Abstract
St. Louis encephalitis virus is a complex zoonoses. In 2005, 47 laboratory-confirmed and probable clinical cases of SLEV infection were reported in Córdoba, Argentina. Although the causes of 2005 outbreak remain unknown, they might be related not only to virological factors, but also to ecological and environmental conditions. We hypothesized that one of the factors for SLE reemergence in Córdoba, Argentina, was the introduction of a new SLEV genotype (SLEV genotype III), with no previous activity in the area. In order to evaluate this hypothesis we carried out a molecular characterization of SLEV detections from mosquitoes collected between 2001 and 2004 in Córdoba city. A total of 315 mosquito pools (11,002 individuals) including 12 mosquitoes species were analyzed. Overall, 20 pools (8 mosquitoes species) were positive for SLEV. During this study, genotypes II, V and VII were detected. No mosquito pool infected with genotype III was detected before the 2005 outbreak. Genotype V was found every year and in the 8 sampled sites. Genotypes II and VII showed limited temporal and spatial activities. We cannot dismiss the association of genotype II and V as etiological agents during the outbreak. However, the silent circulation of other SLEV strains in Córdoba city before the 2005 outbreak suggests that the introduction of genotype III was an important factor associated to this event. Not mutually exclusive, other factors such as changes in avian hosts and mosquitoes vectors communities, driven by climatic and environmental modifications, should also be taken into consideration in further studies.
Author Summary
The St. Louis encephalitis is a complex zoonoses in the New World. In South America (Argentina and Brazil), SLE is an emerging arbovirosis. SLEV reemerged in Argentina during 2002 and, since then, outbreaks have been reported in 2005, 2006, 2010 and 2011. During the 2005 outbreak two SLEV genotype III strains were isolated. Although the causes of the 2005 outbreak remain unknown, they might be related not only to virological factors, but also to changes in the structure and dynamics of vectors and/or avian amplifying hosts' populations and environmental conditions. We hypothesized that one of the factors for SLE reemergence in Córdoba, Argentina, was the introduction of a new SLEV genotype, with no previous activity in the area. No mosquitoes were detected infected with genotype III during this four-year study, even 10 months before the outbreak. The silent circulation of other SLEV strains in Córdoba city before the 2005 outbreak suggests that the introduction of genotype III was an important factor associated to this event. Not mutually exclusive, other factors such as changes in avian hosts and mosquitoes vectors communities, driven by climatic and environmental modifications, should also be taken into consideration in further studies.
Introduction
The St. Louis encephalitis (SLE), caused by the homonymous virus (SLEV, genus Flavivirus, family Flaviviridae), is a complex zoonoses in the New World [1]. In South America, SLE is an emerging arbovirosis, with febrile illness and encephalitis cases reported in Argentina and Brazil [2], [3]. SLEV reemerged in the central region of Argentina during 2002 [2] and, since then, outbreaks have been reported in Córdoba (2005) [4], Entre Rios (2006), Buenos Aires (2010) and San Juan provinces (2011) [5].
SLEV shows biological and molecular variability among strains isolated throughout its geographic distribution [6]–[8]. Based on complete Envelope gene sequencing, SLEV strains can be classified into 8 lineages or genotypes (I–VIII) [9]. According to a phylogeographic analyses, genotypes I and II are prevalent in the USA, while the others were found only in countries from Central and South America [9]–[11]. Exceptionally, genotype V strains were recently isolated in Florida [12]. SLEV strains circulating in Argentina were clustered with genotype III (79V-2533 –year 1978-, CbaAr-4005, CbaAr-4006 – year 2005-), V (78V-6507 – year 1978-), both isolated from Culex spp. Mosquitoes, and VII (CorAn-9124, CorAn-9275 – year 1966-), isolated from rodents [9], [13].
In USA, the transmission cycles of SLEV are maintained by Culex mosquito vector species (Culex quinquefasciatus, Cx. tarsalis and Cx. nigripalpus) and Columbiformes (Mourning doves - Zenaida macroura) and Passeriformes (House finches - Carpodacus mexicanus, House sparrows - Passer domesticus) bird host species. Humans and mammals represent a dead-end host for the virus. Although it is widely distributed in the American continent, its ecology is scarcely known outside the USA [1].
Available data in Argentina suggests that Cx. quinquefasciatus mosquito would act as a main vector, while Picui ground dove (Columbina picui) and Eared dove (Zenaida auriculata) would be important hosts in urban and rural environments [13]–[16]. However, an alternative transmitting rodent-mosquito cycle was postulated in Argentina for genotype VII SLEV strains (CorAn-9124, CorAn-9275), isolated from rodents [14].
SLEV reemerged in the central region of Argentina in 2002 [2]. In 2005, 47 laboratory-confirmed and probable clinical cases of SLEV infection, including nine fatalities, were reported in the central Córdoba Province [4]. During this outbreak two SLEV genotype III strains were isolated [13]. This was the first SLEV-induced encephalitis outbreak reported in South America. Although the causes of 2005 outbreak remain unknown, they might be related not only to virological factors, but also to changes in the structure and dynamics of vectors and/or avian amplifying hosts populations, and environmental conditions. We hypothesized that one of the factors for SLE reemergence in Córdoba, Argentina was the introduction of a new SLEV genotype, with no previous activity in the area. In order to evaluate this hypothesis we carried out a molecular characterization of SLEV detections from mosquitoes collected between 2001 and 2004 in Córdoba city.
Methods
Collection sites
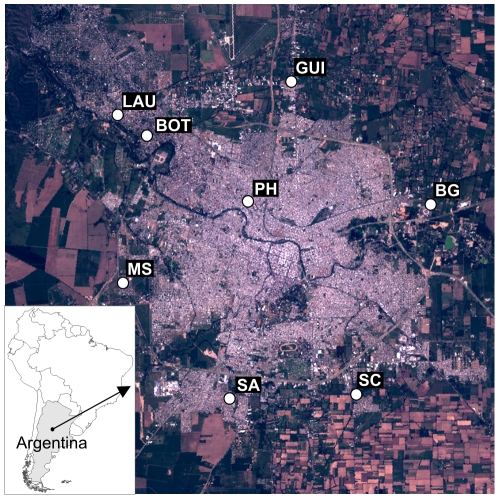
Mosquito collections were carried out during 2001 to 2004 in the city of Córdoba (31°24′30″S, 64°11′02″O) (Córdoba Province, Argentina), just before the 2005 SLE local outbreak. The city is situated at 450 m over sea level and its extension covers 576 km2, of which 37.2% is urbanized. The population is 1,330,023 inhabitants (http://200.51.91.231/censo2010/). The area belongs to the phytogeographic region of the Espinal, Chaqueño Domain, intensively modified by human activities (urbanization, agriculture, cattle, and industry). The city is surrounded by land crops (soy, fruit tree), industries and autochthonous shrubs patches isolated. The climate is mild without warm winters and with water deficit, in spite of its relatively high precipitation levels (750 and 800 mm), due to high evapotranspiration. A total of 8 collection sites were selected based on accessibility, owners' authorizations and previous evidence of mosquitoes abundance; most of them are located in peripheral areas of the city of Córdoba (Figure 1).
Figure 1. Sampling sites where mosquito collections were carried out during 2001–2004.
LAU: Libre del Ambiente University; BOT: Botanic Garden; GUI: Guiñazú; PH: Pediatric Hospital; BG: Bajo Grande; MS: Military School; SA: San Antonio; SC: San Carlos.
Sample collection
Mosquitoes were collected using CDC light traps (supplemented with dry ice) and chicken and rabbit baited can traps. Three light traps and 2 baited traps were set up and maintained in each site during 2 nights per season. Traps remained active during 18:00 until 09:00. The trapping schedule is detailed in Table 1. Collected mosquitoes were transported alive in refrigerated containers to the laboratory. Individuals were identified on a chill table and sorted by species, sex, collection date and site, and non-engorged and engorged status. Mosquitoe pools were homogenized using pestles and mortars in minimum essential medium (MEM) supplemented with 10% fetal bovine sera (FBS), 1% gentamicine and 1% Fungizone. Pools containing 1–25 mosquitoes were homogenized in 1 ml of MEM and those containing 25–50 mosquitoes were added 2 ml of MEM. Homogenates were centrifuged at 11,400 g during 30 min at 4°C for decontamination. Supernatants were stored in tubes at −70°C until utilization.
Table 1. Mosquito collection carried out in Córdoba during summer and fall season between 2001–2004.
| Year | Month | Site | Trap |
| 2001 | November | Bajo Grande | CDC light trap |
| December | Botanical Garden | Chicken can trap | |
| C. San Antonio | Rabbit can trap | ||
| Aviation School | |||
| Pediatric Hospital | |||
| Guiñazú | |||
| U. Libre del Ambiente | |||
| 2002 | January | Bajo Grande | CDC light trap |
| February | Botanical Garden | Chicken can trap | |
| March | C. San Antonio | Rabbit can trap | |
| April | C. San Carlos | ||
| November | Aviation School | ||
| December | Pediatric Hospital | ||
| Guiñazú | |||
| U. Libre del Ambiente | |||
| 2003 | January | Bajo Grande | CDC light trap |
| February | Botanical Garden | Chicken can trap | |
| March | C. San Carlos | ||
| April | Pediatric Hospital | ||
| Guiñazú | |||
| 2004 | January | Bajo Grande | CDC light trap |
| February | Pediatric Hospital | ||
| March | C. San Carlos | ||
| April | Guiñazú |
Flavivirus and SLEV molecular detection and sequencing analyses
RT-Nested PCR
Viral RNA was extracted from 150 µl of the mosquito pool homogenates using 750 µl of Trizol Reagent (Invitrogen BRL, Life Technologies, Rockville, MD), following the manufacturer indications. Reverse transcription was carried out with Moloney Murine Leukemia virus reverse transcriptase (MMLV, Promega, Madison, WI, USA) with Random Hexamer Primers (Promega, Madison, WI, USA) following the manufacturer indications. The cDNAs obtained from pools were used to amplified a 143 bp fragment from NS5 Flavivirus protein and 234 pb fragment from the Envelope gene of SLEV using Green Go Taq Reagents (Promega, Madison, WI, USA), as described previously [17], [18]. Positive RT-Nested SLEV specific PCR mosquito homogenates were subjected to viral isolation attempts. VERO (Green monkey kidney cells) cells monolayer were inoculated with 100 µl mosquito homogenates and checked daily for cytopathic effect.
Sequencing and phylogenetic analyses
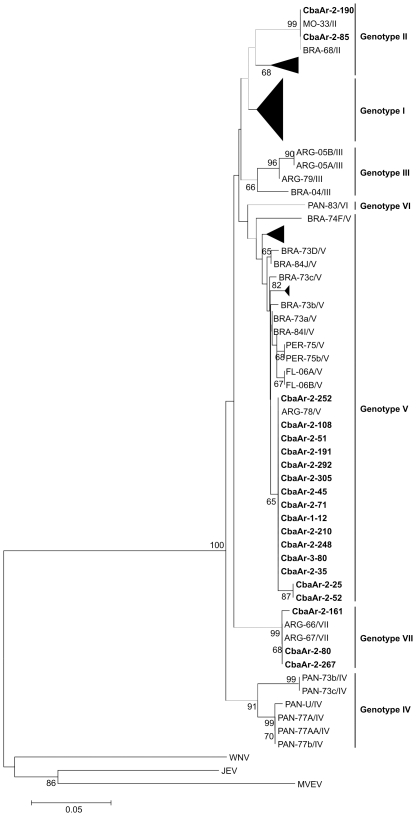
All PCR products obtained from mosquito pools were purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA, USA), submitted to direct nucleotide sequencing reaction using Big Dye Terminator v3.1 Cycle kit (Applied Biosystem), and analyzed in an automatic sequencer. The obtained sequences were submitted to BLASTn 2.2.19 -Basic Local Alignment Search Tool- (http://blast.ncbi.nlm.nih.gov) to detect homologies among Flaviviruses and SLEV sequences available in the data base. A phylogenetic analysis of the SLEV alignment including 134 partial Envelope gene sequences was performed using neighbor joining (NJ) in MEGA v5 [19]; the proportion of differences between the sequences was considered as the distance measure (p distance). The NJ analysis was bootstrapped 10,000 replications.
Viral isolation attempts
RT PCR SLEV positive mosquito pools were subjected to viral isolation. One hundred µl of mosquito homogenate were inoculated onto 24 hs VERO cells monolayers, incubated for 60 min at 37°C, and observed daily for cythopatic effect. After 7 day post inoculation, blind passages were realized.
Results
A total of 315 mosquito pools (11,002 individuals) including 12 mosquitoes species were analyzed. Overall, 20 pools (8 mosquitoes species) were positive for SLEV (Table 2). The BLASTn search and the phylogenetic analyses were performed using a 212 bp fragment of the Envelope protein. Viral isolation attempts were unsuccessful due to probable multiple freeze/thaws cycles during the process of mosquito homogenates.
Table 2. SLEV RT-PCR positive mosquito pools collected in Córdoba city, Argentina between 2001 and 2004.
| Pool | Especie | GenBank | Month/Year | Site | Genotype |
| CbaAr 1-12 | Cx. quinquefasciatus | FJ361863 | 02/02 | Bajo Grande | V |
| CbaAr 2-45 | Ae. scapularis | FJ361867 | 11/02 | Botanic Garden | V |
| CbaAr 2-25 | Cx. quinquefasciatus | FJ361872 | 11/02 | Bajo Grande | V |
| CbaAr 2-51 | An. albitarsis | FJ361877 | 12/02 | Bajo Grande | V |
| CbaAr 2-71 | Cx. apicinus | FJ361861 | 12/02 | Pediatric Hosp. | V |
| CbaAr 2-80 | Cx. interfor | FJ361880 | 12/02 | C. San Carlos | VII |
| CbaAr 2-52 | Cx. quinquefasciatus | FJ361873 | 12/02 | Bajo Grande | V |
| CbaAr 2-35 | Ps. ferox | FJ361876 | 12/02 | Guiñazú | V |
| CbaAr 2-108 | Ae. aegypti | FJ361869 | 01/03 | Botanic Garden | V |
| CbaAr 2-85 | Ae. albifasciatus | FJ361881 | 01/03 | Bajo Grande | II |
| CbaAr 2-161 | Ae. scapularis | FJ361878 | 02/03 | Bajo Grande | VII |
| CbaAr 2-252 | Cx. quinquefasciatus | FJ361862 | 02/03 | Bajo Grande | V |
| CbaAr 2-248 | Cx. quinquefasciatus | FJ361866 | 02/03 | Pediatric Hosp. | V |
| CbaAr 2-191 | Ae. albifasciatus | FJ361860 | 03/03 | Guiñazú | V |
| CbaAr 2-267 | Ae. albifasciatus | FJ361879 | 03/03 | Guiñazú | VII |
| CbaAr 2-210 | Cx. interfor | FJ361864 | 03/03 | Pediatric Hosp. | V |
| CbaAr 2-190 | Cx. quinquefasciatus | FJ361875 | 03/03 | Guiñazú | II |
| CbaAr 2-292 | Ae. albifasciatus | FJ361874 | 04/03 | C. San Carlos | V |
| CbaAr 2-305 | Ae. albifasciatus | FJ361865 | 04/03 | C. San Carlos | V |
| CbaAr 3-80 | Ae. scapularis | FJ361868 | 02/04 | C. San Carlos | V |
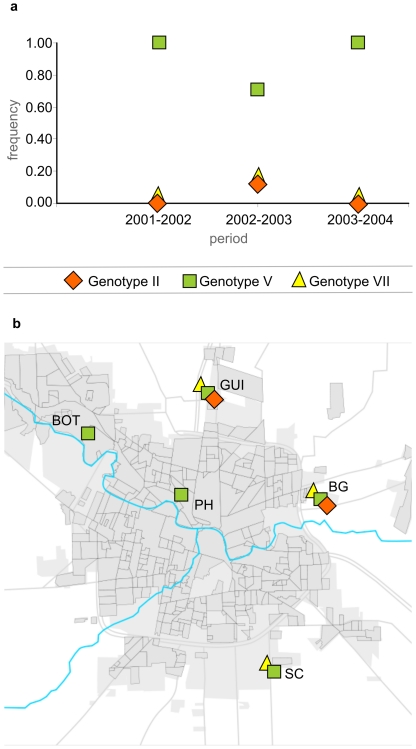
Based on the classification proposed by Kramer and Chandler [9], three different SLEV genotypes (II, V, and VII) were detected in Córdoba city before the encephalitis human outbreak associated to SLEV genotype III (Figure 2). Genotype V was detected in 5 of the 8 collection sites during the 4 years (Figure 3) and it was found in the 8 infected mosquito species collected in our study (Aedes aegypti, Anopheles albitarsis, Ae. albifasciatus, Ae. scapularis, Culex apicinus, Cx. interfor, Cx. quinquefasciatus, Psoropohora ferox) (Table 2). Sequences belonging to genotype V share a 100% of homology with SLEV strain 78V6507 (AF205481). This genotype was previously isolated from Cx. quinquefasciatus mosquitoes in Santa Fe province in 1978 [20].
Figure 2. SLEV genotype activity in mosquitoes collected in Córdoba city between 2001–2004.
a) SLEV genotypes temporal distribution. b) SLEV genotypes geographical distribution. LAU: Libre del Ambiente University; BOT: Botanic Garden; GUI: Guiñazú; PH: Pediatric Hospital; BG: Bajo Grande; MS: Military School; SA: San Antonio; SC: San Carlos. White circles represent sampled sites without SLEV activity detected during the study period.
Figure 3. Phylogenetic Neighbor Joining analysis of SLEV.
In bold strains detected in mosquitoes during our study in Córdoba, Argentina. Percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10 000 replicates) is shown next to the branches. The scale bar indicates substitutions per site.
Genotypes II and VII were detected only sporadically during the study. However, this is the first report of SLEV genotype II in Argentina. The nucleotide sequence corresponding to this genotype share 100% homology with SLEV strains SPAn-11916 (EF117302), was isolated in 1968 from rodents collected in Sao Paulo (Brazil), and Parton-MSI-7 (EF158070) was isolated from ill humans in Missouri (USA) in 1933. Only 2 positive pools (Ae. albifasciatus and Cx. quinquefasciatus mosquitoes) collected in Bajo Grande and Guiñazú in 2003 proved to be infected with this genotype in our study (Figure 3). Sequences clustering with Genotype VII have a 100% of homology with SLEV strains CorAn-9124 (EF158063) and CorAn-9275 (EF158068) isolated in 1966. Although genotype VII has been isolated from small rodents (Calomys musculinus, Mus musculus) in the province of Córdoba [14], there was no previous report of its activity neither in mosquitoes nor in the capital city.; it was detected in 3 sites (Guiñazú, Bajo Grande, San Carlos) during 2002 and 2003 (Table 2, Figure 3). Mosquitoes infected with genotype VII were identified as Ae. albifasciatus, Ae. scapularis and Cx. interfor.
Discussion
Three SLEV genotypes circulated simultaneously in Córdoba city between 2001 and 2004. None of these genotypes are related to the SLEV Genotype III CbaAr-4005 strain isolated during the encephalitis human outbreak that occurred later in 2005 [13].
The most prevalent variant was genotype V, genetically related to SLEV 78V-6507 strain isolated in Santa Fe province in 1978 [20]. Our results confirm the endemicity of SLEV in Córdoba city during 2001–2005. The sustained activity of genotype V in Bajo Grande, Botanical Garden, Guiñazú, Pediatric Hospital, and San Carlos between 2001 and 2004 (Figure 1) indicates local genotype persistence and probable overwintering of SLEV activity in this temperate area. Culex mosquitoe abundance decreases drastically during winter, so vector transmission to vertebrates would not be maintained during this season. Supporting endemicity of SLEV, Flores et al. [21] detected vertical transmission of SLEV genotype V in local Culex mosquito populations under laboratory conditions. We believe that this mechanism would satisfactory explain the maintenance of some viral variants until the next favorable season for mosquito vector proliferation.
The presence of a predominant genotype could be evidence for a higher viremogenic capacity of this strain in avian hosts. Higher viremias would enhance the transmission by mosquitoes, increasing viral circulation and expanding the distribution of this variant. In fact, SLEV genotype V (strain 78V-6507) developed higher viremias in birds than genotype III and VII strains [7]. SLEV not only requires the ability to infect avian hosts with highly enough viremias to be deemed infectious to mosquitoes, but it also needs to be able to disseminate the mosquito mid-gut and enter the salivary glands. This ecological and evolutionary feature should not be unattended, indeed.
In a different way, genotypes II and VII showed limited temporal and spatial activities (Figure 1). Powers et al. [22] pointed out that arboviruses maintained by a rodents-mosquitoes cycle show limited and constrained geographic distributions, which agree with the limited dispersion capacity observed in rodents. In fact, Genotype VII SLEV strains (CorAn-9124, CorAn-9275) were isolated only from small rodents in Córdoba province [14], which suggests that mammals are the actual hosts for these strains. Moreover, the 3 mosquito species (Ae. albifasciatus, Ae. scapularis and Cx. interfor) infected with Genotype VII frequently feed on mammals hosts [20], [23]–[25]. Our data support the hypothesis that some SLEV strains are being maintained through alternative rodents-mosquitoes transmission cycles [14].
Analyzing temporal and geographic patterns of SLEV in Texas by molecular techniques, Chandler et al. [26] detected the presence of dominant and limited strains fluctuating in space and time. This activity pattern could be the result of multiple intermittent virus introductions by birds from neighboring regions [27]. Other flavivirus such as Japanese encephalitis (JEV) and West Nile virus (WNV) showed similar dynamics [28], [29]. These dynamics are characterized by the introduction/persistence of certain genotypes and the presence/absence of clinical cases in the study area. Another possibility is that SLEV genotype III had been silently present in Córdoba prior to 2001. Although a Dengue virus has different ecological requirements compared with JEV, SLEV and WNV, it has been demonstrated that some serotypes can remain silent for many years causing periodic epidemics [30].
During 2005, a human encephalitis outbreak was caused by SLEV in Córdoba city [4], and two genotype III strains were isolated [13]. Molecular characterization determined that both variants are closely related to SLEV strain 79V-2533, isolated 27 years ago in Santa Fe province [13]. This evidence supports the hypothesis that the introduction of a more virulent genotype could have caused the mentioned outbreak. In a recent study, Diaz et al. [31] compared epidemic (CbaAr-4005) and non-epidemic (79V-2533) genotype III SLEV strains. The epidemic variant produced higher viremias in House sparrows (Passer domesticus) than the non-epidemic strain. According to this, the epidemic strain (CbaAr-4005) appears to broaden the number of avian species that are likely to be competent amplifying hosts relative to the non-epidemic 79V-2533 strain [31].
The introduction of new strains and the extension of their geographic distributions are factors that can cause the emergence and reemergence of flaviviruses in different regions [32]. For example, the introduction, spread and establishment of WNV in America and Japanese Encephalitis Virus in Australasia and the annual introduction of SLEV strains in the USA (California state) [27]. During our four-year study a non-genotype III SLEV strain was detected. One year prior to the outbreak a total of 2,093 mosquitoes were analyzed and only one mosquito pool was positive (genotype V) (Table 1). The results here exposed suggest that genotype III was introduced in Córdoba city a few months before the outbreak and it could be one of the factors contributing to the outbreak.
SLEV strains show biological and molecular variability [6]–[9]. Strains belonging to genotypes I, II, III, and V showed pathogenicity in mice and Rhesus monkeys [6]. During SLEV human encephalitis outbreaks in the USA, genotypes I and II were frequently isolated [9]. Spinsanti et al. [2] confirmed one human encephalitis case in Córdoba concomitant with our detection of Genotype V in the same area during the same year (Pediatric Hospital, 2002 – Figure 1). Although genotype III was detected in mosquitoes collected around the encephalitis human cases during the 2005 outbreak, we cannot dismiss the association of genotype II and V as etiological agents during the outbreak. However, the silent circulation of other SLEV strains in Córdoba city before the 2005 outbreak suggests that the introduction of genotype III was an important factor associated to this event. Not mutually exclusive, other factors such as changes in avian hosts and mosquitoes vectors communities, driven by climatic and environmental modifications, should also be taken into consideration in further studies.
Acknowledgments
The authors thank Analía Boris, Agustín Zarco, Noelia Villafañes, Andrea Cortes, Cecilia Castilla, Sabino Ortiz Bergia and Romina Reinoso for technical support in mosquito collection, and Adrian Farias and Viviana Re for molecular detection.
Footnotes
The authors have declared that no competing interests exist.
This study was supported by grants from FONCYT-PICT 38060 (Fondo Nacional de Ciencia y Técnica – Ministerio de Ciencia y Tecnología de Argentina), CONICET PIP No: 11220090100882 (Consejo Nacional de Investigaciones Científicas y Técnicas), Agencia Córdoba Ciencia: 0279-005490/2006. Resol. 1210/2007, SECYT-UNC (Secretaría de Ciencia y Técnica – Universidad Nacional de Córdoba) and from ISCIII PI07/1308 (Instituto de Salud Carlos III), RD06/0021 (RICET–Red de Investigación Cooperativa en Enfermedades Tropicales) and DGSP/ISCIII (Dirección General de Salud Pública del Ministerio de Sanidad, Política Social e Igualdad, Spain), with the technical collaboration of the RIVE – CYTED laboratories (Red Iberoamericana de Virosis Emergentes). L.A.D. is an assistant research investigator with CONICET, Argentina. G.A.L. is a recipient of a postdoctoral scholarship from CONICET, Argentina. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Reisen W. Epidemiology of St. Louis Encephalitis virus. Adv Virus Res. 2003;61:139–183. doi: 10.1016/s0065-3527(03)61004-3. [DOI] [PubMed] [Google Scholar]
- 2.Spinsanti LI, Basquiera A, Bulacio S, Somale V, Kim SC, et al. St. Louis Encephalitis in Argentina: the First Case Reported in the Last Seventeen Years. Emerg Infect Dis. 2003;9:271–273. doi: 10.3201/eid0902.020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocco IM, Santos CL, Bisordi I, Petrella SM, Pereira LE, et al. St. Louis encephalitis virus: first isolation from a human in São Paulo State, Brazil. Rev Inst Med Trop Sao Paulo. 2005;47:281–285. doi: 10.1590/s0036-46652005000500008. [DOI] [PubMed] [Google Scholar]
- 4.Spinsanti LI, Díaz LA, Glatstein N, Arselán S, Morales MA, et al. Human outbreak of St. Louis encephalitis detected in Argentina, 2005. J Clin Virol. 2008;42:27–33. doi: 10.1016/j.jcv.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Seijo A, Morales A, Poustis G, Romer Y, Efron E, et al. Outbreak of St. Louis encephalitis in the Metropolitan Buenos Aires Area. Medicina (B Aires) 2011;71:211–217. [PubMed] [Google Scholar]
- 6.Monath TP, Cropp CB, Bowen GS, Kemp GE, Mitchell CJ, et al. Variation in virulence for mice and rhesus monkeys among St. Louis encephalitis virus strains of different origin. Am J Trop Med Hyg. 1980;29:948–962. doi: 10.4269/ajtmh.1980.29.948. [DOI] [PubMed] [Google Scholar]
- 7.Bowen GS, Monath TP, Kemp GE, Kerschner JH, Kirk LK. Geographic variation among St. Louis encephalitis virus strains in the viremic responses of avian hosts. Am J Trop Med Hyg. 1980;29:1411–1419. doi: 10.4269/ajtmh.1980.29.1411. [DOI] [PubMed] [Google Scholar]
- 8.Trent DW, Monath TP, Bowen GS, Vorndam AV, Cropp BC, et al. Variation among strains of St. Louis encephalitis virus: basis for a genetic, pathogenetic and epidemiological classification. Annals NY Acad Sci. 1980;354:219–237. doi: 10.1111/j.1749-6632.1980.tb27969.x. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues SG, Nunes MR, Casseb SM, Prazeres AS, Rodrigues DS, et al. Molecular epidemiology of Saint Louis encephalitis virus in the Brazilian Amazon: genetic divergence and dispersal. J Gen Virol. 2010;91:2420–2427. doi: 10.1099/vir.0.019117-0. [DOI] [PubMed] [Google Scholar]
- 10.Kramer LD, Chandler LJ. Phylogenetic analysis of the Envelope gene of St. Louis encephalitis virus. Arch Virol. 2001;146:2341–2355. doi: 10.1007/s007050170007. [DOI] [PubMed] [Google Scholar]
- 11.Auguste AJ, Pybus OG, Carrington CV. Evolution and dispersal of St. Louis encephalitis virus in the Americas. Infect Genet Evol. 2009;9:709–715. doi: 10.1016/j.meegid.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Ottendorfer CL, Ambrose JH, White GS, Unnasch TR, Stark LM. Isolation of genotype V St. Louis encephalitis virus in Florida. Emerg Infect Dis. 2009;15:604–606. doi: 10.3201/eid1504.081094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz LA, Ré V, Almirón WR, Farías A, Vázquez A, et al. Genotype III Saint Louis encephalitis virus outbreak, Argentina, 2005. Emerg Infect Dis. 2006;12:1752–1754. doi: 10.3201/eid1211.060486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabattini MS, Avilés G, Monath TP. Historical, epidemiological and ecological aspects of arbovirus in Argentina: Flaviviridae, Bunyaviridae and Rhabdoviridae. In: Travassos da Rosa APA, Vasconcelos PFC, Travassos da Rosa JFS, editors. An overview of arbovirology in Brazil and neighboring countries. Belem (Brazil): Instituto Evandro Chagas; 1998. pp. 113–134. [Google Scholar]
- 15.Diaz LA, Occelli M, Almeida FL, Almirón WR, Contigiani MS. Eared dove (Zenaida auriculata, Columbidae) as host for St. Louis encephalitis virus (Flaviviridae, Flavivirus). Vector Borne Zoonotic Dis. 2008;8:277–282. doi: 10.1089/vbz.2007.0168. [DOI] [PubMed] [Google Scholar]
- 16.Diaz LA. Patrones de actividad y estacionalidad del virus St. Louis encephalitis en Córdoba, Argentina. Doctoral Thesis. Faculty of Exact, Physics and Natural Sciences. 2009. 193 National University of Córdoba.
- 17.Sánchez-Seco MP, Rosario D, Domingo C, Hernández L, Valdés K, et al. Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Methods. 2005;126:101–109. doi: 10.1016/j.jviromet.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Ré V, Spinsanti L, Farías A, Díaz A, Vázquez A, et al. Reliable detection of St. Louis encephalitis virus by RT-nested PCR. Enferm Infecc Microbiol Clin. 2008;26:10–15. doi: 10.1157/13114389. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr121. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell CJ, Monath TP, Sabattini MS, Cropp CB, Daffner JF, et al. Arbovirus investigations in Argentina, 1977–1980. II. Arthropod collections and virus isolations from argentine mosquitoes. Am J Trop Med Hyg. 1985;34:945–955. [PubMed] [Google Scholar]
- 21.Flores FS, Díaz LA, Batallán GP, Almirón WR, Contigiani MS. Vertical transmission of St. Louis encephalitis virus in Culex quinquefasciatus (Diptera: Culicidae) in Córdoba, Argentina. Vector Borne Zoonotic Dis. 2010;10:999–1002. doi: 10.1089/vbz.2009.0136. [DOI] [PubMed] [Google Scholar]
- 22.Powers AM, Aguilar PV, Chandler LJ, Brault AC, Meakins TA, et al. Genetic relationships among Mayaro and Una viruses suggest distinct patterns of transmission. Am J Trop Med Hyg. 2006;75:461–469. [PubMed] [Google Scholar]
- 23.Mitchell CJ, Darsie RF, Jr, Monath TP, Sabattini MS, Daffner J. The use of an animal-baited net trap for collecting mosquitoes during western equine encephalitis investigations in Argentina. J Am Mosq Control Assoc. 1985;1:43–47. [PubMed] [Google Scholar]
- 24.Mitchell CJ, Monath TP, Sabattini MS, Christensen HA, Darsie RF, Jr, et al. Host-feeding patterns of Argentine mosquitoes collected during and after an epizootic of Western Equine encephalitis. J Med Entomol. 1987;24:260–267. doi: 10.1093/jmedent/24.2.260. [DOI] [PubMed] [Google Scholar]
- 25.Almirón WR, Brewer ME. Preferencia de hospedadores de Culicidae (Diptera) recolectados en el centro de Argentina. Rev Saude Pública. 1995;29:108–114. doi: 10.1590/s0034-89101995000200004. [DOI] [PubMed] [Google Scholar]
- 26.Chandler LJ, Parsons R, Randle Y. Multiple genotypes of St. Louis encephalitis virus (Flaviviridae: Flavivirus) circulate in Harris County, Texas. Am J Trop Med Hyg. 2001;64:12–19. doi: 10.4269/ajtmh.2001.64.1.11425155. [DOI] [PubMed] [Google Scholar]
- 27.Kramer LD, Presser SB, Hardy JL, Jackson AO. Genotypic and phenotypic variation of selected St. Louis encephalitis viral strains isolated in California. Am J Trop Med Hyg. 1997;57:222–229. doi: 10.4269/ajtmh.1997.57.222. [DOI] [PubMed] [Google Scholar]
- 28.Sotelo E, Fernández-Pinero J, Llorente F, Vázquez A, Moreno A, et al. Phylogenetic relationships of Western Mediterranean West Nile virus strains (1996–2010) using full-length genome sequences: single or multiple introductions? J Gen Virol. 2011;92:2512–2522. doi: 10.1099/vir.0.033829-0. [DOI] [PubMed] [Google Scholar]
- 29.Takhampunya R, Kim HC, Tippayachai B, Kengluecha A, Klein TA, et al. Emergence of Japanese encephalitis virus genotype V in the Republic of Korea. Virol J. 2011;8:449–456. doi: 10.1186/1743-422X-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dash PK, Parida MM, Saxena P, Kumar M, Rai A, et al. Emergence and continued circulation of dengue-2 (genotype IV) virus strains in northern India. J Med Virol. 2004;74:314–322. doi: 10.1002/jmv.20166. [DOI] [PubMed] [Google Scholar]
- 31.Díaz LA, Nemeth NM, Bowen RA, Almirón WR, Contigiani MS. Comparison of Argentinean Saint Louis Encephalitis Virus Non-Epidemic and Epidemic Strain Infections in an Avian Model. PLoS Negl Trop Dis. 2011;5(5):e1177. doi: 10.1371/journal.pntd.0001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and Dengue viruses. Nat Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]