Abstract
Diagnosis of dengue virus (DENV) infection in fatal cases is challenging because of the frequent unavailability of blood or fresh tissues. For formalin-fixed, paraffin-embedded (FFPE) tissues immunohistochemistry (IHC) can be used; however, it may not be as sensitive and serotyping is not possible. The application of reverse transcription-polymerase chain reaction (RT-PCR) for the detection of DENV in FFPE tissues has been very limited. We evaluated FFPE autopsy tissues of 122 patients with suspected DENV infection by flavivirus and DENV RT-PCR, sequencing, and DENV IHC. The DENV was detected in 61 (50%) cases by RT-PCR or IHC. The RT-PCR and sequencing detected DENV in 60 (49%) cases (DENV-1 in 16, DENV-2 in 27, DENV-3 in 8, and DENV-4 in 6 cases). No serotype could be identified in three cases. The IHC detected DENV antigens in 50 (40%) cases. The RT-PCR using FFPE tissue improves detection of DENV in fatal cases and provides sequence information useful for typing and epidemiologic studies.
Introduction
Dengue virus (DENV), a member of the family Flaviviridae, genus Flavivirus, consists of four serologically related but antigenically distinct serotypes designated DENV-1, 2, 3, and 4.1 These viruses are transmitted to humans primarily by Aedes aegypti and Aedes albopictus mosquitoes.2 Infection with DENV generally causes a mild, febrile illness or classic dengue fever that can progress to the severe disease forms, dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), which can be fatal.3,4 The prevalence of DENV infection has increased in recent decades and dengue has emerged as the most important arboviral infection in humans. Over the past 50 years, because of rapid uncontrolled urbanization, modulating climatic factors, expansion of Ae. aegypti in urban environments, and increasing use of inter-continental air travel, DENV infection has expanded its geographical distribution to almost all tropical and subtropical countries and has become endemic in more than 100 countries in Africa, the Americas, the Eastern Mediterranean, Asia, and the Western Pacific, with as many as 2.5 billion people at risk of infection.5–7 The World Health Organization (WHO) estimated that 50–100 million dengue infections occur annually worldwide resulting in 500,000 cases of DHF/DSS and about 25,000 deaths.6
Effective surveillance and efficient control depend on timely and accurate laboratory diagnosis and serotyping. Currently, the main direct and indirect methods that are used to diagnose DENV infections are virus isolation, detection of dengue specific antibodies and antigens, and amplification of viral RNA.8 Virus isolation using culture followed by indirect fluorescent antibody staining is often regarded as the “gold standard” in dengue diagnostics.9 However, it is tedious, time-consuming, and requires cell culture and bio-containment facilities that are costly and difficult to maintain. Furthermore, it is not always successful because of small amounts of viable virus in specimens.10 Conventional serologic methods usually require acute and convalescent-phase serum samples. Immunoglobulin M (IgM) enzyme-linked immunosorbent assay can be performed on a single serum sample but does not provide information about the serotype of the virus. The plaque reduction neutralization technique allows typing using paired sera but extensive cross-reactivity among the flaviviruses and dengue serotypes makes the identification difficult, particularly where multiple flaviviruses are circulating.10,11 Recently, DENV NS1 antigen detection assays have also been applied for the diagnosis of DENV in serum,12–15and one very recent study also evaluated its usefulness for postmortem fresh tissues.16 All these studies have shown that this can be a valuable approach, especially in the early phase of infection; however, NS1 assays may not be as sensitive as reverse transcription-polymerase chain reaction (RT-PCR), particularly for secondary infections in which pre-existing NS1 antibodies in the serum could inhibit the detection of NS1 antigen.12,14,15 The RT-PCR is a rapid, sensitive, and specific technique and a number of PCR-based assays using serum and fresh tissue specimens have been described previously.10,17–19 One report also showed the detection of DENV by RT-PCR in formalin-fixed, paraffin-embedded (FFPE) autopsy tissues of seven children20; however, there is no study that has systematically evaluated the usefulness of RT-PCR for the detection of DENV in FFPE tissues of a large number of fatal cases.
In fatal cases, diagnosis of dengue can be difficult, often caused by the lack of acute and convalescent serum and fresh or frozen tissue specimens. Tissue specimens obtained at autopsy are routinely stored in formalin or as FFPE blocks. Tissue-based techniques such as histopathology and immunohistochemistry (IHC) are often performed on FFPE tissue specimens.21 Dengue IHC can be used for diagnosis and localization of viral antigens in the tissues but it cannot correctly identify serotypes because of cross-reactivity among the serotypes.21–24 Identification of serotypes in fatal cases is particularly important to better understand the pathogenic potential of different serotypes and serotypes information can also be used for epidemiologic studies.25–27 Therefore, a clear need exists for a rapid, sensitive, and specific assay such as RT-PCR for use with FFPE tissue to facilitate the clinical detection and typing of DENV in fatal cases.
In this study, we optimized an extraction method to isolate RNA from FFPE archived autopsy tissue specimens and performed detection and serotype identification of DENV by using RT-PCR and sequencing. In addition, we also compared the RT-PCR results with DENV IHC. To our knowledge, this is the first study to use both RT-PCR assay and IHC to evaluate a large number of autopsy tissues of fatal suspect dengue cases. This approach could significantly expand the opportunity for the diagnosis and typing of fatal dengue disease and can have important implications for phylogenetic and epidemiologic studies.
Materials and Methods
Clinical specimens.
The FFPE autopsy tissue specimens of 122 case-patients with clinical suspicion of DHF or DSS were evaluated by flavivirus and DENV-specific RT-PCR and DENV IHC assays. The cases were submitted to the Infectious Diseases Pathology Branch (IDPB), Centers for Disease Control and Prevention (CDC) from 1995 to 2010 for diagnostic consultation and comprise cases from Puerto Rico (N = 78), Costa Rica (17), Paraguay (8), Continental United States (5), El Salvador (4), U.S. Virgin Islands (3), Somalia (3), Micronesia (2), Venezuela (1), and Ecuador (1). Clinical and demographic information and other relevant laboratory results were collected when available. Tissue specimens tested by RT-PCR and IHC included FFPE liver, kidney, spleen, lung, heart, gastrointestinal (GI), and central nervous system (CNS).
Histopathology and immunohistochemical analysis.
Routine hematoxylin-eosin stains were performed for histopathological evaluation. The IHC assay for DENV was performed on 3-μm sections of FFPE tissues using a polyclonal mouse anti-dengue antibody (Viral Special Pathogens Branch, CDC, Atlanta, GA) and the protocol that we described previously for other viruses28; appropriate positive and negative controls were run in parallel.
RNA extraction.
The RNA was extracted from FFPE tissue specimens of all study case-patients and controls using a phenol-chloroform extraction protocol. Briefly, one 10-μm paraffin section of tissue was placed directly into a sterile 1.5-mL microcentrifuge tube for each extraction. The section was deparaffinized by addition of 1.2-mL xylene and followed by two 100% ethanol washes to remove residual xylene. After the final wash, the ethanol was aspirated and the tissue pellet was air-dried for 15–20 min. The dried tissue pellet was resuspended in 150 μL of proteinase K digestion (PKD) buffer (QIAGEN, Valencia, CA) containing 10 μL of proteinase K (20 mg/mL) and incubated at 55°C for 15 min and then at 45°C overnight. The sample was then incubated at 80°C for 10 min and 550 μL of guanidium-based RLT lysis buffer (QIAGEN) was added to the digested sample. The RNA was separated from other cellular components using 700 μL of acid phenol: chloroform and spinning at 13,000 rpm for 15 min. The aqueous phase was transferred to a fresh tube and after the addition of 2 μL of linear acrylamide (Ambion-Applied Biosystems, Carlsbad, CA) as a carrier, one volume of isopropanol (Sigma-Aldrich, St. Louis, MO) was added and the sample was incubated at −20°C for 3 hours. The sample was then centrifuged at 13,000 rpm at 4°C for 30 min. The supernatant was removed and the pellet was dried out after washing with 75% ethanol. The RNA was resuspended in 20 μL of RNA storage solution (Ambion-Applied Biosystems, Inc.) and stored at −80°C until used.
RT-PCR assays.
All patient samples were tested by a flavivirus-specific RT-PCR assay targeting the NS5 gene to detect DENV serotypes 1–4 and any other medically important flavivirus such as West Nile virus, yellow fever virus, Japanese encephalitis virus, St. Louis encephalitis virus in the samples. A DENV specific RT-PCR assay that can detect all 4 serotypes and targets the capsid (C) gene was also performed on the same samples for comparison. Primer sequences, gene targets, and amplification product sizes are summarized in Table 1. Primer sequences for RT-PCR assays were published previously,29,30 and the primers were synthesized by Biotechnology Core Facility, CDC. The RT-PCR assays were modified to use with FFPE specimens. The RT-PCR assays were performed using the OneStep RT-PCR Kit (QIAGEN) and 5 μL of RNA template following the manufacturer’s instructions. Amplification was carried out on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Carlsbad, CA) and thermocycling conditions used for flavivirus RT-PCR assays were as follows: 1 cycle at 50°C for 30 min, 1 cycle at 95°C for 15 min, and then 40 cycles of incubation at 94°C, 54°C, and 72°C for 1 min each, followed by one cycle of final extension at 72°C for 10 min. Thermocycling conditions used for dengue RT-PCR were the same, except the annealing temperature was adjusted to 55°C.
Table 1.
Oligonucleotide primers used in the study
| Primer sequence (5′–3′) | Gene target | Product size | |
|---|---|---|---|
| Flavivirus RT-PCR29 | |||
| mFU1 | TACAACATGATGGGAAAGCGAGAGAAAAA | NS5 | 250–300 bp |
| CFD2 | GTGTCCCAGCCGGCGGTGTCATCAGC | NS5 | |
| Dengue virus RT-PCR30 | |||
| DN-F | CAATATGCTGAAACGCGAGAGAAA | C | 171 bp |
| DN-R | CCCCATCTATTCAGAATCCCTGCT | C | |
RT-PCR = reverse transcription-polymerase chain reaction; NS5 = Non-Structural 5 gene; C = Capsid gene.
For RT-PCR assays, positive controls consisted of RNA extracted from FFPE blocks of cultured cells infected with DENV of known serotypes and with other flaviviruses (West Nile virus, yellow fever virus, Japanese encephalitis virus, and St. Louis encephalitis virus). The negative controls for dengue RT-PCR included RNA extracted from FFPE tissues of patients with culture and/or IHC-confirmed infections with West Nile virus, yellow fever virus, Japanese encephalitis virus, St. Louis encephalitis virus, and influenza A virus. Negative controls for flavivirus included RNA extracted from FFPE tissues of confirmed Enterovirus, Eastern and Western equine encephalitis virus, influenza A virus, Leptospira, and Rickettsia rickettsii infection cases. To monitor the quality of extraction and presence of PCR inhibitors, each sample was also tested for the amplification of the housekeeping gene 18S rRNA as described.31
Sequencing of PCR amplification products.
The amplification products were analyzed by electrophoresis in a 1.8% agarose gel containing ethidium bromide. All positive amplicons, targeting both the capsid and NS5 region, were extracted from the gel by using the QIAquick gel extraction kit (QIAGEN) and cycle sequenced by GenomeLab Dye Terminator Cycle Sequencing Quick Start Kit (Beckman Coulter, Fullerton, CA). The samples were sequenced on a CEQ 8000 XL sequencer (Beckman Coulter) and a search for homologies to known sequences was done using the nucleotide database of the Basic Local Alignment Search Tool (BLAST).
Results
Dengue virus RNA or antigens were detected in 61 (50%) of 122 cases by RT-PCR or IHC or both tests. The characteristics of dengue-positive patients are summarized in Table 2. The median age was 28 years (range 4 months to 68 years), 49% were males and 27% of patients were pediatric cases (≤ 17 years of age). The median duration from illness onset to death was 6 days (range 1–25 days). All 4 DENV-positive cases in the continental United States had travel history to dengue-endemic countries including Mexico (2), Ecuador (1), and Saint Kitts islands (1).
Table 2.
Characteristics of 61 dengue-positive case-patients
| Characteristic | No. (%) |
|---|---|
| Male | 30 (49) |
| Age Group* | |
| 0–17 | 17 (27) |
| 18–24 | 6 (9) |
| 25–64 | 36 (59) |
| 65 or old | 2 (3) |
| Duration of illness, median days† | 6 days |
| Clinical features known | 46 (75) |
| Fever | 45 (98) |
| Rash | 14 (30) |
| Low platelets (≤ 100,000/mm3) | 27 (59) |
| Hemorrhage manifestations | 26 (57) |
Median 28 years (range 4 months–68 years).
Range 1–25 days.
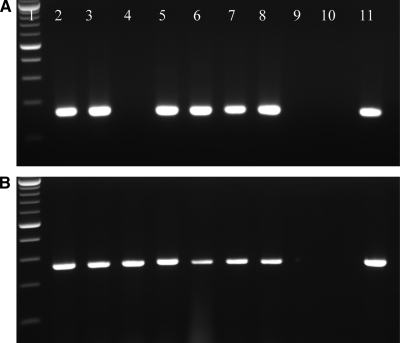
Of 122 cases, 60 cases (49%) were positive by flavivirus RT-PCR assay, whereas 45 (36%) were positive by dengue RT-PCR assay. Dengue antigens were detected by IHC in 50 (40%) cases. As described in Table 3, DENV was detected in 37 of 61 (60%) positive cases by all three assays, whereas in 57 (93%) cases DENV was detected by at least two assays. Forty-nine (80%) of cases were positive by both RT-PCR and IHC. In 11 (18%) of 61 positive cases, DENV was detected only by RT-PCR assays and 1 case was found to be positive only by IHC. All RT-PCR positive cases generated correct sized amplicons as shown in Figure 1 and sequence analysis of amplicons confirmed the presence of dengue virus. Sequence analysis of the positive amplicons identified DENV-1 in 16 (26%) cases, DENV-2 in 27 (45%) cases, DENV-3 in 8 (13%) cases, and DENV-4 in 6 (10%) cases. No serotype could be identified in three cases because concentrations of amplified PCR products were not adequate for sequencing. The serotype results of the cases divided on the basis of geographical distribution are summarized in Table 4. The correct serotypes were identified in all control DENV isolates RNA samples extracted from the tissue blocks by both DENV and flavivirus RT-PCR assays. No amplification product was detected in any negative control. The 18S rRNA housekeeping gene was amplified in all the samples, confirming the presence of amplifiable nucleic acids.
Table 3.
Comparison of different assays in 61 dengue-positive case-patients
| Dengue-positive cases | |||
|---|---|---|---|
| Flavivirus RT-PCR | Dengue RT-PCR | Dengue IHC | Total no. of cases |
| Positive | Positive | Positive | 37 |
| Positive | Positive | Negative | 8 |
| Positive | Negative | Negative | 3 |
| Positive | Negative | Positive | 12 |
| Negative | Negative | Positive | 1 |
RT-PCR = reverse transcription-polymerase chain reaction; IHC = immunohistochemistry.
Figure 1.
Agarose gel analysis of polymerase chain reaction (PCR) products: Gel A (Dengue virus [DENV] reverse transcription [RT]-PCR) and B (Flavivirus RT-PCR) Lane 1: 100-bp ladder; Lanes 2–8: patients samples; Lane 9: no template water control; Lane 10: negative control tissue; Lane 11: positive control. Note negative DENV PCR result in lane 4 of gel A. The same case sample was positive by flavivirus PCR as shown in lane 4 of gel B.
Table 4.
Geographical distribution
| Territory/country | RT-PCR positive for dengue | Serotypes | |||
|---|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | ||
| Puerto Rico* | 36 | 14 | 13 | 4 | 3 |
| Costa Rica | 5 | – | 5 | – | – |
| Paraguay | 5 | 2 | – | 3 | – |
| El Salvador | 4 | – | 4 | – | – |
| Continental U.S. | 3 | – | 1 | 1 | 1 |
| U.S. Virgin Islands | 3 | – | 3 | – | – |
| Micronesia | 2 | – | – | – | 2 |
| Venezuela | 1 | – | 1 | – | – |
| Ecuador† | 1 | – | – | – | – |
Serotype could not be identified in two cases.
Serotype could not be identified.
RT-PCR = reverse transcription-polymerase chain reaction; DENV = Dengue virus.
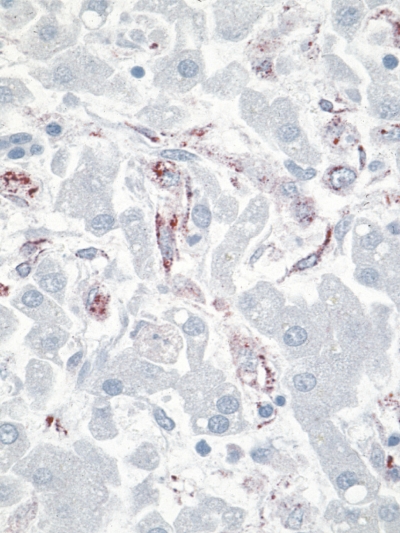
Ninety-four percent (51 of 54) of liver, 91% (22 of 24) of lung, 86% (13 of 15) of spleen, and 73% (17 of 23) of kidney tissue specimens from DENV-positive cases were positive for DENV by RT-PCR. No tissue specimens of heart, GI, and CNS tested positive by RT-PCR. By IHC, DENV antigens were primarily localized in the liver, kidney, spleen, and lung. Figure 2 is a photomicrograph of IHC assay showing positive immunostaining of viral antigens predominantly within sinusoidal Kupffer cells of liver. Eighty percent (45 of 56) of liver, 72% (26 of 36) of lung, 48% (15 of 31) of kidneys, and 40% (9 of 22) of spleen tissues from DENV-positive cases were found to be positive for DENV by IHC, whereas all the heart, GI, and CNS tissues tested were DENV IHC negative.
Figure 2.
Photomicrograph of immunohistochemical assay using a polyclonal mouse anti-dengue antibody showing immunostaining of liver demonstrating viral antigens (red color) predominantly within sinusoidal Kupffer cells. Original magnification×158.
Discussion
With the recent increase in dengue outbreaks in various parts of the world, DENV infection continues to be a major public health problem. According to a WHO report, an estimated 500,000 people with DHF require hospitalization each year and about 2.5% of those affected die, a very large proportion of whom are children.6 Fatalities can be higher in some countries caused by inadequate disease management facilities and without proper treatment DHF fatality rates can exceed 20%.6 In addition, fatality rates reported in hospitalized patients can reach up to 50–60% in dengue patients with complications (acute renal failure, fulminant hepatitis, liver failure, and encephalopathy).32–34
The increased prevalence of dengue infections worldwide in recent decades and the high mortality caused by DHF and DSS highlights the need for more sensitive and specific diagnostic assays such as RT-PCR for the detection and typing of DENV in fatal cases. Diagnosis of DENV infection in fatal cases often can be challenging because of the unavailability of serum and fresh or frozen specimens. Furthermore, in patients who die during the first week of illness, serology may have limited use because of low levels of IgM antibodies that cannot be detected by serological assays.35,36 On the other hand, in the early viremic stage of DENV infections patients may have higher viral loads.37–39 Therefore, RT-PCR analysis of FFPE tissues, often the only specimens available for fatal cases, can be a useful adjunct to conventional diagnostic techniques, particularly in patients who die relatively soon after the disease onset. However, the application of RT-PCR for the detection of DENV in FFPE tissues has been very limited because of difficulties in extracting good quality RNA.
In the current study, we recovered RNA from archived FFPE autopsy tissues (some of which had been stored for more than 15 years) and showed that RT-PCR was a sensitive and valuable diagnostic tool for the detection and serotyping of DENV in FFPE tissues of fatal cases. Although RT-PCR and sequencing detected DENV in 60 (49%) cases of this series, IHC was able to detect DENV antigens only in 49 of these PCR-positive cases. Thus, in 11 (18%) cases dengue diagnosis was confirmed by RT-PCR only. The IHC negative results of these 11 cases may be due to less sensitivity of assay or clearance of viral antigens by the host immune response.40 Therefore, this study underscores the importance of postmortem tissue analysis by combination of RT-PCR with IHC for the diagnosis of DENV and shows that this approach improves the detection of DENV in fatal cases.
Our data also showed that 15 out of 60 PCR-positive cases were negative by DENV RT-PCR but positive by flavivirus RT-PCR assay. Interestingly, 14 of these cases were recent DENV-1 cases from Puerto Rico. This may indicate that the flavivirus RT-PCR that targets the more conserved NS5 gene was able to detect this more recent and maybe variant strain of DENV-1, whereas DENV RT-PCR that targets the capsid region was not. Similarly, DENV antibody, which is also a broad spectrum polyclonal antibody and reacts with different dengue strains, also detected DENV in 12 of the dengue RT-PCR negative cases. Previous studies have shown that many variant strains exist within each DENV serotype.41,42 Furthermore, DENV, being a positive-strand RNA virus, has a high potential for mutation, resulting in nucleotide differences between genotypes and also within a genotype.42 Therefore, to reduce the rates of false negatives, we recommend using more than one gene target for the PCR assays and selecting primers in the conserve regions of the viral genome for the detection of DENV. Additionally, using pan-flavivirus RT-PCR may also be useful for the identification of other flaviviruses in the samples, particularly from the countries where multiple flavivirus circulate.
Infecting serotype was identified for 95% (57 of 60) of DENV tissue RT-PCR-positive cases in our series. In 45% of the cases (27 of 60), DENV-2 was identified to be the cause of fatality. One possible reason for this may be that 13 out of 27 (48%) DENV-2 cases were from Puerto Rico where DENV-2 was the most prevalent serotype before the recent epidemic. On the other hand, some previous studies have also indicated that DENV-2 can cause more severe disease outcome.43,44 Moreover, the genotypes originating in Southeast Asia and the Indian subcontinent, which belongs to serotype 2 and 3, respectively, have been identified to cause more outbreaks of severe dengue disease.41,45–47 Although the host immune status can also play an important role in determining the outcome of infection,35 the determination of the virus nucleotide sequences in the fatal cases can help to identify the origin and serotype of the infecting virus and its association with severe disease and fatality. Furthermore, we also observed that out of 36 dengue-positive cases from Puerto Rico, all the cases that occurred from 1998 to 2003 were DENV-3 cases. Other studies also reported the re-introduction of DENV-3 in 1998 in Puerto Rico after a 20-year absence.48 Interestingly, all the cases that occurred from 2009 to early 2010 were DENV-1 cases and in more recent 2010 cases, DENV-4 was identified. This shows that even though the epidemiologic impact of retrospective tissue analysis may not be immediate and direct, such as for the identification of the circulating serotype during the outbreak, this approach can help to link some of the previously undiagnosed cases to the particular outbreaks and may be helpful to assess the true burden of the outbreak, and to a certain extent, in the phylogenetic analysis.
In this study, we detected both dengue viral antigens and RNA predominately in the liver and in lung, spleen, and kidney. Several previous studies also recognized liver as the major target organ in DENV pathogenesis.17,49,50 Interestingly, in 28% (17 of 61) of DENV-negative cases, organism-specific IHC and/or PCR detected other pathogens (IDPB, CDC, unpublished data) including Leptospira, Streptococcus pneumoniae, Staphylococcus aures, 2009 pandemic H1N1 influenza A virus, and West Nile virus, which shows that clinical differential diagnosis of dengue-like syndrome can be extremely difficult and emphasizes the importance of postmortem tissue analysis of cases suspect to have DENV infection by histopathologic evaluation and other tissue-based techniques. However, availability of postmortem tissue for analysis is usually very limited because autopsies are not often performed, particularly in dengue-endemic regions, because of religious and cultural beliefs and bio-safety issues.
In conclusion, the data presented in this study shows that RT-PCR is a more sensitive and specific assay than IHC for the detection of dengue viruses in formalin-fixed tissue of fatal cases and provides sequence information that can be useful to facilitate typing and phylogenetic analysis, and may be also helpful to better characterize the pathogenic potential of distinct DENV serotypes. The RT-PCR on FFPE tissues can be a particularly valuable diagnostic tool in patients who die relatively soon after disease onset and for whom serology may be negative, and also when the FFPE tissues are the only specimens available. Thus, this approach can have important implications for dengue diagnosis and epidemiologic studies. In addition, combination of RT-PCR and IHC analysis of pathological specimens allows determination of tissue tropism and provides an insight into the pathogenesis of severe disease outcome.
ACKNOWLEDGMENTS
We gratefully thank John Roehrig and Robert Lanciotti, Arboviral Diseases Branch, Division of Vector-Borne Infectious Diseases, CDC, Fort Collins, Colorado for providing cell culture controls of DENV and other flaviviruses for PCR and IHC assays. We also thank Thomas G. Ksiazek (University of Texas Medical Branch, Galveston, Texas; formerly at Viral Special Pathogens Branch, CDC, Atlanta, Georgia) for providing antibody used in IHC assay. We greatly acknowledge Kay Tomashek, Jorge L. Munoz-Jordan, Harold Margolis, and other colleagues in Dengue Branch, Division of Vector-Borne Infectious Diseases, CDC, San Juan, Puerto Rico and all the public health departments, laboratories, the pathologists, and medical examiners that submitted specimens to the IDPB.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Authors’ addresses: Julu Bhatnagar, Dianna M. Blau, Wun-Ju Shieh, Christopher D. Paddock, Clifton Drew, Lindy Liu, Tara Jones, Mitesh Patel, and Sherif R. Zaki, Infectious Diseases Pathology Branch, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention (CDC), Atlanta, GA, E-mails: JBhatnagar@cdc.gov, DBlau@cdc.gov, WShieh@cdc.gov, CPaddock@cdc.gov, CDrew1@cdc.gov, LLiu1@cdc.gov, TJones5@cdc.gov, MPatel1@cdc.gov, and SZaki@cdc.gov.
Reprint requests: Julu Bhatnagar, Infectious Diseases Pathology Branch, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention (CDC), Bldg.18/SB-105, MS-G32, Atlanta, GA, Tel: 404-639-2826, Fax: 404-639-3043, E-mail: JBhatnagar@cdc.gov.
References
- 1.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. In: The Arboviruses: Epidemiology and Ecology. Monath TP, editor. Volume 2. Boca Raton, FL: CRC Press; 1988. pp. 223–260. (Dengue). [Google Scholar]
- 3.Centers for Disease Control and Prevention Dengue Clinical Guidance. 2010. http://www.cdc.gov/dengue/clinicalLab/clinical.html Available at. Accessed April 31, 2011.
- 4.Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 5.Gubler DJ. The global pandemic of dengue/dengue hemorrhagic fever: current status and prospects for the future. Ann Acad Med Singapore. 1998;27:227–234. [PubMed] [Google Scholar]
- 6.World Health Organization Dengue and Dengue Hemorrhagic Fever. 2009. http://www.who.int/mediacentre/factsheets/fs117/en/index.html Fact sheet no. 117. Available at. Accessed March 31, 2011.
- 7.Wilder-Smith A, Gubler DJ. Geographic expansion of dengue: the impact of international travel. Med Clin North Am. 2008;92:1377–1390 x. doi: 10.1016/j.mcna.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. Geneva, Switzerland: World Health Organization; 2009. [PubMed] [Google Scholar]
- 9.Gubler DJ, Kuno G, Sather GE, Velez M, Oliver A. Mosquito cell cultures and specific monoclonal antibodies in surveillance for dengue viruses. Am J Trop Med Hyg. 1984;33:158–165. doi: 10.4269/ajtmh.1984.33.158. [DOI] [PubMed] [Google Scholar]
- 10.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino Y, Tadano M, Saito M, Maneekarn N, Sittisombut N, Sirisanthana V, Poneprasert B, Fukunaga T. Studies on serological cross-reaction in sequential flavivirus infections. Microbiol Immunol. 1994;38:951–955. doi: 10.1111/j.1348-0421.1994.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 12.Chaterji S, Allen JC, Jr, Chow A, Leo YS, Ooi EE. Evaluation of the NS1 rapid test and the WHO dengue classification schemes for use as bedside diagnosis of acute dengue fever in adults. Am J Trop Med Hyg. 2011;84:224–228. doi: 10.4269/ajtmh.2011.10-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessoff K, Phoutrides E, Delorey M, Acosta LN, Hunsperger E. Utility of a commercial nonstructural protein 1 antigen capture kit as a dengue virus diagnostic tool. Clin Vaccine Immunol. 2010;17:949–953. doi: 10.1128/CVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tricou V, Vu HT, Quynh NV, Nguyen CV, Tran HT, Farrar J, Wills B, Simmons CP. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viremia and antibody responses. BMC Infect Dis. 2010;10:142. doi: 10.1186/1471-2334-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez AH, Moros Z, Comach G, Zambrano J, Bravo L, Pinto B, Vielma S, Cardier J, Liprandi F. Evaluation of dengue NS1 antigen detection tests with acute sera from patients infected with dengue virus in Venezuela. Diagn Microbiol Infect Dis. 2009;65:247–253. doi: 10.1016/j.diagmicrobio.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Lima Mda R, Nogueira RM, Schatzmayr HG, de Filippis AM, Limonta D, dos Santos FB. A new approach to dengue fatal cases diagnosis: NS1 antigen capture in tissues. PLoS Negl Trop Dis. 2011;e1147:5. doi: 10.1371/journal.pntd.0001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Araujo JM, Schatzmayr HG, de Filippis AM, dos Santos FB, Cardoso MA, Britto C, Coelho JM, Nogueira RM. A retrospective survey of dengue virus infection in fatal cases from an epidemic in Brazil. J Virol Methods. 2009;155:34–38. doi: 10.1016/j.jviromet.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Rosen LL, Drouet MM, Deubel VV. Detection of dengue virus RNA by reverse transcription-polymerase chain reaction in the liver and lymphoid organs but not in the brain in fatal human infection. Am J Trop Med Hyg. 1999;61:720–724. doi: 10.4269/ajtmh.1999.61.720. [DOI] [PubMed] [Google Scholar]
- 19.Johnson BW, Russell BJ, Lanciotti RS. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sariol CA, Pelegrino JL, Martinez A, Arteaga E, Kouri G, Guzman MG. Detection and genetic relationship of dengue virus sequences in seventeen-year-old paraffin-embedded samples from Cuba. Am J Trop Med Hyg. 1999;61:994–1000. doi: 10.4269/ajtmh.1999.61.994. [DOI] [PubMed] [Google Scholar]
- 21.Zaki SR, Peters CJ. In: Diagnostic Pathology of Infectious Diseases. Connor DH, Chandler FW, Schawartz DA, Manz HJ, Lack EE, editors. Stamford, CT: Appleton and Lange; 1997. pp. 347–364. (Viral hemorrhagic fevers). [Google Scholar]
- 22.de Macedo FC, Nicol AF, Cooper LD, Yearsley MM, Pires AR, Nuovo GJ. Histologic, viral, and molecular correlates of dengue fever infection of the liver using highly sensitive immunohistochemistry. Diagn Mol Pathol. 2006;15:223–228. doi: 10.1097/01.pdm.0000213462.60645.cd. [DOI] [PubMed] [Google Scholar]
- 23.Miagostovich MP, Ramos RG, Nicol AF, Nogueira RM, Cuzzi-Maya T, Oliveira AV, Marchevsky RS, Mesquita RP, Schatzmayr HG. Retrospective study on dengue fatal cases. Clin Neuropathol. 1997;16:204–208. [PubMed] [Google Scholar]
- 24.Bhoopat L, Bhamarapravati N, Attasiri C, Yoksarn S, Chaiwun B, Khunamornpong S, Sirisanthana V. Immunohistochemical characterization of a new monoclonal antibody reactive with dengue virus-infected cells in frozen tissue using immunoperoxidase technique. Asian Pac J Allergy Immunol. 1996;14:107–113. [PubMed] [Google Scholar]
- 25.Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 26.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–809. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aquino VH, Anatriello E, Goncalves PF, DA Silva EV, Vasconcelos PF, Vieira DS, Batista WC, Bobadilla ML, Vazquez C, Moran M, Figueiredo LT. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002–2004. Am J Trop Med Hyg. 2006;75:710–715. [PubMed] [Google Scholar]
- 28.Guarner J, Shieh WJ, Hunter S, Paddock CD, Morken T, Campbell GL, Marfin AA, Zaki SR. Clinicopathologic study and laboratory diagnosis of 23 cases with West Nile virus encephalomyelitis. Hum Pathol. 2004;35:983–990. doi: 10.1016/j.humpath.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Chao DY, Davis BS, Chang GJ. Development of multiplex real-time reverse transcriptase PCR assays for detecting eight medically important flaviviruses in mosquitoes. J Clin Microbiol. 2007;45:584–589. doi: 10.1128/JCM.00842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu PY Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 32.Lee IK, Liu JW, Yang KD. Clinical characteristics, risk factors, and outcomes in adults experiencing dengue hemorrhagic fever complicated with acute renal failure. Am J Trop Med Hyg. 2009;80:651–655. [PubMed] [Google Scholar]
- 33.Seneviratne SL, Malavige GN, de Silva HJ. Pathogenesis of liver involvement during dengue viral infections. Trans R Soc Trop Med Hyg. 2006;100:608–614. doi: 10.1016/j.trstmh.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Nimmannitya S, Thisyakorn U, Hemsrichart V. Dengue hemorrhagic fever with unusual manifestations. Southeast Asian J Trop Med Public Health. 1987;18:398–406. [PubMed] [Google Scholar]
- 35.Halstead SB. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 36.Bhatnagar J, Guarner J, Paddock CD, Shieh WJ, Lanciotti RS, Marfin AA, Campbell GL, Zaki SR. Detection of West Nile virus in formalin-fixed, paraffin-embedded human tissues by RT-PCR: a useful adjunct to conventional tissue-based diagnostic methods. J Clin Virol. 2007;38:106–111. doi: 10.1016/j.jcv.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J Clin Microbiol. 1999;37:2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh K, Lale A, Eong Ooi E, Chiu LL, Chow VT, Tambyah P, Koay ES. A prospective clinical study on the use of reverse transcription-polymerase chain reaction for the early diagnosis of Dengue fever. J Mol Diagn. 2006;8:613–616. doi: 10.2353/jmoldx.2006.060019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tricou V, Minh NN, Farrar J, Tran HT, Simmons CP. Kinetics of viremia and NS1 antigenemia are shaped by immune status and virus serotype in adults with Dengue. PLoS Negl Trop Dis. 2011;5:e1309. doi: 10.1371/journal.pntd.0001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shieh WJ, Blau DM, Denison AM, Deleon-Carnes M, Adem P, Bhatnagar J, Sumner J, Liu L, Patel M, Batten B, Greer P, Jones T, Smith C, Bartlett J, Montague J, White E, Rollin D, Gao R, Seales C, Jost H, Metcalfe M, Goldsmith CS, Humphrey C, Schmitz A, Drew C, Paddock C, Uyeki TM, Zaki SR. Pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2009;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rico-Hesse R. Dengue virus evolution and virulence models. Clin Infect Dis. 2007;44:1462–1466. doi: 10.1086/517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lourenco J, Recker M. Viral and epidemiological determinants of the invasion dynamics of novel dengue genotypes. PLoS Negl Trop Dis. 2010;4:e894. doi: 10.1371/journal.pntd.0000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balmaseda AA, Hammond SN, Pérez LL, Tellez YY, Saborío SI, Mercado JC, Cuadra RR, Rocha JJ, Pérez MA, Silva SS, Rocha CC, Harris EE. Serotype-specific differences in clinical manifestations of dengue. Am J Trop Med Hyg. 2006;74:449–456. [PubMed] [Google Scholar]
- 44.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 45.Nisalak AA, Endy TP, Nimmannitya SS, Kalayanarooj SS, Thisayakorn UU, Scott RM, Burke DS, Hoke CH, Innis BL, Vaughn DW. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am J Trop Med Hyg. 2003;68:191–202. [PubMed] [Google Scholar]
- 46.Kumaria R. Correlation of disease spectrum among four Dengue serotypes: a five years hospital based study from India. Braz J Infect Dis. 2010;14:141–146. [PubMed] [Google Scholar]
- 47.Sumarmo, Wulur H, Jahja E, Gubler DJ, Suharyono W, Sorensen K. Clinical observations on virologically confirmed fatal dengue infections in Jakarta, Indonesia. Bull World Health Organ. 1983;61:693–701. [PMC free article] [PubMed] [Google Scholar]
- 48.Tomashek KM, Rivera A, Munoz-Jordan JL, Hunsperger E, Santiago L, Padro O, Garcia E, Sun W. Description of a large island-wide outbreak of dengue in Puerto Rico, 2007. Am J Trop Med Hyg. 2009;81:467–474. [PubMed] [Google Scholar]
- 49.Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–1418. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- 50.Hall WC, Crowell TP, Watts DM, Barros VL, Kruger H, Pinheiro F, Peters CJ. Demonstration of yellow fever and dengue antigens in formalin-fixed paraffin-embedded human liver by immunohistochemical analysis. Am J Trop Med Hyg. 1991;45:408–417. doi: 10.4269/ajtmh.1991.45.408. [DOI] [PubMed] [Google Scholar]