Summary
Ars2 is a component of the nuclear cap-binding complex that contributes to microRNA biogenesis and is required for cellular proliferation. Here we expand on the repertoire of Ars2-dependent microRNAs and determine that Ars2 regulates a number of mRNAs, the largest defined subset of which code for histones. Histone mRNAs are unique among mammalian mRNAs because they are not normally polyadenylated, but rather cleaved following a 3′ stem loop. A significant reduction in correctly processed histone mRNAs was observed following Ars2 depletion, concurrent with an increase in polyadenylated histone transcripts. Furthermore, Ars2 physically associated with histone mRNAs and the non-coding RNA 7SK. Knockdown of 7SK led to an enhanced ratio of cleaved to polyadenylated histone transcripts, an effect dependent on Ars2. Together, the data demonstrate that Ars2 contributes to histone mRNA 3′ end formation and expression and these functional properties of Ars2 are negatively regulated by interaction with 7SK RNA.
Introduction
Arsenic resistance protein 2 (Ars2) is a nuclear protein encoded by the human SRRT gene that contains multiple putative RNA binding domains including an arginine-rich domain, an RNA recognition motif and a zinc-finger. Genes homologous to SRRT are found in plants, metazoans and fission yeast, but not budding yeast (Wilson et al., 2008). Ars2 is highly expressed during cellular proliferation, and is rapidly down-regulated upon removal of growth signals. In vitro deletion of Ars2 or expression of dominant-negative Ars2 results in proliferative arrest of immortalized cells (Gruber et al., 2009; Rossman and Wang, 1999). Biochemical and genetic data from multiple organisms indicate that Ars2 is a component of the nuclear cap-binding complex (CBC) that binds the 7-methylguanosine (7mG) cap structure of nuclear RNA polymerase II transcripts (Gruber et al., 2009; Laubinger et al., 2008; Sabin et al., 2009). Ars2 may therefore play a critical role in regulating RNA polymerase II (RNAPII) transcripts that are required for cellular proliferation.
Recent evidence suggests that microRNAs represent a class of Ars2-regulated RNAPII transcripts that can significantly alter cellular proliferation. In Arabidopsis, mutations in SERRATE, a homolog of SRRT, cause decreased levels of mature microRNAs and increased levels of primary microRNA transcripts (Grigg et al., 2005; Lobbes et al., 2006; Yang et al., 2006). Serrate protein acts as a scaffold to stimulate the efficiency and accuracy of primary microRNA processing by the Dcl-1/Hyl-1 complex (Dong et al., 2008; Machida et al., 2011). In mammalian systems, depletion of Ars2 from cell lines caused decreased levels of mature miR-21 and let-7 microRNAs, and diminished primary miR-21 transcripts (Gruber et al., 2009). These data indicate that as a component of the nuclear CBC, Ars2 may contribute to the stability and delivery of primary microRNA transcripts to the microprocessor complex in the nucleus.
Replication-dependent histone mRNAs are rapidly induced and turned over during progression through the cell cycle in order to produce sufficient histone proteins to properly package newly synthesized DNA in chromatin. Unlike other metazoan mRNAs, replication-dependent histone mRNAs are not 3′ polyadenylated, but rather cleaved following a conserved 25–26 nucleotide sequence within their 3′ UTR. This sequence consists of a 16 nucleotide stem loop surrounded by 5 conserved upstream nucleotides and 4–5 conserved downstream nucleotides that act to recruit stem-loop binding protein (SLBP) and the U7 snRNP complex responsible for 3′ end cleavage (Marzluff et al., 2008). Recent evidence suggests that several other proteins, including cyclin dependent kinase 9 (CDK9) and CBC components, are required for proper co-transcriptional regulation of 3′ end cleavage and that in the absence of this regulation polyadenylated replication-dependent histone mRNAs are produced (Narita et al., 2007; Pirngruber et al., 2009).
The current study was undertaken to determine the contribution of Ars2 to mammalian gene expression. Levels of microRNA and mRNA were analyzed by microarray in Ars2-depleted cell lines, leading to the delineation of a subset of microRNAs that are dependent on Ars2 for high levels of expression. In contrast to cells depleted of DiGeorge syndrome critical region 8 (DGCR8), cells depleted of Ars2 also had alterations in the level of expression of a number of mRNAs. Specifically, Ars2 was required for expression of replication-dependent histone mRNAs with properly processed 3′ termini. Additional studies demonstrated that Ars2 bound to histone mRNA transcripts and that its ability to promote proper histone mRNA 3′ end formation was negatively regulated by its interaction with 7SK RNA. These data suggest Ars2 contributes to the proper processing of multiples classes of nuclear transcripts, including microRNAs and histone mRNAs.
Results
Ars2 regulates a subset of microRNAs
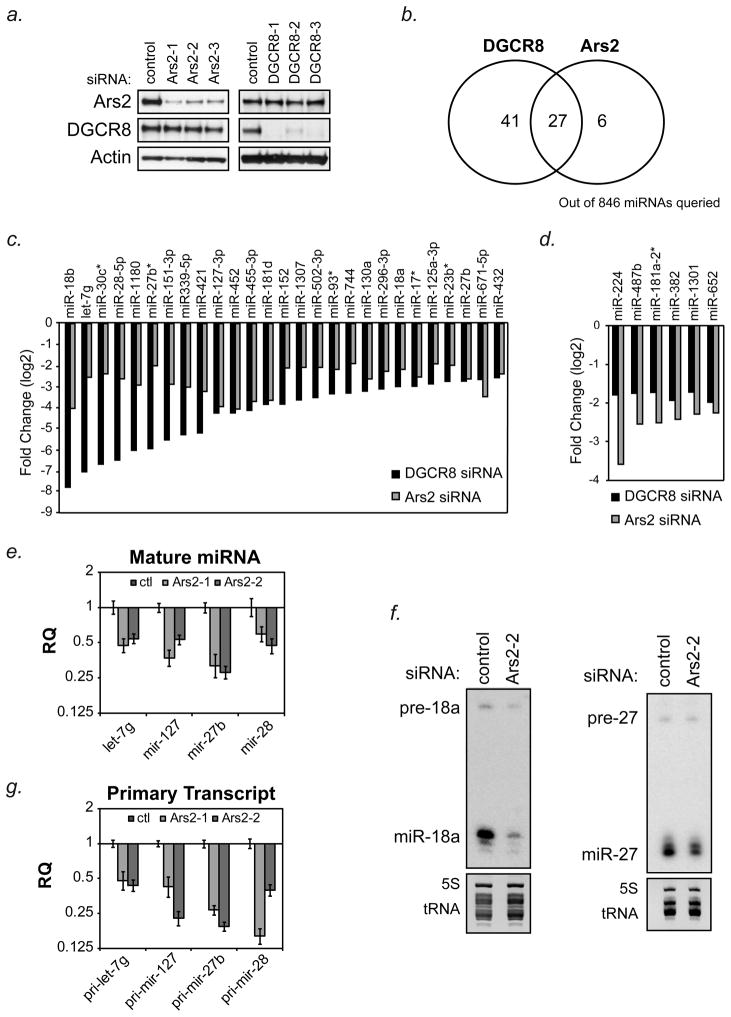
To determine the complement of Ars2-dependent microRNAs, array-based profiling experiments were performed on Ars2-depleted HeLa cells. In order to determine which microRNAs were expressed and have half-lives short enough to be significantly down-regulated over the course of the experiment, the microprocessor component DGCR8 was knocked-down with three independent siRNAs. Western blotting showed that the three siRNAs targeted to Ars2 and the three siRNAs targeted to DGCR8 effectively decreased the protein levels of Ars2 or DGCR8, respectively (Figure 1a). For microRNA array analysis, each siRNA to Ars2, DGCR8, or control was transfected into independent cultures of HeLa cells and three days later RNA was extracted and hybridized separately to Affymetrix microRNA arrays. Expression of individual microRNAs were considered to be significantly increased or decreased by siRNA depletion of Ars2 or DGCR8 if their levels changed by 2-fold or more for all three independent siRNAs compared to the control siRNA. This analysis defined a set of 68 microRNAs (out of 846 human microRNAs considered) whose levels were decreased by ≥2-fold by all three siRNAs targeted to DGCR8, compared to control siRNAs. Of these microRNAs, 27 were decreased ≥2-fold by siRNAs targeted to Ars2 (Figure 1b, c). Interestingly, there were 6 microRNAs whose levels diminished by ≥2-fold after knockdown of Ars2 that were not as robustly affected by DGCR8 depletion (Figure 1b, d). These data demonstrate that Ars2 regulates roughly half of all microRNAs that depend on the microprocessor for their expression.
Figure 1. Ars2 regulates a subset of microRNAs.
(A) HeLa cells were transfected with control siRNA or 3 siRNAs targeted to either Ars2 (Ars2-1, Ars2-2, Ars2-3) or DGCR8 (DGCR8-1, DGCR8-2, DGCR8-3). Protein was harvested three days following transfection to confirm specific depletion of Ars2 or DGCR8 by Western blot.
(B) HeLa cells were transfected with the siRNAs as in (A) and RNA was isolated three days later and analyzed separately on Affymetrix GeneChip® miRNA Arrays. The number of microRNAs decreased at least 2-fold (log2) following transfection of all three siRNAs per gene are depicted by Venn Diagram.
(C) Bar graphs showing 27 microRNAs determined by microarray to decrease 2-fold (log2) or more following depletion of DGCR8 or Ars2. Bars represent the average of the three siRNAs targeting DGCR8 or Ars2 shown in (A).
(D) Bar graphs showing 6 microRNAs determined by microarray to decrease 2-fold (log2 ) or more following depletion of Ars2 but not DGCR8. Bars represent the average of the three siRNAs targeting DGCR8 or Ars2 shown in (A).
(E) HeLa cells were transfected with two siRNAs targeted to Ars2 (Ars2-1, Ars2-2) or a control siRNA (ctl). Three days later RNA was isolated and TaqMan® microRNA assays were used to detect the mature microRNAs indicated. Bars represent relative quantification using the ΔΔCt method +/− 95% confidence interval of three replicates. U6 snRNA was used as an endogenous control.
(F) HeLa cells were transfected with siRNA to Ars2 (Ars2-2) or a control siRNA (ctl) and three days later RNA was isolated. Precursor and mature microRNAs indicated were detected by Northern blot. Ethidium bromide staining of 5S rRNA and tRNAs are shown as loading controls.
(G) RNA from (E) was reverse transcribed and TaqMan®-based qPCR was performed for the indicated primary microRNA transcripts. Bars represent relative quantification using the ΔΔCt method +/− 95% confidence interval of three replicates. U6 snRNA was used as an endogenous control.
MicroRNA changes detected by microarray analysis were independently confirmed by quantitative real-time PCR (qPCR) and/or Northern blot (Figs. 1e,f). Our previous data showed that the primary miR-21 transcript was decreased by depletion of Ars2 (Gruber et al., 2009). We therefore checked the primary transcript levels of let-7g, miR-127, miR-27b and miR-28 by qPCR following Ars2 depletion and found that depletion of Ars2 led to decreased levels of primary microRNA transcripts for all microRNAs tested (Figure 1g). These data suggest that Ars2, in contrast to what has been observed for DGCR8 (Gregory et al., 2004), likely regulates microRNA expression at the level of the primary transcript.
Detection of histone mRNAs increases after Ars2 depletion
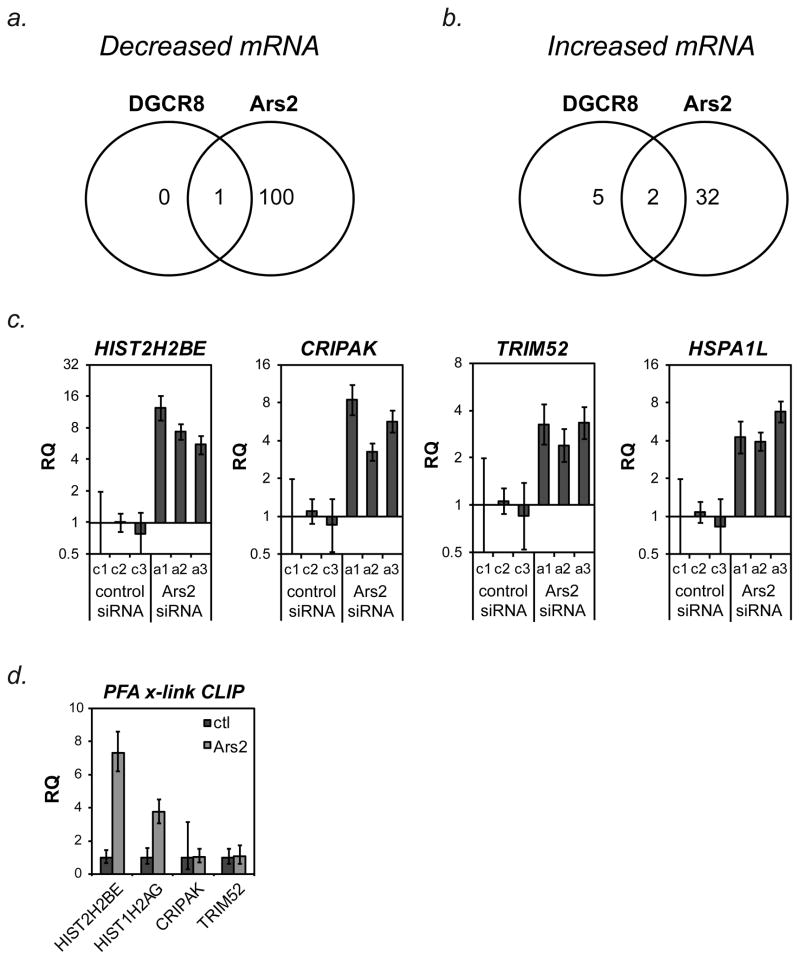
Gene expression profiling was performed to determine changes in mRNA expression caused by depletion of Ars2 or DGCR8. RNA used for microRNA array analysis in Figure 1 was reverse-transcribed and hybridized to Affymetrix Human Gene 1.0ST mRNA arrays. In all, 102 mRNAs (including SRRT) were found to be significantly decreased (> 2-fold) following Ars2 depletion compared to control, of which only 1 mRNA (FN1) decreased after DGCR8 depletion (Figure 2a, Supplemental Table 1). In contrast, 34 mRNAs were increased by at least 2-fold following depletion of Ars2 compared to control cells, of which only two (CLDN1 and PPPDE1) were increased after DGCR8 depletion (Figure 2b). Five additional mRNAs increased following depletion of DGCR8, none of which were significantly affected by Ars2 knockdown. These data suggest that most of the changes seen in mRNA expression following Ars2 knockdown were not secondary to decreased microRNA expression, since DGCR8 depletion did not affect similar changes.
Figure 2. Ars2 regulates expression of numerous mRNAs and associates with replication-dependent histone mRNAs.
(A) RNA used for the miRNA array in Figure 1b was reverse-transcribed and hybridized to Affymetrix GeneChip® Human Gene 1.0ST Arrays. The number of mRNAs decreased at least 2-fold (log2) following transfection of all three siRNAs targeting Ars2 or DGCR8 are depicted by Venn Diagram.
(B) The number of mRNAs increased at least 2-fold (log2) following transfection of all three siRNAs Ars2 or DGCR8 are depicted by Venn Diagram.
(C) To confirm microarray results, HeLa cells were transfected independently with control siRNAs (c1, c2, c3) or three siRNAs to Ars2 (a1, a2, a3) and the indicated mRNA transcript levels were measured by TaqMan®-based qPCR. Bars represent relative quantification using the ΔΔCt method normalized to c1 +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(D) To determine if Ars2-containing protein complexes could associate with genes found to increase after Ars2 depletion, paraformaldehyde crosslinking followed by immunoprecipitation (PFA-CLIP) with a monoclonal antibody to Ars2 (2G10) or control antibody (SP/0) was performed. RNA isolated from the precipitates was reverse transcribed with random hexamer primers and used for qPCR with TaqMan® primer/probe sets to the indicated genes. Bars represent relative quantification using the ΔΔCt method normalized to control antibody CLIP +/- 95% confidence interval of three replicates. Non-specifically bound 18S rRNA was used as an endogenous control.
Table 1 lists the 34 genes found to be increased by depletion of Ars2. qPCR was used to confirm increased expression of the genes CRIPAK, HIST2H2BE, HSPA1L, and TRIM52 following depletion of Ars2 (Figure 2c). In addition to HIST2H2BE, several other replication-dependent histone genes increased following Ars2 depletion including HIST1H3H, HIST3H2A, HIST1H2AG, HIST2H2BF and HIST1H2AI (Table 1). Additionally, H1F0, a non-replication-dependent histone gene that accumulates in growth-inhibited cultured cells (Pehrson and Cole, 1980) was also found to increase following Ars2 knockdown. In contrast, histone mRNAs were not increased following depletion of DGCR8. Therefore, Ars2 may play a direct role in controlling the expression of histone mRNAs.
Table 1.
Genes increased at least 2-fold (log2) following Ars2 depletion from HeLa cells.
| Gene Symbol | RefSeq | Fold Change |
|---|---|---|
| HIST2H2BE* | NM_003528 | 4.31 |
| HIST1H3H* | NM_003536 | 3.27 |
| PIGW | NM_178517 | 3.15 |
| CRIPAK* | NM_175918 | 2.92 |
| DRAM | NM_018370 | 2.86 |
| HIST3H2A* | NM_033445 | 2.86 |
| TRIM52* | NM_032765 | 2.83 |
| HIST1H2AG* | NM_021064 | 2.65 |
| ABCB1 | NM_000927 | 26 |
| H0XA7 | NM_006896 | 2.59 |
| NUDCD2 | NM_145266 | 2.58 |
| PPPDE1† | NM_016076 | 2.55 |
| KCTD6 | NM_153331 | 2.53 |
| BRMS1L | NM_032352 | 2.48 |
| CROT | NM_001143935 | 2.44 |
| VPS4B | NM_004869 | 242 |
| CYLD | NM_015247 | 2.41 |
| HSPA1L* | NM_005527 | 2.41 |
| CLDN1† | NM_021101 | 2.39 |
| HIST2H2BF | NM_001024599 | 2.38 |
| C5orf54 | NM_022090 | 2.37 |
| ATP8B1 | NM_005603 | 229 |
| H1F0* | NM_005318 | 227 |
| HPDL | NM_032756 | 2.26 |
| CRYAB | NM_001885 | 2.25 |
| SATB2 | NM_015265 | 2.25 |
| TRAPPC6B | NM_001079537 | 224 |
| TSPYL4 | NM_021648 | 222 |
| PIK3R3 | NM_003629 | 2.14 |
| HIST1H2AI | NM_003509 | 2.11 |
| ACRC | NM_052957 | 2.09 |
| AP1AR | NM_018569 | 2.04 |
| ZNF251 | NM_138367 | 2.04 |
| MRPL50 | NM_019051 | 2.02 |
indicates that the transcript has been confirmed to be increased after Ars2 depletion by qPCR.
indicates that the transcript also increased after DGCR8 depletion.
Ars2 binds histone mRNAs
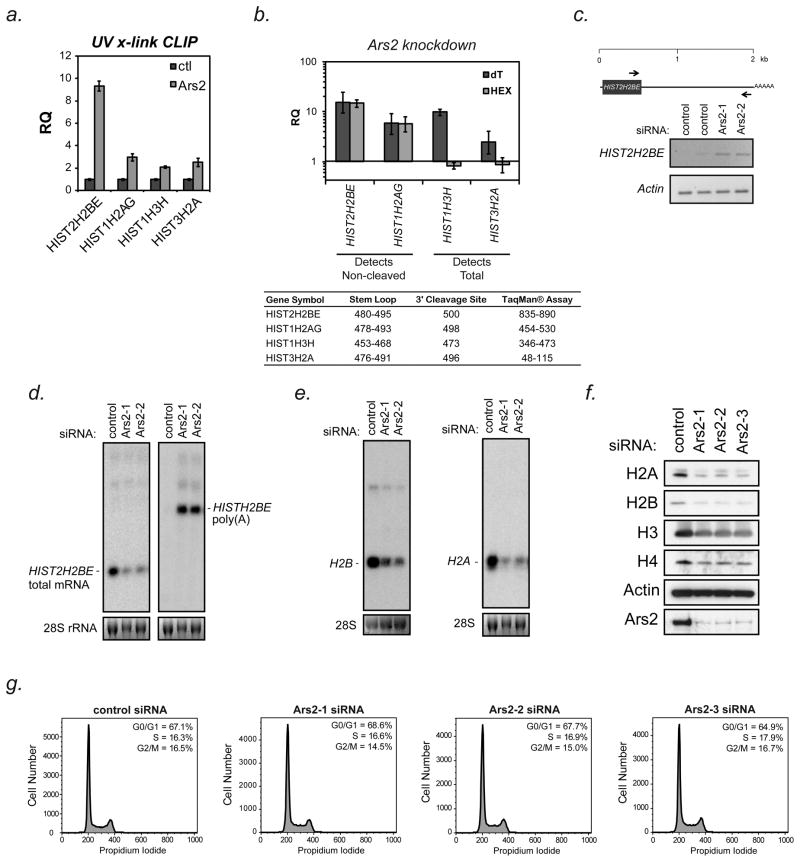
The transcript found to increase the most following Ars2 depletion was HIST2H2BE, an unusual histone transcript that produces both a replication-dependent 3′ cleaved transcript and a constitutively expressed polyadenylated transcript (Collart et al., 1991; Collart et al., 1992). The relative level of each of these two transcripts is likely regulated by co-transcriptional processing of the pre-mRNA. Co-transcriptional regulation of replication-dependent histone mRNA 3′ end processing is facilitated by the nuclear CBC (Narita et al., 2007). Since Ars2 binds the CBC, we sought to determine if the Ars2/CBC complex is associated with histone mRNA. Paraformaldehyde crosslinking immunoprecipitation (PFA-CLIP) was performed using antibodies to Ars2 or a nonspecific control antibody. Two replication-dependent histone transcripts, HIST2H2BE and HIST1H2AG, whose expression increased following Ars2 depletion, were enriched in Ars2 immunoprecipitates when compared to control (Figure 2d). The transcripts CRIPAK and TRIM52 were not enriched in Ars2 PFA-CLIP, despite increased expression in Ars2 depleted cells. To determine if Ars2 directly interacts with histone mRNAs, ultraviolet (UV) crosslinking followed by Ars2 immunoprecipitation (UV-CLIP) was performed. Unlike formaldehyde, UV light does not readily cross-link closely associated proteins. RNA isolated by Ars2 UV-CLIP was found to be enriched in histone mRNA (Figure 3a), confirming that Ars2 directly binds histone mRNAs. These data suggest that Ars2 contributes to the CBC complex’s ability to interact with histone mRNAs, while other observed changes in gene expression following Ars2 knockdown may result from indirect effects.
Figure 3. Ars2 binds to and regulates the expression and processing of replication-dependent histone transcripts.
(A) To determine if Ars2 could directly associate with replication-dependent histone mRNAs, ultraviolet light crosslinking followed by immunoprecipitation (UV-CLIP) with a monoclonal antibody to Ars2 (9A12) or control antibody (SP/0) was performed. RNA isolated from the precipitates was reverse transcribed with random hexamer primers and used for qPCR with TaqMan® primer/probe sets to the indicated genes. Bars represent relative quantification using the ΔΔCt method normalized to control antibody CLIP +/− 95% confidence interval of three replicates. Non-specifically bound 18S rRNA was used as an endogenous control.
(B) HeLa cells were transfected with siRNA to Ars2 (Ars2-3) or a control siRNA and 3 days later RNA was isolated. cDNA synthesis was performed using either oligo(dT) or random hexamer primers. Transcript levels were measured by TaqMan®-based qPCR using the assays outlined in the inset table. Because the TaqMan® assays used for HIST2H2BE and HIST2H2AG amplified a region downstream of their 3′ cleavage sites they only detected non-cleaved transcripts (i.e. those that extend beyond the normal 3′ cleavage sites), whereas the assays for HIST1H3H and HIST3H2A were upstream of their 3′ cleavage sites and could therefore differentiate between polyadenylated transcripts (dT) and total mRNA expression (HEX). Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(C) Top - Schematic of the human HIST2H2BE gene with scale in kilobases (kb). The arrows indicate the position of primers used for PCR analysis. Bottom - HeLa cells were transfected with siRNAs to Ars2 (Ars2-1, Ars2-2) or control siRNAs and three days later RNA was isolated. PCR was performed following oligo(dT) reverse-transcription using the primers indicated in the schematic. Actin was amplified as a control.
(D) HeLa cells were transfected with control or Ars2 siRNAs (Ars2-1, Ars2-2), RNA was isolated after 3 days and Northern blotting was performed for the HIST2H2BE transcript. Left - Total HIST2H2BE mRNA was detected with a 34 nucleotide probe designed to hybridize selectively to the HIST2H2BE 3′UTR upstream of the stem-loop. Right - To detect HIST2H2BE transcripts that were not properly cleaved, the membrane was stripped and re-probed with a probe generated by nick translation of a cDNA of the HIST2H2BE 3′UTR.
(E) Additional Northern blots were performed on RNA isolated from HeLa cells transfected with Ars2 siRNAs (Ars2-1, Ars2-2) or control siRNA. Blots were probed with oligonucleotide probes within the open reading frames of histone H2B (left) or H2A (right) genes and therefore detect multiple conserved transcripts.
(F) HeLa cells were transfected with control or three different Ars2 siRNAs (Ars2-1, Ars2-2, Ars2-3) and proteins were harvested after three days. Western blotting was performed for the four core histone proteins. Knockdown was confirmed by probing for Ars2 and Actin was used as a loading control.
(G) Cell cycle analysis was performed on HeLa cells three days after transfection with control or three independent siRNAs targeting Ars2 (Ars2-1, Ars2-2, Ars2-3). Quantification of cells in G0/G1 (1N), G2 (2N) and S-phase (intermediate) of the cell cycle is shown in the upper right corner of each histogram. In agreement with these data the amount of DNA per cell remained unaltered following Ars2 knockdown (43.3 pg/cell in Ars2 siRNA vs. 42 pg/cell in control).
Ars2 promotes histone mRNA maturation and expression
Replication-dependent histone mRNAs undergo 3′ end cleavage downstream of a 14 nucleotide stem-loop structure within their 3′ UTR to yield mRNA lacking a poly(A) tail (Marzluff et al., 2008). However, Affymetrix microarray hybridization was performed following an oligo(dT) reverse transcription step, raising the possibility that the histone mRNAs detected in this way were polyadenylated. To test if polyadenylated histone mRNAs were produced by cells following Ars2 depletion, TaqMan®-based qPCR was performed. For all of the histone genes tested (HIST2H2BE, HIST1H2AG, HIST1H3H, and HIST3H2A), increased transcript levels were detected following reverse transcription with oligo(dT) primers in Ars2 depleted cell (Figure 3b; dT). Detection of increased levels of histone mRNAs under oligo(dT) reverse-transcribed conditions further suggests that depletion of Ars2 leads to an increase in polyadenylated histone mRNAs, which occurs under conditions where 3′ end processing is defective. To determine if histone mRNAs were properly cleaved, TaqMan® primer/probe sets were used to either amplify a region downstream of or spanning the 3′ cleavage site (HIST2H2BE and HIST1H2AG, respectively) or amplify a region upstream of the 3′ cleavage site (HIST1H3H and HIST3H2A) from RNA reverse transcribed using random hexamer primers. As expected, increased amplification only occurred when the region amplified was downstream of the 3′ cleavage site (Figure 3b; HEX), confirming that depletion of Ars2 led to an error in histone mRNA 3′ end processing. Additional TaqMan® primer/probe sets that amplify sequences within the open reading frame of HIST1H2AG or the downstream region of HIST1H3H were used to further demonstrate increased detection of non-cleaved histone transcripts following Ars2 knock-down (Supplementary Figure 1).
The HIST2H2BE gene contains a canonical polyadenylation site roughly 2 kilobases downstream of the stem-loop structure in the 3′ UTR as depicted in Figure 3c (top panel). To determine if this polyadenylation site was used in cells upon Ars2 depletion, RT-PCR was performed with specific primers designed to amplify the entire 3′UTR of HIST2H2BE mRNA. In cells depleted of Ars2, a PCR product was detected of the appropriate size corresponding to the lengthened 3′UTR generated by usage of the distal polyadenylation site (Figure 3c).
To further confirm that a longer HIST2H2BE transcript was generated after Ars2 depletion Northern blotting was performed on RNA isolated from Ars2 depleted cells. Hybridization of a probe that recognizes the long 3′UTR of HIST2H2BE confirmed that long HIST2H2BE transcripts accumulated when Ars2 was depleted (Figure 3d, right). Somewhat surprisingly, hybridization of a probe designed to the HIST2H2BE 3′UTR upstream of the cleavage site revealed that Ars2 depletion led to reduced levels of normally processed HIST2H2BE mRNA (Figure 3d, left). Additionally, little long polyadenylated HIST2H2BE transcript was detected by this probe, suggesting that only a small portion of total HIST2H2BE mRNA was improperly processed in the absence of Ars2. Thus, the main effect of Ars2 depletion is a reduction in the accumulation of correctly cleaved histone mRNA.
Additional Northern blots were performed on RNA isolated from Ars2 depleted cells using probes that recognize the open reading frames (ORFs) of H2B or H2A. In each case, histone transcripts were markedly reduced when Ars2 was depleted (Figure 3e). Since probes to histone ORFs recognize multiple transcripts, these data suggest that the overall levels of histone mRNAs decreased following Ars2 depletion. To test if decreased histone mRNA levels in Ars2 knockdown cells affect histone protein production, Western blots were performed. Diminished H2A, H2B, H3, and H4 protein levels were detected in cells depleted of Ars2 when compared to control cells (Figure 3f) without significant change in the ratio of total protein to DNA in these cells (8.64 in control vs. 8.8 in Ars2 depleted). These data are consistent with the notion that the proliferative defect seen following Ars2 depletion occurs because cells lose their ability to produce sufficient histone proteins to support cell division. Unlike depletion of stem-loop binding protein (SLBP), an established component of histone mRNA biogenesis (Salzler et al., 2009; Sullivan et al., 2009), Ars2 depleted cells do not accumulate in the S phase of the cell cycle. Ars2 depleted cells arrested throughout the cell cycle and did not show signs of S phase accumulation as the proportion of cells in S phase varied by less than 2% from control cells (Figure 3g).
7SK RNA binds Ars2
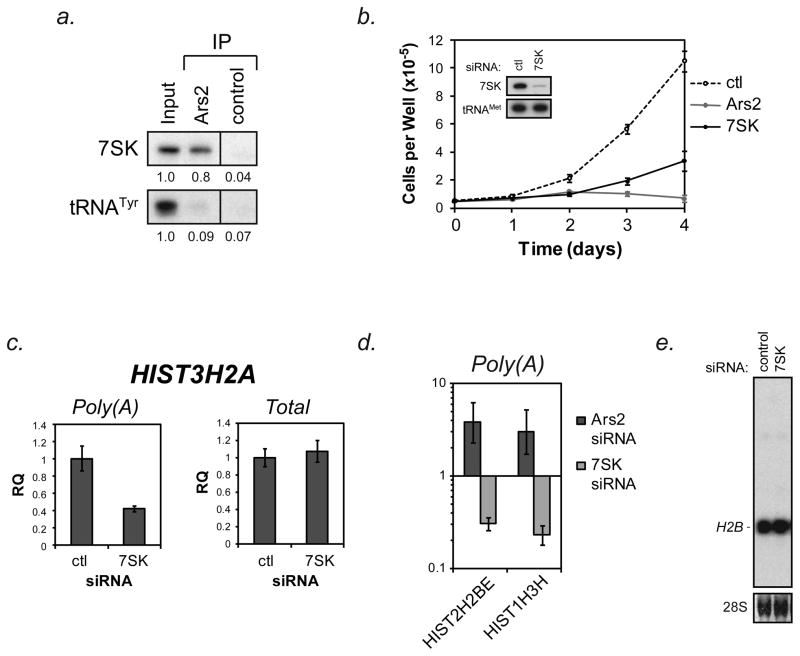
Ars2 and other CBC components (CBP80 and CBP20) are required for proper histone mRNA biogenesis. Additionally, components of the CBC interact with histone loci (Narita et al., 2007), suggesting that the Ars2/CBC complex may act to coordinate transcription of histone mRNAs with their proper processing. In agreement with this hypothesis, capping enzyme is the earliest protein to associate with RNAPII transcripts, likely via recruitment to stalled transcription complexes prior to initiation of elongation (Mandal et al., 2004). Initiation of elongation is regulated by the positive transcription elongation factor b (P-TEFb) component cyclin-dependent kinase 9 (CDK9), which has been reported to be required for proper 3′ end processing of replication-dependent histone mRNAs (Pirngruber et al., 2009). CDK9 kinase activity is controlled by association with the non-coding RNA 7SK (Diribarne and Bensaude, 2009). When CDK9 is associated with 7SK, its kinase activity is low, but upon release from 7SK the CDK9 kinase is activated (Nguyen et al., 2001; Yang et al., 2001). To determine if Ars2 binds the 7SK-CDK9 complexes, Ars2 was immunoprecipitated from HeLa cell extracts. Specific interactions between Ars2 and CDK9 or 7SK were determined by Western or Northern blot, respectively. 7SK RNA (Figure 4a), but not CDK9 (data not shown) was specifically enriched in Ars2 immunoprecipitates.
Figure 4. The Ars2 bound RNA 7SK promotes proliferation yet opposes proper replication-dependent histone mRNA 3′ end processing.
(A) HeLa cells were crosslinked and immunoprecipitation was performed using control or Ars2 antibodies. RNA was isolated from the precipitate and Northern blotting was performed with a probe specific for 7SK RNA. tRNATyr was probed to demonstrate the specificity of the Ars2-7SK interaction. Relative quantification of bands is displayed below each blot with input set to 1. 100 ng of total RNA was used for input.
(B) HeLa cells were transfected with siRNAs targeting Ars2 (Ars2-2), 7SK, or a control siRNA. Two days later cells were collected, counted and re-seeded in triplicate into 6 well plates at equal densities. Cell counts were performed over the course of the next four days (day 0–4). Data points represent the average number of cells per well from two independent experiments +/− standard deviation. Inset - HeLa cells were transfected with siRNA targeting 7SK RNA or control siRNA and three days later RNA was isolated. Northern blotting was performed to confirm knockdown of 7SK. tRNAMet was probed as a loading control.
(C) HeLa cells were transfected with siRNA targeting 7SK or control siRNA and RNA was isolated three days later. cDNA synthesis was performed using either oligo(dT) or random hexamer primers and TaqMan®-based qPCR was used to determine changes in the levels of polyadenylated (poly(A)) and total HIST3H2A mRNA. Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(D) Hela cells were transfected with siRNA targeting Ars2 (Ars2-2), 7SK or a control siRNA and RNA was isolated three days later. cDNA synthesis was performed using oligo(dT) primers and TaqMan®-based qPCR was used to determine changes in the levels of polyadenylated HIST2H2BE (H2BE) and HIST1H3H (H3H) mRNA. Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(E) Northern blotting was used to determine the effect of 7SK depletion on total levels of histone H2B transcript.
7SK supports proliferation yet opposes histone 3′ end formation
Since the most striking function of Ars2 is to promote cellular proliferation, we tested the effect of 7SK RNA on proliferation. We found that siRNA against Ars2 or 7SK RNA could significantly inhibit the proliferation of cultured cells (Fig 4b). Knockdown of 7SK was confirmed by Northern blot (Figure 4b,inset). These data demonstrate that similar to Ars2, 7SK RNA is necessary for cellular proliferation.
Because Ars2 could interact with the 7SK RNA and both were found to be necessary for cellular proliferation, the contribution of 7SK to histone mRNA maturation was tested. Cells depleted of 7SK showed reduced levels of polyadenylated, but not total, HIST3H2A transcripts compared to control transfected cells (Figure 4c). A reciprocal effect of Ars2 and 7SK depletion on polyadenylation of histone transcripts was also seen for HIST2H2BE and HIST1H3H (Figure 4d), suggesting that 7SK may inhibit histone mRNA processing. Additionally, depletion of 7SK from cells slightly increased the level of properly processed H2B transcript in cells as detected by Northern blot (Figure 4e). These data demonstrate that the proliferative defect of cultured cells following siRNA knock-down of Ars2 or 7SK does not result from a shared ability to promote histone mRNA expression and 3′ end processing.
Reciprocal interaction of 7SK with Ars2 and CDK9
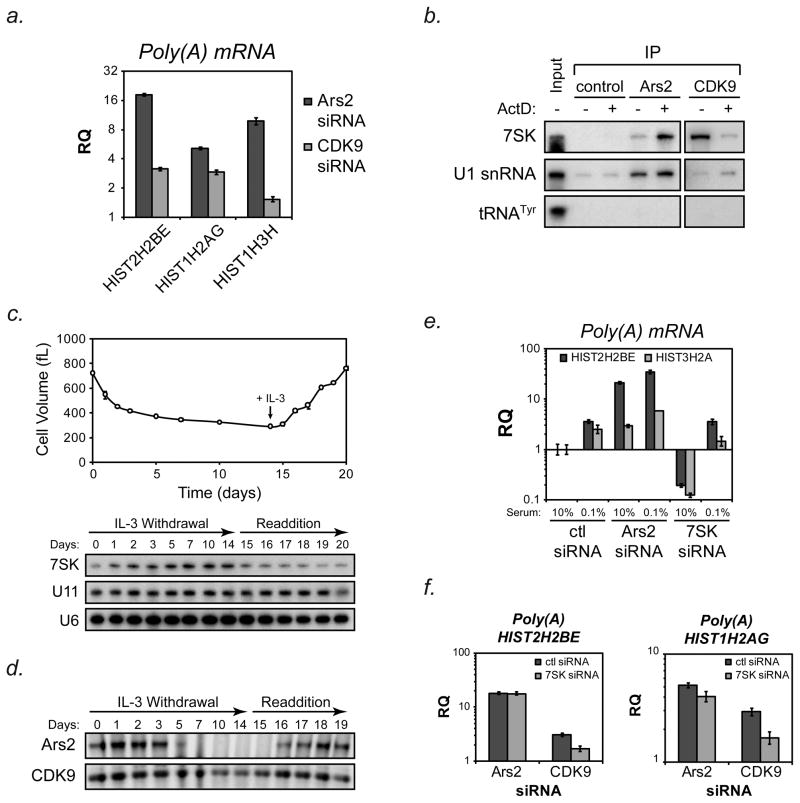
Similar to what we have found for Ars2, CDK9 has been reported to bind 7SK RNA (Diribarne and Bensaude, 2009) and regulate 3′ end processing of histone mRNAs (Pirngruber et al., 2009). We first confirmed that depletion of CDK9 resulted in increased polyadenylation of the same histone transcripts as Ars2 (Figure 5a). To further test the interaction of Ars2 with 7SK-CDK9 complexes, we took advantage of the fact that the 7SK-CDK9 interaction can be disrupted by treatment with low-doses of actinomycin D (Nguyen et al., 2001; Yang et al., 2001). Immunoprecipitation of Ars2 from extracts of HeLa cells treated with 1 μg/mL actinomycin D for 1 hour uncovered an approximately 6-fold increase in the association of 7SK RNA with Ars2 when compared to untreated cells (Figure 5b). In contrast, actinomycin D treatment led to a 7-fold decrease in the amount of 7SK RNA bound to CDK9.
Figure 5. Reciprocal interactions of 7SK RNA with Ars2 and CDK9.
(A) Hela cells were transfected with siRNA targeting Ars2 (Ars2-2), CDK9 or a control siRNA and RNA was isolated three days later. cDNA synthesis was performed using oligo(dT) primers and TaqMan®-based qPCR was used to determine changes in the levels of polyadenylated HIST2H2BE (H2BE) HIST1H2AG (H2AG) and HIST1H3H (H3H) mRNA. Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(B) HeLa cells were treated with 1μg/mL actinomycin D or DMSO control for one hour and then crosslinked. Immunoprecipitation with antibodies against Ars2, CDK9 or a control antibody followed by Northern blotting for 7SK was performed. U1 snRNA, which binds Ars2, was probed to insure Ars2 RNA binding was not compromised by actinomycin D treatment. Detection of 7SK in Ars2 immunoprecipitates increased 5.7 fold following actinomycin D treatment while detection of U1 snRNA only increased 1.4 fold, similar to the increase in non-specific U1 binding seen in control immunoprieipitates (1.3 fold increase). Detection of 7SK in CDK9 immunoprecipitates decreased 7.1 fold following actinomycin D treatment. A shorter exposure time was used to retain linearity of 7SK signal in CDK9 immunoprecipitates (right blot). tRNATyr was probed to demonstrate the specificity of the Ars2-7SK and CDK9-7SK interactions. 100ng of total RNA was used as input.
(C) IL-3-dependent Bax−/−Bak−/− cells were grown in the presence of IL-3 until day 0, at which point media was removed, cells were washed with PBS and resuspended in media without IL-3. Cells were maintained for 14 days in IL-3 deficient media, followed by restimulation with IL-3 at day 14.5. At the time points indicated aliquots were removed from the culture and RNA was extracted for Northern blotting with a probe for 7SK. The membrane was stripped and reprobed for U6 and U11 as loading controls.
(D) IL-3-dependent Bax−/−Bak−/− cells were subjected to IL-3 withdrawal and restimulation as in (B). Aliquots of the culture were removed at the indicated time points and protein was extracted in RIPA buffer. An equal amount of total protein was run on SDS-PAGE gels and Western blotting was performed for Ars2 and CDK9.
(E) HeLa cells were transfected with control, Ars2 or 7SK siRNAs. Two days later cells were washed with PBS and fresh media was added containing 10% serum or 0.1% serum. Twenty-four hours later RNA was extracted, reverse transcribed with oligo(dT) primers and TaqMan®-based qPCR was performed to determine relative levels of polyadenylated HIST2H2BE and HIST3H2A. Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(F) Hela cells were co-transfected with siRNAs targeting Ars2 or CDK9 plus siRNAs targeting 7SK or control siRNA. Three days later RNA was extracted, reverse transcribed with oligo(dT) primers and TaqMan®-based qPCR was performed to determine relative levels of polyadenylated HIST2H2BE and HIST1H2AG. Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
Since Ars2 is selectively expressed in proliferating cells, we examined the expression of 7SK RNA and CDK9 in proliferating versus quiescent cells. Bax−/−Bak−/− hematopoietic cells dependent on the growth factor IL-3 for growth and proliferation were subjected to withdrawal of IL-3 for 14 days, during which time they become quiescent and engage in macroautophagy to maintain viability (Lum et al., 2005). Northern blotting showed that 7SK RNA levels were induced relative to control non-coding RNAs during the period of IL-3 withdrawal (Figure 5c). At day 14.5 IL-3 was added back to the medium and over the course of 5 days the cells recovered and resumed cell division on approximately day 18. During this period of growth factor re-addition levels of 7SK RNA were gradually reduced compared to control RNAs (Figure 5c). A similar time course of IL-3 withdrawal and re-stimulation was performed to examine protein levels of Ars2 and CDK9. As previously shown (Gruber et al., 2009), Ars2 levels were rapidly decreased upon withdrawal of IL-3 and recovered upon IL-3 re-stimulation. In contrast, CDK9 remained robustly expressed during days 5 through 14 of IL-3 withdrawal when cells were fully quiescent (Figure 5d). These data suggest that in proliferating cells 7SK-Ars2 complexes and 7SK-CDK9 complexes may exist, whereas quiescent cells are likely enriched for 7SK-CDK9 complexes.
To determine the role of 7SK RNA in histone mRNA 3′ end processing during cellular quiescence, HeLa cells were transfected with siRNAs and then two days later exposed to 0.1% serum-containing medium for 24 hours to induce quiescence. Depletion of 7SK by siRNA was unaffected by serum starvation (Supplemental Figure 2). When 7SK RNA was depleted from cells grown in 10% serum, the levels of polyadenylated HIST2H2BE and HIST3H2A transcripts were diminished compared to control transfected cells. However, when cells were cultured in low serum 7SK RNA depletion was not able to block the increase in polyadenylated histone transcripts (Figure 5e). We have previously reported that exposure of cells to low serum resulted in decreased Ars2 levels (Gruber et al., 2009). Taken together with the inability of 7SK RNA knock-down to repress histone mRNA polyadenylation during serum starvation, we hypothesized that the effect of 7SK RNA on histone mRNA processing was dependent on Ars2. Consistent with this hypothesis, knockdown of 7SK did not reduce the level of polyadenylated histone mRNA in Ars2-depleted cells but retained the ability to decrease the accumulation of polyadenylated histone transcripts in CDK9-depleted cells (Figure 5f). Co-transfection of 7SK siRNA with Ars2 or CDK9 siRNA did not reduce the effectiveness of any of the siRNAs (Supplemental Figure 3).
Discussion
The Ars2 protein is a component of the nuclear cap binding complex (CBC) that is highly expressed and required only in proliferating cells, whereas other components of the nuclear CBC remain expressed in quiescent cells. Our previous work established that Ars2 contributes to the biogenesis of microRNAs that participate in the regulation of cellular proliferation, including let-7, miR-21 and miR-155. The current study expands on the repertoire of microRNAs known to be regulated by Ars2. Among the microRNAs confirmed to be dependent on Ars2 for expression (Figure 1), several have been implicated in oncogenesis. miR-18a is expressed from the c-myc induced miR-17-92 cluster that contributes to lymphomagenesis (He et al., 2005; O’Donnell et al., 2005) and miR-127 has been suggested to function as a tumor suppressor through repression of Bcl-6 (Saito et al., 2006). Over-expression of miR-27b is capable of promoting cancer cell invasion (Wang et al., 2009), while miR-28 over-expression is associated with neoplastic transformation in myeloproliferative disease (Girardot et al., 2010). These data suggest that Ars2 may specifically regulate a subset of microRNAs that are associated with cellular proliferation and oncogenesis.
MicroRNA biogenesis begins with co-transcriptional regulation of primary RNA polymerase II (RNAPII) transcript cleavage by Drosha (Morlando et al., 2008). Ars2 associates with Drosha and is required for expression of microRNA primary transcripts in all cases examined to date ((Gruber et al., 2009) and Figure 1g). This suggests that Ars2 may act early during co-transcriptional Drosha processing of primary microRNA transcripts. Coupling of transcription and pre-mRNA processing is a common feature among RNAPII transcripts (Pawlicki and Steitz, 2010). If Ars2 regulates this coupling, the expression of numerous mRNAs would likely be affected by Ars2 depletion. Microarray profiling led to the discovery of 101 microRNA pathway-independent mRNAs whose expression decreased and 32 whose expression increased after depletion of Ars2 from HeLa cells. Among the increased transcripts, replication-dependent histone mRNAs were highly represented (Table 1). In mammals, replication-dependent histone mRNAs are the only protein-coding transcripts that do not contain a poly(A) tail, but instead contain a stem-loop structure in the 3′UTR that directs endonucleolytic cleavage of the transcript. As microarray profiling was dependent on the transcripts containing a poly(A) tail, this led to the possibility that Ars2 depletion could interfere with histone mRNA 3′ end processing, leading to histone mRNA polyadenylation. Indeed, depletion of Ars2 led to bypass of the proper 3′ processing site in replication-dependent histone mRNA 3′UTRs and utilization of downstream polyadenylation sites, leading to an increase in polyadenylated histone transcripts (Figure 3).
Several proteins known to interact with Ars2 play roles in replication-dependent histone mRNA 3′ end processing. Most strikingly, the nuclear CBC has been shown interact with the negative elongation factor (NELF) complex to regulate replication-dependent histone 3′ end processing (Narita et al., 2007). Furthermore, Ars2 has been shown to interact with the nuclear protein FLASH and the interaction between Ars2 and FLASH was found to be essential for S-phase progression (Kiriyama et al., 2009). FLASH was identified as a protein that binds the U7 snRNP, which recruits the cleavage factor CPSF-73 to the proper histone mRNA 3′ cleavage site. In drosophila, loss of FLASH expression inhibited proper 3′ end processing of replication-dependent histone mRNAs and resulted in their polyadenylation (Yang et al., 2009). The fact that Ars2 promotes proper replication-dependent histone 3′ end processing and is associated with several other proteins shown to have the same function suggests that Ars2 may be either involved in a cascade of protein-protein interactions or in a protein complex that is required for histone mRNA processing during cellular proliferation.
Another protein shown to be required for histone mRNA 3′ end processing is CDK9, which phosphorylates serine 2 of the RNAPII C-terminal domain (CTD) and NELF to stimulate transcriptional elongation of paused transcripts. CDK9 is a component of the P-TEFb complex and was recently shown to associate with the nuclear CBC to link transcription elongation to pre-mRNA processing (Lenasi et al., 2011). We tested whether the CBC component Ars2 could co-immunoprecipitate CDK9 and found that the two proteins did not physically associate (data not shown). However, we were able to show that Ars2 bound the RNA 7SK (Figure 4), an endogenous regulator of CDK9 kinase activity. Interestingly, the Ars2-7SK interaction increased when the CDK9-7SK complex was disrupted by low-dose actinomycin D suggesting a reciprocal interaction of 7SK with Ars2 and CDK9. Our data further demonstrate that depleting cells of 7SK could partially reverse the increase in polyadenylation resulting from loss of CDK9, but not Ars2 (Figure 5f). Although we confirmed previous reports that CDK9 depletion resulted in increased polyadenylation of histone mRNA, its effect was always a fraction of the increase achieved by Ars2 depletion. Depletion of both Ars2 and CDK9 did not result in a further increase in polyadenylated histone mRNA when compared to Ars2 depletion (data not shown), suggesting that Ars2 and CDK9 function in the same pathway to promote proper replication-dependent histone mRNA 3′ end processing. Importantly, no changes in U7 snRNA expression were observed when Ars2, CDK9 or 7SK were depleted from cells. Together these data suggest that Ars2-containing CBCs are primary contributors to the regulation of histone mRNA 3′ end cleavage. The ability of Ars2 to contribute to this process is negatively affected by 7SK. In contrast, our data suggest the previously reported role for CDK9 in histone mRNA 3′ end processing is indirect and results from CDK9’s ability to sequester 7SK thus relieving 7SK’s repression of Ars2.
The co-transcriptional recruitment of factors to the cap structure of pre-RNAs may provide a point of regulation in histone and microRNA biosynthesis during cellular proliferation. Our data, when placed in the context of previous studies, suggest that Ars2 may play a central role in coordinating interactions between transcription and pre-mRNA processing machinery to facilitate proper RNA biogenesis. Interestingly, the two classes of transcripts thus far identified as regulated by Ars2, histone mRNA and microRNA, share stem-loop structures and require recruitment of specialized processing machineries for proper gene expression. This may be coincidental or may reflect a secondary structure requirement for Ars2 binding to its RNA targets. Continued work will undoubtedly refine our understanding of the role of Ars2 in RNA biogenesis and cellular proliferation.
Experimental Protocols
Cell lines and transfection
Adherent cell lines were maintained in DMEM supplemented with 10% FBS, HEPES, and L-Glutamine in a humidified incubator at 37°C. For serum starvation experiments cells were washed with PBS, then 0.1% serum-containing DMEM media was added for 24 hours. Transfection of siRNAs was performed with RNAiMAX reagent (Invitrogen). Sequences are provided in Supplemental Experimental Procedures and Allstars siRNA negative control (Qiagen) was used as a control. Growth and manipulation of the IL-3-dependent Bax−/−Bak−/− cell line was performed as previously described (Lum et al., 2005).
RNA immunoprecipitation
Cells were cross linked in PBS with 0.2% formaldehyde with agitation for 10 minutes, then quenched with 0.15 M glycine for 5 minutes or under ultraviolet light for 3 minutes and then washed with PBS. Protein-RNA complexes were extracted with RSB buffer with 250 mM NaCl, 1% empigen and 0.5% Triton-X-100, IP was performed with specific antibodies and RNA was recovered by proteinase K digestion, phenol-choloform extraction and ethanol precipitation.
Western blots, Northern blots and RT-PCR
RT-PCR, Western and Northern blots were performed using standard protocols (see Supplementary Experimental Procedures for details).
Microarrays
Total RNA was hybridized to Affymetrix microRNA microarrays, and Affymetrix Human Gene 1.0ST mRNA microarrays by the University of Pennsylvania microarray core. All protocols were conducted as described in the Affymetrix GeneChip Expression Analysis Technical Manual. Prior to hybridization total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter.
Supplementary Material
Highlights.
Ars2 binds histone mRNAs and facilitates their 3′ end processing and expression
7SK RNA binds Ars2 and negatively affects histone mRNA 3′ end processing
In addition to histone mRNAs, Ars2 regulates a sub-set of microRNAs
Acknowledgments
The authors would like to thank members of the Thompson and Dreyfuss labs for their support and critical review of data and this manuscript, Isabela Oliva for help with antibody production, Eric Lai and Celia Andreu-Agullo for discussions on the function of Ars2 and the University of Pennsylvania microarray core facility for technical assistance. This work was funded in part by grants from the NCI and the Abramson Family Cancer Research Institute.
Footnotes
Accession Numbers
Data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE34681 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34681).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Collart D, Ramsey-Ewing A, Bortell R, Lian J, Stein J, Stein G. Isolation and characterization of a cDNA from a human histone H2B gene which is reciprocally expressed in relation to replication-dependent H2B histone genes during HL60 cell differentiation. Biochemistry. 1991;30:1610–1617. doi: 10.1021/bi00220a024. [DOI] [PubMed] [Google Scholar]
- Collart D, Romain PL, Huebner K, Pockwinse S, Pilapil S, Cannizzaro LA, Lian JB, Croce CM, Stein JL, Stein GS. A human histone H2B.1 variant gene, located on chromosome 1, utilizes alternative 3′ end processing. J Cell Biochem. 1992;50:374–385. doi: 10.1002/jcb.240500406. [DOI] [PubMed] [Google Scholar]
- Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot M, Pecquet C, Boukour S, Knoops L, Ferrant A, Vainchenker W, Giraudier S, Constantinescu SN. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood. 2010;116:437–445. doi: 10.1182/blood-2008-06-165985. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama M, Kobayashi Y, Saito M, Ishikawa F, Yonehara S. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol Cell Biol. 2009;29:4729–4741. doi: 10.1128/MCB.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Peterlin BM, Barboric M. Cap-binding Protein Complex Links Pre-mRNA Capping to Transcription Elongation and Alternative Splicing through Positive Transcription Elongation Factor b (P-TEFb) J Biol Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Machida S, Chen HY, Adam Yuan Y. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J, Cole RD. Histone H10 accumulates in growth-inhibited cultured cells. Nature. 1980;285:43–44. doi: 10.1038/285043a0. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman TG, Wang Z. Expression cloning for arsenite-resistance resulted in isolation of tumor-suppressor fau cDNA: possible involvement of the ubiquitin system in arsenic carcinogenesis. Carcinogenesis. 1999;20:311–316. doi: 10.1093/carcin/20.2.311. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Salzler HR, Davidson JM, Montgomery ND, Duronio RJ. Loss of the histone pre-mRNA processing factor stem-loop binding protein in Drosophila causes genomic instability and impaired cellular proliferation. PLoS One. 2009;4:e8168. doi: 10.1371/journal.pone.0008168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rathinam R, Walch A, Alahari SK. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J Biol Chem. 2009;284:23094–23106. doi: 10.1074/jbc.M109.012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Wang D, Wagner R, Breyssens H, Gertsenstein M, Lobe C, Lu X, Nagy A, Burke RD, Koop BF, et al. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol Cell Biol. 2008;28:1503–1514. doi: 10.1128/MCB.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.