl H, Hatz C, Alifrangis M, Burchard G, McWhinney P, Schulze M, Kollaritsch H, da Cunha S, Beran J, Kern P, Gjorup I, Cuadros J. Molecular surveillance of drug resistance through imported isolates of Plasmodium falciparum in Europe. Malar J. 2002;1:11. doi: 10.1186/1475-2875-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabak F, Mert A, Celik AD, Ozaras R, Altiparmak MR, Ozturk R, Aktuglu Y. Fever of unknown origin in Turkey. Infection. 2003;31:417–420. doi: 10.1007/s15010-003-3040-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhiyong Z, Bingjun L, Xiaoju L, Xinjian F, Ping F, Wenya W. Fever of unknown origin: a report from China of 208 cases. Int J Clin Pract. 2003;57:592–596. [PubMed] [Google Scholar]
- 5.Leelarasamee A, Chupaprawan C, Chenchittikul M, Udompanthurat S. Etiologies of acute undifferentiated febrile illness in Thailand. J Med Assoc Thai. 2004;87:464–472. [PubMed] [Google Scholar]
- 6.Manock SR, Jacobsen KH, de Bravo NB, Russell KL, Negrete M, Olson JG, Sanchez JL, Blair PJ, Smalligan RD, Quist BK, Espin JF, Espinoza WR, MacCormick F, Fleming LC, Kochel T. Etiology of acute undifferentiated febrile illness in the Amazon basin of Ecuador. Am J Trop Med Hyg. 2009;81:146–151. [PubMed] [Google Scholar]
- 7.Arthur JD, Bodhidatta L, Echeverria P, Phuphaisan S, Paul S. Diarrheal disease in Cambodian children at a camp in Thailand. Am J Epidemiol. 1992;135:541–551. doi: 10.1093/oxfordjournals.aje.a116321. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland JE, Avant RF, Franz WB, 3rd, Monzon CM, Stark NM. Indochinese refugee health assessment and treatment. J Fam Pract. 1983;16:61–67. [PubMed] [Google Scholar]
- 9.Srey VH, Sadones H, Ong S, Mam M, Yim C, Sor S, Grosjean P, Reynes JM, Grosjean P, Reynes JM. Etiology of encephalitis syndrome among hospitalized children and adults in Takeo, Cambodia, 1999–2000. Am J Trop Med Hyg. 2002;66:200–207. doi: 10.4269/ajtmh.2002.66.200. [DOI] [PubMed] [Google Scholar]
- 10.Chhour YM, Ruble G, Hong R, Minn K, Kdan Y, Sok T, Nisalak A, Myint KS, Vaughn DW, Endy TP. Hospital-based diagnosis of hemorrhagic fever, encephalitis, and hepatitis in Cambodian children. Emerg Infect Dis. 2002;8:485–489. doi: 10.3201/eid0805.010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boivin G, Hardy I, Tellier G, Maziade J. Predicting influenza infections during epidemics with use of a clinical case definition. Clin Infect Dis. 2000;31:1166–1169. doi: 10.1086/317425. [DOI] [PubMed] [Google Scholar]
- 12.Navarro-Mari JM, Perez-Ruiz M, Cantudo-Munoz P, Petit-Gancedo C, Jimenez-Valera M, Rosa-Fraile M. Influenza-like illness criteria were poorly related to laboratory-confirmed influenza in a sentinel surveillance study. J Clin Epidemiol. 2005;58:275–279. doi: 10.1016/j.jclinepi.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson KG. Clinical features of influenza. Semin Respir Infect. 1992;7:26–37. [PubMed] [Google Scholar]
- 14.Porter KR, Tan R, Istary Y, Suharyono W, Sutaryo, Widjaja S, Ma’Roef C, Listiyaningsih E, Kosasih H, Hueston L, McArdle J, Juffrie M. A serological study of Chikungunya virus transmission in Yogyakarta, Indonesia: evidence for the first outbreak since 1982. Southeast Asian J Trop Med Public Health. 2004;35:408–415. [PubMed] [Google Scholar]
- 15.Graf PC, Chretien JP, Ung L, Gaydos JC, Richards AL. Prevalence of seropositivity to spotted fever group rickettsiae and Anaplasma phagocytophilum in a large, demographically diverse US sample. Clin Infect Dis. 2008;46:70–77. doi: 10.1086/524018. [DOI] [PubMed] [Google Scholar]
- 16.Richards AL, Soeatmadji DW, Widodo MA, Sardjono TW, Yanuwiadi B, Hernowati TE, Baskoro AD, Roebiyoso, Hakim L, Soendoro M, Rahardjo E, Putri MP, Saragih JM, Strickman D, Kelly DJ, Dasch GA, Olson JG, Church CJ, Corwin AL. Seroepidemiologic evidence for murine and scrub typhus in Malang, Indonesia. Am J Trop Med Hyg. 1997;57:91–95. doi: 10.4269/ajtmh.1997.57.91. [DOI] [PubMed] [Google Scholar]
- 17.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynes JM, Ong S, Mey C, Ngan C, Hoyer S, Sall AA. Improved molecular detection of dengue virus serotype 1 variants. J Clin Microbiol. 2003;41:3864–3867. doi: 10.1128/JCM.41.8.3864-3867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry KM, Jiang J, Rozmajzl PJ, Azad AF, Macaluso KR, Richards AL. Development of quantitative real-time PCR assays to detect Rickettsia typhi and Rickettsia felis, the causative agents of murine typhus and flea-borne spotted fever. Mol Cell Probes. 2007;21:17–23. doi: 10.1016/j.mcp.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J, Chan TC, Temenak JJ, Dasch GA, Ching WM, Richards AL. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am J Trop Med Hyg. 2004;70:351–356. [PubMed] [Google Scholar]
- 21.Aitichou M, Saleh SS, McElroy AK, Schmaljohn C, Ibrahim MS. Identification of Dobrava, Hantaan, Seoul, and Puumala viruses by one-step real-time RT-PCR. J Virol Methods. 2005;124:21–26. doi: 10.1016/j.jviromet.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Seriwatana J, Shrestha MP, Scott RM, Tsarev SA, Vaughn DW, Myint KS, Innis BL. Clinical and epidemiological relevance of quantitating hepatitis E virus-specific immunoglobulin M. Clin Diagn Lab Immunol. 2002;9:1072–1078. doi: 10.1128/CDLI.9.5.1072-1078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blair PJ, Wierzba TF, Touch S, Vonthanak S, Xu X, Garten RJ, Okomo-Adhiambo MA, Klimov AI, Kasper MR, Putnam SD. Influenza epidemiology and characterization of influenza viruses in patients seeking treatment for acute fever in Cambodia. Epidemiol Infect. 2010;138:199–209. doi: 10.1017/S095026880999063X. [DOI] [PubMed] [Google Scholar]
- 24.Ol HS, Bjoerkvoll B, Sothy S, Van Heng Y, Hoel H, Husebekk A, Gutteberg T, Larsen S, Husum H. Prevalence of hepatitis B and hepatitis C virus infections in potential blood donors in rural Cambodia. Southeast Asian J Trop Med Public Health. 2009;40:963–971. [PubMed] [Google Scholar]
- 25.Berlioz-Arthaud A, Guillard B, Goarant C, Hem S. Hospital-based active surveillance of human leptospirosis in Cambodia. Bull Soc Pathol Exot. 2010;103:111–118. doi: 10.1007/s13149-010-0043-2. [DOI] [PubMed] [Google Scholar]
- 26.Laras K, Cao BV, Bounlu K, Nguyen TK, Olson JG, Thongchanh S, Tran NV, Hoang KL, Punjabi N, Ha BK, Ung SA, Insisiengmay S, Watts DM, Beecham HJ, Corwin AL. The importance of leptospirosis in southeast Asia. Am J Trop Med Hyg. 2002;67:278–286. doi: 10.4269/ajtmh.2002.67.278. [DOI] [PubMed] [Google Scholar]
- 27.Blacksell SD, Smythe L, Phetsouvanh R, Dohnt M, Hartskeerl R, Symonds M, Slack A, Vongsouvath M, Davong V, Lattana O, Phongmany S, Keolouangkot V, White NJ, Day NP, Newton PN. Limited diagnostic capacities of two commercial assays for the detection of Leptospira immunoglobulin M antibodies in Laos. Clin Vaccine Immunol. 2006;13:1166–1169. doi: 10.1128/CVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henttonen H, Buchy P, Suputtamongkol Y, Jittapalapong S, Herbreteau V, Laakkonen J, Chaval Y, Galan M, Dobigny G, Charbonnel N, Michaux J, Cosson JF, Morand S, Hugot JP. Recent discoveries of new hantaviruses widen their range and question their origins. Ann N Y Acad Sci. 2008;1149:84–89. doi: 10.1196/annals.1428.064. [DOI] [PubMed] [Google Scholar]
- 29.Reynes JM, Soares JL, Hue T, Bouloy M, Sun S, Kruy SL, Flye Sainte Marie F, Zeller H. Evidence of the presence of Seoul virus in Cambodia. Microbes Infect. 2003;5:769–773. doi: 10.1016/s1286-4579(03)00149-7. [DOI] [PubMed] [Google Scholar]
- 30.Lin JN, Lai CH, Chen YH, Lin HL, Huang CK, Chen WF, Wang JL, Chung HC, Liang SH, Lin HH. Sphingomonas paucimobilis bacteremia in humans: 16 case reports and a literature review. J Microbiol Immunol Infect. 2010;43:35–42. doi: 10.1016/S1684-1182(10)60005-9. [DOI] [PubMed] [Google Scholar]
- 31.Lopez Rodriguez E, Tomashek KM, Gregory CJ, Munoz J, Hunsperger E, Lorenzi OD, Irizarry JG, Garcia-Gubern C. Co-infection with dengue virus and pandemic (H1N1) 2009 virus. Emerg Infect Dis. 2010;16:882–884. doi: 10.3201/eid1605.091920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suttinont C, Losuwanaluk K, Niwatayakul K, Hoontrakul S, Intaranongpai W, Silpasakorn S, Suwancharoen D, Panlar P, Saisongkorh W, Rolain JM, Raoult D, Suputtamongkol Y. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann Trop Med Parasitol. 2006;100:363–370. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]