Abstract
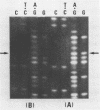
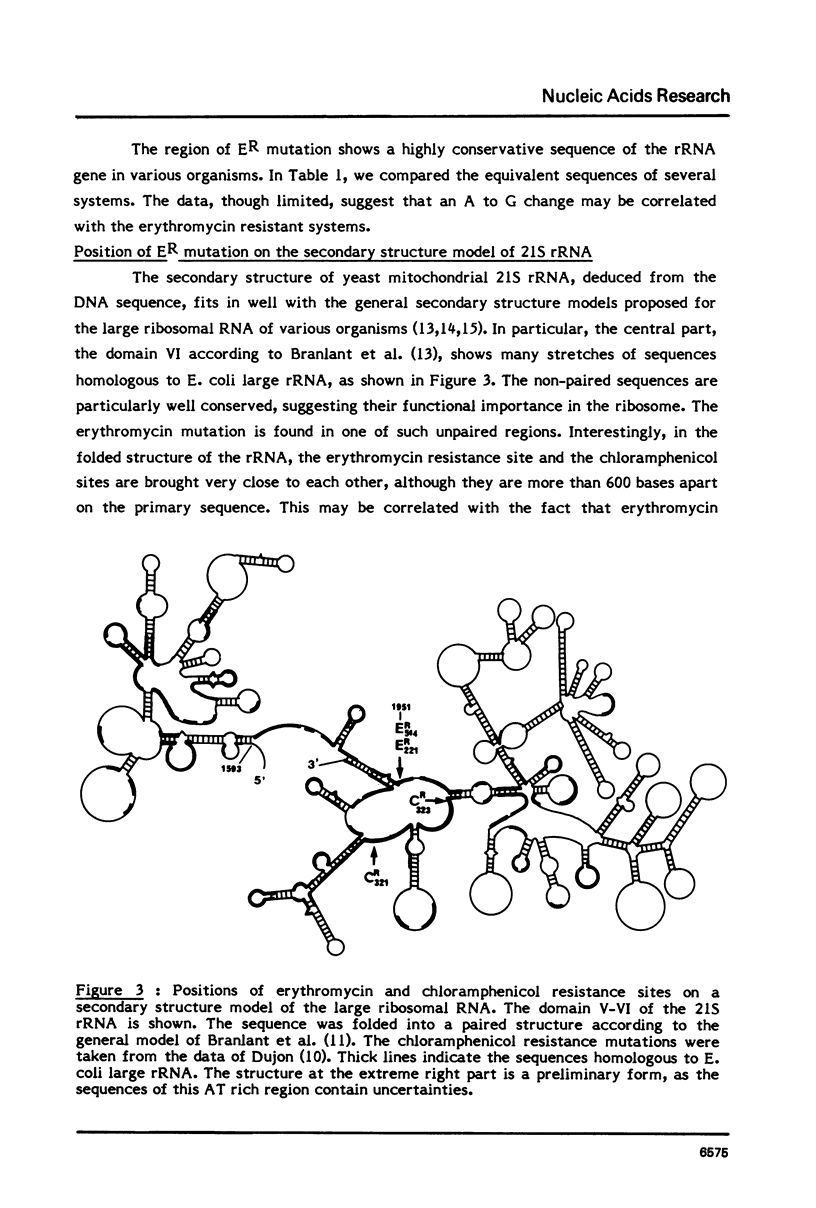
Two independent erythromycin resistance mutations, ER514 and ER221, have been identified in the mitochondrial gene coding for the 21S ribosomal RNA. The two mutations were found to be identical, corresponding to a A to G transition at the nucleotide position 1951 of the ribosomal RNA gene. In the secondary structure model of the ribosomal RNA, the ER resistance site is found at the proximity of the chloramphenicol resistance sites located about 500 bases downstream.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M., Fay G., Fukuhara H. Temperature-sensitive respiratory-deficient mitochondrial mutations: isolation and genetic mapping. Mol Gen Genet. 1977 Apr 29;152(3):295–305. doi: 10.1007/BF00693083. [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. Mitochondrial and nuclear mutations that affect the biogenesis of the mitochondrial ribosomes of yeast. I. Genetics. Mol Gen Genet. 1979;177(1):39–46. doi: 10.1007/BF00267251. [DOI] [PubMed] [Google Scholar]
- Borst P., Grivell L. A. Mitochondrial ribosomes. FEBS Lett. 1971 Feb 19;13(2):73–88. doi: 10.1016/0014-5793(71)80204-1. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Faye G., Fukuhara H., Grandchamp C., Lazowska J., Michel F., Casey J., Getz G. S., Locker J., Rabinowitz M., Bolotin-Fukuhara M. Mitochondrial nucleic acids in the petite colonie mutants: deletions and repetition of genes. Biochimie. 1973;55(6):779–792. doi: 10.1016/s0300-9084(73)80030-6. [DOI] [PubMed] [Google Scholar]
- Fukuhara H., Bolotin-Fukuhara M. Deletion mapping of mitochondrial transfer RNA genes in Saccharomyces cerevisiae by means of cytoplasmic petite mutants. Mol Gen Genet. 1976 Apr 23;145(1):7–17. doi: 10.1007/BF00331551. [DOI] [PubMed] [Google Scholar]
- Fukuhara H., Rabinowitz M. Yeast petite mutants for DNA gene amplification. Methods Enzymol. 1979;56:154–163. doi: 10.1016/0076-6879(79)56017-0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A., Netter P., Borst P., Slonimski P. P. Mitochondrial antibiotic resistance in yeast: ribosomal mutants resistant to chloramphenicol, erythromycin and spiramycin. Biochim Biophys Acta. 1973 Jun 23;312(2):358–367. doi: 10.1016/0005-2787(73)90380-8. [DOI] [PubMed] [Google Scholar]
- Gunatilleke I. A., Scazzocchio C., Arst H. N., Jr Cytoplasmic and nuclear mutations to chloramphenicol resistance in Aspergillus nidulans. Mol Gen Genet. 1975;137(3):269–276. doi: 10.1007/BF00333022. [DOI] [PubMed] [Google Scholar]
- Köchel H. G., Küntzel H. Mitochondrial L-rRNA from Aspergillus nidulans: potential secondary structure and evolution. Nucleic Acids Res. 1982 Aug 11;10(15):4795–4801. doi: 10.1093/nar/10.15.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morimoto R., Lewin A., Hsu H. J., Rabinowitz M., Fukuhara H. Restriction endonuclease analysis of mitochondrial DNA from grande and genetically characterized cytoplasmic petite clones of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3868–3872. doi: 10.1073/pnas.72.10.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netter P., Petrochilo E., Slonimski P. P., Bolotin-Fukuhara M., Coen D., Deutsch J., Dujon B. Mitochondrial genetics. VII. Allelism and mapping studies of ribosomal mutants resistant to chloramphenicol, erythromycin and spiramycin in S. cerevisiae. Genetics. 1974 Dec;78(4):1063–1100. doi: 10.1093/genetics/78.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Nature of an inserted sequence in the mitochondrial gene coding for the 15S ribosomal RNA of yeast. Nucleic Acids Res. 1982 Mar 11;10(5):1625–1633. doi: 10.1093/nar/10.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. The complete nucleotide sequence of a 23-S rRNA gene from tobacco chloroplasts. Eur J Biochem. 1982 May;124(1):13–19. doi: 10.1111/j.1432-1033.1982.tb05901.x. [DOI] [PubMed] [Google Scholar]
- Thomas D. Y., Wilkie D. Recombination of mitochondrial drug-resistance factors in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1968 Feb 26;30(4):368–372. doi: 10.1016/0006-291x(68)90753-5. [DOI] [PubMed] [Google Scholar]
- Vazquez D. Binding of chloramphenicol to ribosomes. The effect of a number of antibiotics. Biochim Biophys Acta. 1966 Feb 21;114(2):277–288. doi: 10.1016/0005-2787(66)90309-1. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]