Abstract
Simian varicella virus (SVV) is closely related to human varicella-zoster virus and causes varicella and zoster-like disease in nonhuman primates. In this study, a mini-F replicon was inserted into a SVV cosmid and infectious SVV was generated by co-transfection of Vero cells with overlapping SVV cosmids. The entire SVV genome, cloned as a bacterial artificial chromosome (BAC), was stably propagated upon serial passage in E. coli. Transfection of pSVV-BAC DNA into Vero cells yielded infectious SVV (rSVV-BAC). The mini-F vector sequences flanked by loxP sites were removed by co-infection of Vero cells with rSVV-BAC and adenovirus expressing Cre-recombinase. Recombinant SVV generated using the SVV-BAC genetic system has similar molecular and in vitro replication properties as wild-type SVV. To demonstrate the utility of this approach, a SVV ORF 10 deletion mutant was created using two-step Red-mediated recombination. The results indicate that SVV ORF 10, which encodes a homolog of the HSV-1 virion VP-16 transactivator protein, is not essential for in vitro replication, but is required for optimal replication in cell culture.
Introduction
Simian varicella virus (SVV, Cercopithicae herpesvirus 9) is a primate herpesvirus that causes epizootics of a varicella-like disease in Old World monkeys [7, 8]. Simian varicella is characterized by fever and vesicular skin rash and clinically resembles childhood chickenpox (varicella) caused by varicella-zoster virus (VZV) [6]. Following resolution of acute disease, SVV establishes latent infection within neural ganglia [15]. Viral reactivation may occur later in life causing a secondary disease analogous to herpes zoster, a VZV disease which generally afflicts the elderly [16].
SVV and VZV are closely related alphaherpesviruses sharing extensive antigenic relatedness [5]. In addition, the SVV and VZV genomes are similar in size and structure with >70% DNA homology [11, 12, 22]. Based on the clinical and pathologic similarities between simian and human varicella and the genetic relatedness between SVV and VZV, simian varicella is a useful experimental model to investigate VZV infections in humans [7].
Molecular studies of SVV are hampered by the cell-associated nature of the virus. The entire DNA sequence of the 124.8 kilobase pair SVV genome has been determined and 68 distinct SVV genes are identified, each with a VZV homolog [13]. Studies to determine the role of SVV genes in viral replication, pathogenesis, latency, and reactivation would be facilitated by generation of SVV mutants. A cosmid-based genetic system was developed and used for mutagenesis to construct SVV mutants defective in specific viral genes and to construct recombinant viruses expressing foreign genes [9, 10, 21, 26]. However, the efficiency of this genetic approach is limited since the technique requires co-transfection of four 35–40 kb cosmids spanning the entire SVV genome into the same eukaryotic cell, several recombination events mediated by overlapping viral sequences, and in vitro replication, taking 9 –12 days to generate viral plaques in cell culture. While several SVV mutants and recombinant viruses have been produced using the SVV cosmid system combined with recA- assisted restriction endonuclease (RARE) cleavage, this mutagenesis approach requires specific restriction endonuclease sites and is dependent on viral replication in cell culture [4, 10, 21, 26].
The genomes of VZV and other herpesviruses have been cloned as infectious bacterial artificial chromosomes (BAC) permitting maintenance and propagation of the viral DNA in Escherichia coli [19, 27]. This advance has facilitated genetic manipulation of large viral genomes without the requirement for in vitro replication in eukaryotic cells. VZV mutants have been generated using the BAC system in combination with techniques commonly used in bacterial genetics, including Red-mediated recombination mutagenesis [23, 28].
In this study, the entire SVV genome was cloned as a BAC for propagation in E. coli. The utility of this recombinant SVV-BAC system for genetic manipulation of the SVV genome was demonstrated by mutagenesis of the SVV open reading frame (ORF) 10, which encodes a homolog of the herpes simplex virus type 1 (HSV-1) virion VP-16 transactivator protein.
Materials and methods
Cells and virus
Vero and CV-1 (African green monkey kidney) cells were grown in Eagle’s minimal essential medium (EMEM) supplemented with 5% newborn bovine serum, penicillin (5,000 U/ml), and streptomycin (5,000 U/ml). SVV cosmids were derived from the Delta herpesvirus strain of SVV originally isolated from a patas monkey at the Tulane National Primate Research Center [2]. SVV genome nucleotide (nt) numbers refer to Gene Bank accession number AF275348.
Construction of transfer vector
A gfp expression cassette was generated using pEGFP-M [29] as a template and PCR primers gfpF: 5′-TTTTGTCGACAGTAATCAATTACGGGGTCATTAGTTC-3′ and gfp: 5′-TTTCTCGAGAAGATACATTGATGAGTTTGGACAA-3′, where the underlined sequences are SalI and XhoI restriction sites, respectively. This cassette was digested with XhoI and SalI and inserted into the SalI site of pMBO131 [29], which includes mini-F sequences, to obtain pMBG. A synthesized double-stranded DNA with Loxp-AscI-LoxP sequence was inserted into the SalI site of pMBG to generate BAC vector pMBGA. The SVV cosmid A, which is bracketed by NotI sites and includes a unique AscI site in the intergenic region between ORFs 12 and 13 (SVV nt 19,148) [13, 20], was digested with AscI and NotI. The two SVV fragments were isolated and inserted into the AscI site of pMBGA to generate pMBGA-Cos A.
Reconstitution of SVV with BAC vector sequence from cosmids
The pMBGA-Cos A and SVV cosmids B, C, and D (1.25 μg each) were digested with NotI and transfected into Vero cells using the Superfect transfection reagent and protocol (Qiagen Corp.) as previously described [10]. Viral plaques were detected by day ten post-transfection. After propagation in Vero cells, circular viral DNA was isolated from infected cells by the Hirt method [14] and transferred to competent E. coli DH10B by electroporation using a Bio-Rad E. coli Pulsar at 2.5 kV in 0.2 cm cuvettes. Bacterial clones were selected on LB agar plates including 25 μg/ml chloramphenocol (LB-CAM) and pSVV-BAC DNA was isolated from overnight culture using the Qiagen Plasmid Maxiprep system. Mini-F DNA sequences were confirmed in pSVV-BAC by PCR using BAC specific primers (5′-TACTGACTGGTCAACAGACACCG and 5′-TGCTCATGAAGGTTAGATGCC) to generate an expected 445 bp product or by Southern blot hybridization using the same BAC PCR product as a digoxigenin (DIG)-dUTP-labeled DNA probe.
Generation of infectious SVV
pSVV-BAC DNA was transfected into Vero cells by the Superfect method [10]. Viral plaques were evident by day four post-transfection and were demonstrated to exhibit typical SVV cpe in Vero cell monolayers and green fluorescence upon fluorescent microscopy. Infectious rSVV-BAC was propagated in Vero cells and viral stocks were stored at − 70°C. Cell-free rSVV-BAC was prepared as previously described [25].
The mini-F vector flanked by loxP sites was removed from the rSVV-BAC genome by co-infection of rSVV-BAC infected Vero cells with a recombinant adenovirus (AX-CANCre,) that expresses the Cre recombinase (Vector Biolabs, Philadelphia, PA). Vero cells were infected with AX-CANCre at a multiplicity of 100 pfu/cell. After a 2 hr adsorption, cells were washed with PBS and then incubated with Dulbecco’s minimal essential media (MEM) for 24 hrs. Cells were then superinfected with cell-free rSVV-BAC. Infectious SVV was propagated in Vero cells and microscopy was used to confirm typical SVV cpe without green fluorescence. Removal of mini-F vector sequences was confirmed by DNA sequence analysis of genomic DNA using SVV ORF 12 (5′-GTTTGCTTGATAGCTCTGGC) and ORF 13 (5′-CAGATAACGTTCCAGTTCCG) oligonucleotide primers that flank the BAC insertion site at SVV nt 19,148.
Generation of a SVV ORF 10 mutant
The pSVV-BAC was used to generate a SVV ORF10 mutant with deletion of amino acids 5–79 (SVV nt 14,194 – 14,418) through a two-step Red recombination approach [24]. The aphAI-I-SceI cassette present in pEPkan-S was amplified with SVV10F:5′-gtttcggcctaacggatttttgttgtattgtctttaccatggaacatttcctactagaaacatgtaacgaagataAGGATGA CGACGATAAGTAGGG-3′ and SVV10R:5′-tcattaagaggaaaacaggaaaatatatcttcgttacatgtttctagtaggaaatgttccatggtaaagacaataCAACC AATTAACCAATTCTGATTAG-3′ with italicized sequences corresponding to SVV nt 14,144 – 14,193 and nt 14,419 – 14,468, respectively, lower case sequences corresponding to SVV nt 14,419 – 14,443, and nt 14,193–14,169, respectively, and upper case sequences homologous to both ends of aphAI-I-SceI cassette. The first recombination in E. coli strain GS1783 resulted in insertion of the aphAI-I-SceI cassette into pSVV-BAC, while a second recombination resulted in deletion of SVV nt 14,194 – 14,418. The ORF 10 deletion in pSVV-BACorf10del DNA clones was confirmed by PCR employing primers SVV10F:5′-ATTTGGCGCCTTTATTATGGTATG-3′ (SVV nt 14,039 –14,062) and SVV10R: 5′-ATGTAGACGGTAGCGAAGGAAGAT-3′ (SVV nt 14,602–14,579) and DNA sequence analysis of the amplified product. Infectious rSVVorf10del mutants were reconstituted by transfection of pSVV-BACorf10del DNA into Vero cells.
Assay of in vitro viral replication
Vero cell monolayers (25 cm2) were infected with 500 PFU cell-free wt SVV or SVV mutants. Virus titers at 0, 8, 24, 48, and 72 hr postinfection were calculated by infectious center plaque assay on CV-1 cell monolayers. For plaque size calculations, virus plaques were measured at 72 hours postinfection by light microscopy using a stage micrometer. At least 20 independent viral plaques were measured with a light microscope, and the average diameter of plaques was calculated.
Construction of a self-excisable SVV-BAC
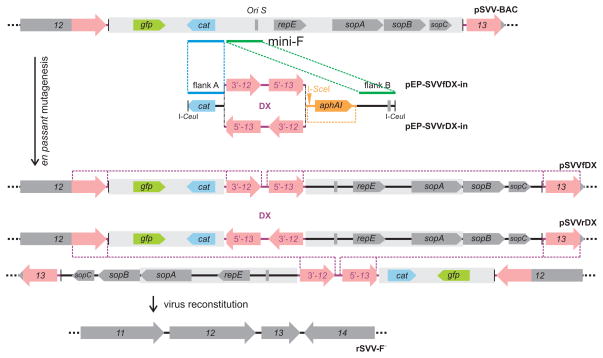
To allow complete excision of mini-F vector sequences upon virus reconstitution in Vero cells, a sequence duplication was inserted into the vector backbone as previously described [23] (Fig. 5). First, SVV sequences (nt 19,002– 20,663) were amplified from the SVV cosmid Cos A clone using primers 5′-GCATGCTAGAATCGTCTATACATGTTTGGTTTTTGTGGG-3′ and 5′-GTCGACGGATCCGGGAATTGTTTTGGTTGTCGTGTTAACTGTAATT-3 and the PCR amplicon was cloned into pCR2.1-TOPO (Invitrogen Corp.) resulting in plasmid p2.1-SVV-DX. To generate transfer constructs with forward or reverse orientation of the duplication within the mini-F vector sequences, p2.1-SVV-DX was transferred into pEPMCS-in-Belo either via SphI and SalI (pEP-SVVfDX-in) or BamHI and SphI (pEP-SVVrDX-in). Finally, two-step en passant mutagenesis was used to introduce the sequence duplications into pSVV-BAC [24]. The linear transfer construct was released by I-CeuI cleavage and inserted into the SVV BAC by a first Red recombination using E. coli GS1783 cells harboring pSVV-BAC. In a second step, the I-SceI-recognition site within the marker cassette was cleaved in E. coli and the kanamycin resistance gene removed via a second Red recombination. The resulting SVV clones pSVVfDX harboring the forward sequence duplication, and pSVVrDX with the inverse duplication were analyzed for integrity and correct modifications by RFLP and nucleotide sequencing. For virus reconstitution, Vero cell monolayers in 6-well plates were transfected with 2 μg of pSVVfDX- or pSVVrDX-DNA per well using FugeneHD (Roche) following the manufacturer’s instructions. Green fluorescing plaques were visible within two days. At 96 hr post-transfection, infected cells from one well were split onto wells of a 96 well plate. After three days, cells from the wells with the highest number of non-fluorescent plaques were used to infect fresh Vero cells seeded in a 6-well plate. After a second round of purification, all SVV plaques obtained after transfection with either BAC construct appeared non-fluorescent. Complete excision of the mini-F backbone was demonstrated by PCR and nucleotide sequencing.
Figure 5.
Schematic presentation of self-excision of mini-F sequences. The transfer constructs pEP-SVVfDX-in and pEP-SVVrDX-in were used to insert a 1.6 kb duplication of viral sequences (red) into the mini-F vector backbone (grey box). In a first step, the linearized constructs were inserted via Red recombination of flanks A and B (blue and green). In a second step, the en passant marker cassette was released by I-SceI cleavage and a second Red recombination (yellow). The resulting BAC clones harbored the sequence duplication in forward (pSVVfDX) or reverse orientation (pSVVrDX). Upon virus reconstitution in eukaryotic cells, mini-F sequences were excised by homologous recombination resulting in non-fluorescing virus progeny virus rSVV-F−.
Results and discussion
Cloning SVV DNA into E. coli
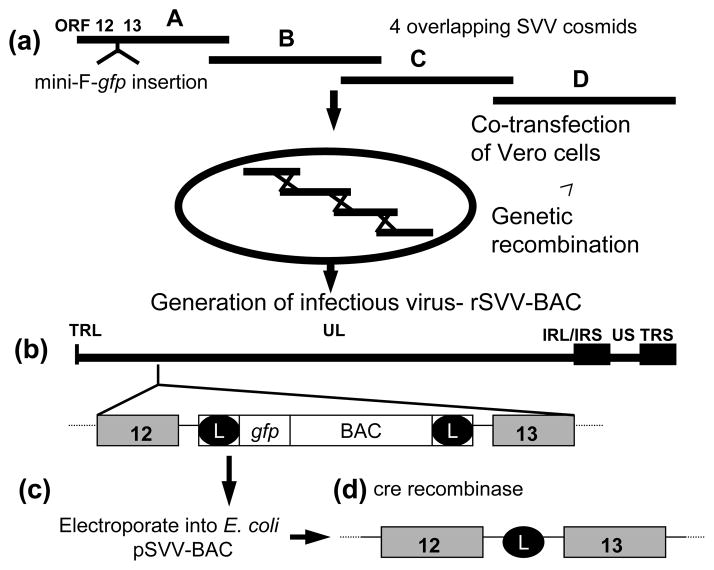
Mini-F replicon sequences, including the green fluorescent protein (gfp) gene, flanked by loxP sites was inserted into SVV cosmid A in a unique AscI site within the intergenic sequence between SVV ORFs 12 and 13 (SVV nt 19,148) [13, 20] to generate pSVV-cosA-F (Fig. 1). Prior studies indicated that insertion of foreign sequences at this site of the SVV genome does not inhibit in vitro viral replication [25]. Vero cells were co-transfected with pSVV-cosA-F and SVV cosmids B, C, and D and viral plaques were observed within ten days. These plaques exhibited green fluorescence upon UV illumination confirming expression of the incorporated gfp gene (data not shown). After further propagation in Vero cells, circular viral DNA was isolated from infected cells, and transferred to competent E. coli DH10B by electroporation.
Figure 1.
Strategy for cloning the SVV genome into E.coli. (a) The mini-F-gfp vector was inserted into a unique AscI site within SVV Cos A. The SVV CosA-mini-F-gfp was co-transfected with Cos B, Cos C, and Cos D into Vero cells and infectious rSVV-BAC was generated by recombination. (b) The rSVV-BAC includes the entire 124 kb SVV genome consisting of a unique long (UL) component covalently linked to a short component composed of a unique short (US) region bracketed by inverted repeat sequences (IRS and TRS). The mini-F-gfp vector sequences are incorporated into the SVV genome within the intergenic region between ORFs 12 and 13. (c) rSVV-BAC DNA was electroporated into E. coli producing pSVV-BAC, which is infectious when transfected back into Vero cells. (D) Removal of BAC vector sequences from rSVV-BAC is mediated by Cre-recombinase leaving the remaining loxP site (L).
Bacterial clones incorporating the SVV genome cloned into the BAC vector (pSVV-BAC) were identified by restriction endonuclease analysis. Upon AscI digestion of pSVV-BAC, high molecular weight SVV DNA fragments were evident along with the 8.0 kb mini-F-gfp vector which was released as confirmed by Southern blot hybridization using a labeled mini-F probe (Fig. 2a, lanes 1 and 2). BamHI digestion of pSVV-BAC revealed SVV BamHI fragments matching those of wild-type (wt) SVV DNA (Fig. 2a, lanes 3 and 5). Southern blot hybridization determined that the upper BamHI band includes the SVV BamHI A DNA fragment plus the 8.0 kb mini-F-gfp vector (Fig 2a, lane 4). The SVV BamHI A and BamHI G (7.1 kb) are terminal fragments of the SVV genome [12]. Due to the circular nature of pSVV-BAC, the SVV BamHI G fragment is missing in the pSVV-BAC BamHI digest and now covalently joined with the BamHI A terminal fragment (Fig. 2a, lanes 3 and 5). These results combined with the following data showing restriction endonuclease profiles of reconstituted infectious rSVV (Fig. 3b) indicate that the pSVV-BAC includes the entire SVV genome.
Figure 2.
RE analysis of pSVV-BAC DNA. (a) AscI digest of pSVV-BAC (lane 1) and Southern blot hybridization using a DIG-labeled BAC DNA probe to detect BAC sequences in pSVV-BAC (lane 2). BamHI digest of pSVV-BAC (lane 3) and Southern blot hybridization using a DIG-labeled BAC DNA probe to detect BAC sequences in pSVV-BAC (lane 4). BamHI digest of wt SVV DNA (lane 5). Arrows indicate pSVV-BAC DNA fragments containing BAC sequences. * indicates the BamHI G fragment missing in pSVV-BAC BamHI digest. (b) Stability of pSVV-BAC DNA upon serial passage in E. coli. BamHI digests of pSVV-BAC passage 1 (lane 1) and passage 10 (lane 2). EcoRI digests of pSVV-BAC passage 1 (lane 3) and passage 10 (lane 4). M- 1 kb molecular size marker.
Figure 3.
(a) Confirmation that rSVV-BAC (lane 1) contains BAC DNA sequences, but the BAC vector is removed from rSVV-BAC− (lane 2) by PCR using a BAC primer set. M- 100 bp marker. (b) BamHI and EcoRI RE analysis comparison of rSVV-BAC− DNA (lanes 1 and 3) and wt SVV DNA (lanes 2 and 4). M- 1 kb DNA marker. (c) Confirmation of the 225 bp deletion in rSVVorf10del DNA (lane 2) compared to rSVV-BAC− DNA (lane 1) by PCR using a SVV ORF 10 primer set. M- 100 bp marker. Arrows indicate expected sizes of PCR products.
Herpesvirus BAC genomes propagated in E. coli DH10B, which carry a rec A mutation to minimize recombination, are usually genetically stable, although recombination events in herpesvirus BACs have been reported [1]. To assess the stability of the pSVV-BAC, E. coli containing the pSVV-BAC were passaged ten times in LB broth with chloramphenocol (CAM, 25 μg/ml) and on LB-CAM plates. pSVV-BAC DNA was isolated from overnight cultures of single bacterial colonies and analyzed by restriction endonuclease and DNA sequence analysis. BamHI and EcoRI profiles of passage ten pSVV-BAC DNA were identical to that of the original pSVV BAC (Fig. 2b), indicating that the pSVV-BAC is stable in E. coli upon multiple passage.
Production of infectious SVV from pSVV-BACDNA
Infectious rSVV-BAC was generated by transfection of Vero cell monolayers with pSVV-BAC DNA. Viral plaques were evident by day four and exhibited typical SVV cytopathic effect (cpe) and green fluorescence upon UV microscopy, confirming expression of the integrated gfp gene. The presence of mini-F sequences in the rSVV-BAC genome was indicated by PCR amplification using BAC primers (Fig. 3a, lane 1) and confirmed by DNA sequence analysis.
The mini-F-gfp vector flanked by loxP sites was excised from the rSVV-BAC genome by co-infection of rSVV-BAC infected Vero cells with a recombinant adenovirus expressing Cre recombinase. The resulting infectious SVV (rSVV-BAC−) had typical SVV cpe on Vero cell monolayers, but did not exhibit green fluorescence upon UV microscopy (data not shown). The removal of the mini-F-gfp vector was confirmed by PCR using mini-F PCR primers (Fig. 3a, lane 2) and DNA sequence analysis which also revealed the presence of the remaining 34 bp loxP sequence in genomic DNA within the intergenic region between SVV ORFs 12 and 13 (data not shown). Restriction endonuclease analysis indicated that the rSVV-BAC− DNA and parental wt SVV gemones share identical BamHI and EcoRI restriction endonuclease profiles (Fig. 3b). These results demonstrate the ability of the SVV BAC system to generate infectious virus with molecular properties similar to wt SVV.
In vitro growth properties of rSVV-BAC− and wt SVV
To compare the in vitro replication properties of rSVV-BAC− and wt SVV, Vero cell monolayers were infected with 500 pfu of cell-free virus. At 8, 24, 48, and 72 hrs postinfection, the number of infected cells was determined by infectious center assay on CV-1 cell monolayers. As indicated in Figure 4a, infectious titers of rSVV-BAC− and wt SVV were similar over the course of the infection. In addition, the median plaque sizes of rSVV-BAC− (0.46 ± 0.06 mm) and wt SVV (0.48 ± 0.8 mm) were similar (Fig. 4b). The findings indicate that the infectious rSVV-BAC−, reconstituted using the SVV-BAC genetic system, replicates as efficiently as wt SVV in Vero cell culture.
Figure 4.
In vitro replication properties of rSVV compared to wt SVV. (a) Vero cell monolayers were infected with cell-free virus (500 pfu) and incubated for 8, 24, 48, or 72 hr. Viral titer for wt SVV, and SVV-BAC− was determined at each time point by plaque assay. (b) Viral plaque diameters of wt SVV, rSVVBAC−, and rSVVorf10del on CV-1 cell monolayers were measured by light microscopy at 72 hr postinfection. At least 20 plaques for each virus were measured.
Generation of a SVV ORF 10 mutant using the rSVV BAC system
To demonstrate the utility of the SVV BAC system for viral genome mutagenesis, an SVV ORF 10 deletion mutant (rSVVorf10del) was generated. While the function of the SVV ORF 10 protein is unknown, it is a homolog of the HSV-1 VP-16 transactivator, and the corresponding VZV ORF 10 encodes a virion tegument protein that stimulates VZV immediate early gene 62 gene expression [3, 18]. The transactivation domain of VZV ORF 10 maps to an amino terminal region including amino acids 5 – 79 [17].
A 225 bp deletion including amino acids 5 – 79 was generated within the 1,218 bp SVV ORF 10 of pSVV-BAC and confirmed by PCR and DNA sequence analysis (Fig. 3c). The pSVVorf10del-BACwas transfected into Vero cells, yielding viral plaques and cpe within five days to generate infectious rSVVorf10del -BAC. The mini-F-gfp vector sequences were subsequently removed by Cre recombinase as described above to generate rSVVorf10del. The ability to produce infectious rSVVorf10del demonstrated that the SVV gene 10 is non-essential for in vitro replication. However, further analysis indicated that ORF 10 is important for efficient viral replication in cell culture. Only low-titer cell-free virus was derived (<500 pfu/ml) and the median diameter of rSVVorf10del plaques (0.20 ± 0.04 mm) in CV-1 cell monolayers was significantly smaller than wt SVV plaques (0.48 ± 0.8 mm) or rSVV-BAC− (0.46 ± 0.06) at 72 hr postinfection (Figure 4b). The VZV ORF 10 is also dispensible for in vitro replication, although a VZV ORF 10 deletion mutant grows to similar titers as wt VZV in MeWo cell culture [3].
Generation of a self-excisable pSVV-BAC
A recent improvement for herpesvirus BAC genetic systems permits complete removal of vector sequences upon reconstitution in eukaryotic cells [23, 24]. A two-step Red-mediated recombination approach was used to insert a 1.6 kb inverted duplication of SVV DNA sequences into the mini-F-gfp vector of pSVV-BAC (Fig. 5). Upon transfection and passage in Vero cells, viral cpe with non-flourescent viral plaques was evident. Viral DNA was isolated and complete excision of mini-F-gfp sequences from the rSVV-F− genome was demonstrated by PCR and nucleotide sequence analysis (data not shown).
The cloning of the entire SVV genome as a BAC, propagation in E. coli, and reconstitution of infectious virus in Vero cells, provides an improved genetic approach to investigate the molecular basis of SVV replication and pathogenesis. The SVV-BAC genetic system effectively generates infectious virus with molecular properties and in vitro replication similar to wt SVV. A study confirming that SVV-BAC generated virus has similar pathogenicity as wt SVV in non-human primates is planned. The ability to efficiently introduce site-specific mutations within the SVV genome and generate SVV mutants will facilitate investigation of the role of specific SVV genes during acute infection, latency, and viral reactivation. These advances will provide new opportunities for study of simian varicella as a model to investigate VZV pathogenesis and antiviral strategies against varicella and herpes zoster, as well as approaches to control simian varicella outbreaks in facilities housing non-human primates.
Acknowledgments
This work was supported by Public Health Service Grant AI052373 of the National Institutes of Health. We thank Dr. Nikolaus Osterrieder (Cornell University) for the pEP-KanS plasmid, Dr. Greg Smith (Northwestern Univ.) for the GS1783 E. coli strain, Dr. Michael O’Connor (Univ. of Minnesota) for the pMBO131 plasmid, Dr. Ravi Mahalingam (Univ. of Colorado Health Sciences Center) for advice and critical review of the manuscript, and Luke Pearson for technical assistance.
References
- 1.Adler H, Messerle M, Wagner M, Koszinowski UH. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen WP, Felsenfeld AD, Wolf RH, Smetana HF. Recent studies on the isolation and characterization of delta herpesvirus. Lab Anim Sci. 1974;24:222–228. [PubMed] [Google Scholar]
- 3.Cohen JI, Seidel K. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J Virol. 1994;68:7850–7858. doi: 10.1128/jvi.68.12.7850-7858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrin LJ, Camerini-Otero RD. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher TM, III, Gray WL. Simian varicella virus: Characterization of virion and infected cell polypeptides and the antigenic cross-reactivity with varicella-zoster virus. J Gen Virol. 1992;73:1209–1215. doi: 10.1099/0022-1317-73-5-1209. [DOI] [PubMed] [Google Scholar]
- 6.Gray WL. Pathogenesis of simian varicella virus. J Med Virol. 2003;70:S4–S8. doi: 10.1002/jmv.10312. [DOI] [PubMed] [Google Scholar]
- 7.Gray WL. Simian varicella: a model for human varicella-zoster virus infections. Rev Med Virol. 2004;14:363–381. doi: 10.1002/rmv.437. [DOI] [PubMed] [Google Scholar]
- 8.Gray WL. Simian varicella in Old World monkeys. Comp Med. 2008;58:22–30. [PMC free article] [PubMed] [Google Scholar]
- 9.Gray WL, Davis K, Ou Y, Ashburn C, Ward TM. Simian varicella virus gene 61 encodes a viral transactivator but is non-essential for in vitro replication. Arch Virol. 2007;152:553–563. doi: 10.1007/s00705-006-0866-0. [DOI] [PubMed] [Google Scholar]
- 10.Gray WL, Mahalingam R. A cosmid-based system for inserting mutations and foreign genes into the simian varicella virus genome. J Virol Meth. 2005;130:89–94. doi: 10.1016/j.jviromet.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Gray WL, Oakes JE. Simian varicella virus DNA shares homology with human varicella-zoster virus DNA. Virology. 1984;136:241–246. doi: 10.1016/0042-6822(84)90263-0. [DOI] [PubMed] [Google Scholar]
- 12.Gray WL, Pumphrey CY, Ruyechan WT, Fletcher TM. The simian varicella virus and varicella zoster virus genomes are similar in size and structure. Virology. 1992;186:562–572. doi: 10.1016/0042-6822(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 13.Gray WL, Starnes B, White MW, Mahalingam R. The DNA sequence of the simian varicella virus genome. Virology. 2001;284:123–130. doi: 10.1006/viro.2001.0912. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infeccted mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Mahalingam R, Smith D, Wellish M, Wolf W, Dueland AN, Cohrs R, Soike K, Gilden D. Simian varicella virus DNA in dorsal root ganglia. Proc Natl Acad Sci USA. 1991;88:2750–2752. doi: 10.1073/pnas.88.7.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahalingam R, Traina-Dorge V, Wellish M, Lorino R, Sanford R, Ribka EP, Alleman SJ, Brazeau E, Gilden DH. Simian varicella reactivation in cynomolgus monkeys. Virology. 2007;368:50–59. doi: 10.1016/j.virol.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Moriuchi H, Moriuchi M, Pichyangkura R, Triezenberg SJ, Straus SE, Cohen JI. Hydrophobic cluster analysis predicts an amino-terminal domain of varicella-zoster virus open reading frame 10 required for transcriptional activation. Proc Natl Acad Sci USA. 1995;92:9333–9337. doi: 10.1073/pnas.92.20.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriuchi H, Moriuchi M, Straus SE, Cohen JI. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J Virol. 1993;67:2739–2746. doi: 10.1128/jvi.67.5.2739-2746.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaike K, Mori Y, Gomi Y, Yoshii H, Takahashi M, Wagner M, Koszinowski U, Yamanishi K. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine. 2004;22:4069–4074. doi: 10.1016/j.vaccine.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 20.Ou Y, Gray WL. The simian varicella virus gene 28 and 29 promoters share a common USF binding site and are induced by IE62 transactivation. J Gen Virol. 2006;87:1501–1508. doi: 10.1099/vir.0.81645-0. [DOI] [PubMed] [Google Scholar]
- 21.Ou Y, Traina-Dorge V, Davis KA, Gray WL. Recombinant simian varicella vaccines induce immune responses to simian immunodeficiency virus (SIV) antigens in imunized vervet monkeys. Virology. 2007;364:291–300. doi: 10.1016/j.virol.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pumphrey CY, Gray WL. The genomes of simian varicella virus and varicella zoster virus are colinear. Virus Res. 1992;26:255–266. doi: 10.1016/0168-1702(92)90017-4. [DOI] [PubMed] [Google Scholar]
- 23.Tischer BK, Kaufer BB, Somer M, Wussow F, Arvin AM, Osterrieder N. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J Virol. 2007;81:13200–13208. doi: 10.1128/JVI.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–196. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 25.Ward TM, Traina-Dorge V, Davis KA, Gray WL. Recombinant simian varicella viruses expressing respiratory syncytial virus antigens are immunogenic. J Gen Virol. 2008;89:741–740. doi: 10.1099/vir.0.83453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward TM, Williams MV, Traina-Dorge V, Gray WL. The simian varicella virus uracil glycosylase and dUTPase genes are expressed in vivo, but are non-essential for replication in cell culture. Virus Res. 2009;142:78–84. doi: 10.1016/j.virusres.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshii H, Somboonthum P, Takahashi M, Yamanishi K, Mori Y. Cloning of full length genome of varicella-zoster virus vaccine strain into a bacterial artificial chromosome and reconstitution of infectious virus. Vaccine. 2007;25:5006–5012. doi: 10.1016/j.vaccine.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Huang Y, Zhu H. A highly efficient protocol of generating and analyzing VZV ORF deletion mutants based on a newly developed luciferase VZV BAC system. J Virol Meth. 2008;148:197–204. doi: 10.1016/j.jviromet.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]