Abstract
Background
CD69 is expressed in several hemopoietic cells and is an early activation marker in chronic lymphocytic leukemia. Chronic lymphocytic leukemia is a clinically heterogeneous disease which needs novel prognostic parameters which can be easily and efficiently managed.
Design and Methods
We investigated CD69 by flow cytometry in a series of 417 patients affected by chronic lymphocytic leukemia and compared this to other biological and clinical prognosticators.
Results
CD69 was associated with Rai stages (P=0.00002), β2-microglobulin (P=0.0005) and soluble CD23 (P<0.0001). CD69 and ZAP-70 (P=0.018) or CD38 (P=0.00015) or immunoglobulin variable heavy chain gene mutations (P=0.0005) were also significantly correlated. Clinically, CD69 positive chronic lymphocytic leukemias received chemotherapy more frequently (74%; P<0.0001), and presented a shorter duration of response after fludarabine plus rituximab (P=0.010) as well as shorter progression free survival and overall survival (P<0.0001). CD69 demonstrated true additive prognostic properties, since the CD69+ plus ZAP-70+ or CD38+ or immunoglobulin variable heavy chain gene unmutated patients had the worst progression free survival and overall survival (P<0.0001). Interestingly, low CD69 expression was necessary to correctly prognosticate the longer progression free survival of patients with a low tumor burden of β2-microglobulin (P=0.002), of soluble CD23 (P=0.020), or of Rai stages 0-I (P=0.005). CD69 was confirmed to be an independent prognostic factor in multivariate analysis of progression free survival (P=0.017) and overall survival (P=0.039).
Conclusions
Our data indicate that CD69 is significantly correlated with poor clinical and biological prognostic factors and is confirmed to be an independent disease prognosticator. This supports its introduction in a routine laboratory assessment and, possibly, in a prognostic scoring system for chronic lymphocytic leukemia, after an adequate standardization process.
Keywords: chronic lymphocytic leukemia, immunophenotyping, prognosis
Introduction
The clinical course of patients with B-cell chronic lymphocytic leukemia (CLL) is quite variable1 and the two major clinical staging systems are unable to prospectively discriminate an indolent or aggressive course within the low and intermediate risk categories.2,3 For this reason, several biological parameters4–9 have been added to the staging systems to differentiate prognostic subsets. Recently, several publications reported the prognostic significance of specific immunoglobulin variable heavy chain (IGHV) gene features in CLL. 10–15 However, since IGHV gene sequencing remains a demanding technique, many studies focused on the identification of alternative markers with similar prognostic value the expression of which is easier to investigate, such as CD38 and T-cell specific zeta-associated protein-70 (ZAP-70).16–22
Literature data confirm that leukemic CLL cells bear the surface membrane phenotype of activated and antigen-experienced B lymphocytes with the overexpression of the activation markers CD23, CD25, CD69 and CD71.23 CD69 identifies a type II integral membrane protein with a single transmembrane domain belonging to the C-type lectin family of surface receptors 24–26 and is expressed very quickly (within 4 h) in several hemopoietic cells upon triggering.23 One preliminary study suggested that CD69 could be a prognostic factor for overall survival (OS) in CLL,27 but its prognostic independency was not evaluated.
By taking advantage of a wide cohort of CLL patients, the aims of the present study were: (i) to propose a reproducible flow cytometric method for testing CD69 in peripheral blood (PB) of CLL samples; (ii) to investigate the relationship between CD69 and other biological prognosticators or markers of tumor burden; and finally, (iii) to test the independent prognostic value of CD69 for progression free survival (PFS) and OS.
Design and Methods
Patients
Approval for this study was obtained from the Institutional Review Board of the Centro di Riferimento Oncologico (IRCCS) of Aviano (PN), Italy. Informed consent was provided according to the Declaration of Helsinki. Four hundred and seventeen consecutive B-CLL patients were enrolled from 1990 to 2009. There were 239 men and 178 women with a median age of 66 years (range 33–89) at the time of diagnosis. Fresh B-CLL cells were available for CD69, ZAP-70, CD38 analyses in all patients and for CD49d in 312 patients. Fresh or frozen serum samples were obtained for sCD23 from 364 patients. According to Rai stages at diagnosis, 127 patients belonged to Rai stage 0, 178 patients to Rai stage I, 94 patients to Rai II, 10 cases to Rai III and 8 cases to Rai IV. Using the modified Rai staging system (mod-Rai), the distribution was as follows: 127 patients stage I (low-risk); 272 patients stage II (intermediate-risk); 18 patients stage III (high-risk). Median follow up was six years with 60 deaths and 357 censored patients. Two hundred and five of the 417 patients (49.2%) received chemotherapy for their disease. Fifty-one patients were treated with a combination of chlorambucil at conventional doses and prednisone. One hundred and nine patients received six courses of fludarabine monophosphate (Fludara; Genzyme, Modena, Italy) at 25 mg/m2 intravenously or 30–40 mg/m2 orally per day for five days every 28 days followed by four courses of rituximab (Mabthera; Roche, Basilea, Switzerland) at 375 mg/m2 for one day every week.28 Finally, thirty-five patients were treated with six courses of fludarabine, cyclophosphamide and rituximab.29
Cellular immunophenotypic analysis
All flow cytometric analyses were performed on a FACSCalibur flow cytometer (BD Biosciences, CA, USA). The instrument was aligned and calibrated daily with the use of a 4-color mixture of CaliBRITE beads (BD Biosciences) with FACSComp Software (BD, Biosciences) according to the manufacturer’s instructions. The beads are used to adjust instrument settings, set fluorescence compensation and check instrument sensitivity in order to monitor instrument performance over time. Expression of CD69, CD38 and CD49d was analyzed by 3-color immunofluorescence,8,9 by combining phycoerythrin (PE)-conjugated anti-CD38 or antiCD49d or anti-CD69 monoclonal antibodies (MoAbs) with peri-dinin-chlorophyll-protein-cyanine-5.5 (PerCP-Cy5.5) or allophy-co-cianine (APC) anti-CD19 MoAbs and fluorescein isothiocyanate (FITC)-conjugated anti-CD5 MoAb. Expression data were reported as percentage of CD5+CD19+ CLL cells displaying specific fluorescence intensity. The threshold of positivity was set at over 30% for all these markers. Estimation of CD69 expression level yelding the best separation of 2 subgroups with different OS and/or PFS probabilities was made by applying different methods, including the maximally selected log rank statistics, Youden’s index, and receiver-operating characteristic analysis.30 Results of all these tests were in agreement in indicating use of the cut off of 30% of positive cells to discriminate between CD69high and CD69low cases when testing the prognostic impact of CD69 on both OS and PFS. To test its reproducibility, expression of CD69 was compared: 1) in fresh versus frozen PB samples (22 cases); and 2) in fresh PB samples delivered by overnight courier and separately analyzed in two different laboratories (i.e. located at the S. Eugenio Hospital, University of Tor Vergata in Rome and at the Clinical and Experimental Onco-Hematology Unit of the Centro di Riferimento Oncologico in Aviano; 55 cases). These sets of experiments yielded almost superimposable results in all cases. To test the stability of CD69 over time, we analyzed sequential samples from 55 patients (range 4–8 samples for each patient) over periods ranging from three to 60 months. In 50 of 55 cases, slight differences (<10%) were detected. A total of 10 cases showed a variation higher than 10%. No patient crossed the 30% cut off used.
Flow cytometric detection of ZAP-70 was performed as previously reported21 and according to previous studies; a cut off of 20% of positive cells was chosen to discriminate ZAP-70 negative and ZAP-70 positive patients.
Soluble CD23 and β2-microglobulin detection
Soluble CD23 (sCD23) immunoenzymometric assay and β2-microglobulin (β2M) determinations were performed as described elsewhere.8 The threshold of positivity was set at 70 U/mL for sCD23 and 2.2 g/L for β2M on the basis of normal laboratory reference values.
Interphase FISH
Separate hybridizations were carried out for loci on chromosome 11, 12, 13 and 17. For chromosomes 11, 13, 12 and 17 specific probes and procedures were used, as reported previously.21,31
IGHV mutation analysis
Total RNAs were extracted and reverse-transcribed. The resulting cDNAs, checked for first strand synthesis,32 were amplified using a mixture of sense primers annealing to either VH1 through VH6 leader sequences or 5’ ends of VH1-VH6 FR1. Purified amplicons were cloned and a minimum of 10–20 colonies for each case were sequenced; CLL specific IGHV transcripts, identified by sharing the same CDR3 in multiple clones were analyzed for percent mutation, as previously described. VH gene sequences deviating more than 2% from the corresponding germline gene were defined as mutated.12
Statistical analysis
Correlations between CD69 percentages and the other biological and clinical variables were based on the two-tailed Fisher’s exact test. The clinical assessment of CLL patients was based on the National Cancer Institute Working Group criteria.33 Progression-free survival (PFS) and overall survival (OS), measured from diagnosis, were estimated according to the Kaplan-Meier method and compared between groups by means of the log rank test. Cox’s proportional hazards regression models were used to assess the independent effect of covariables, treated as dichotomous, on the PFS or OS.
Results
Expression of CD69 and association with other prognostic factors
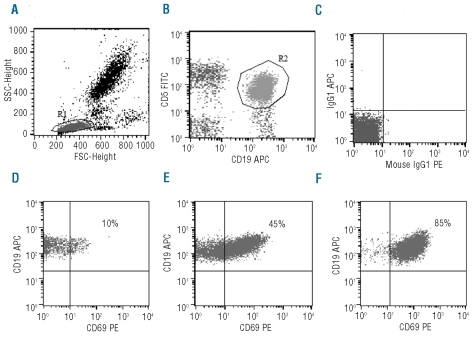
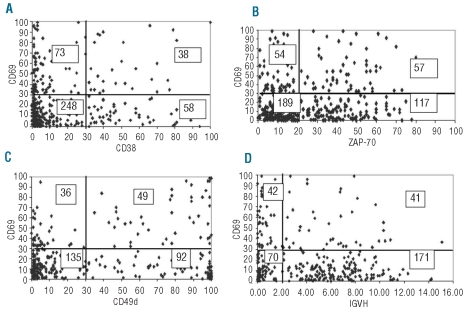
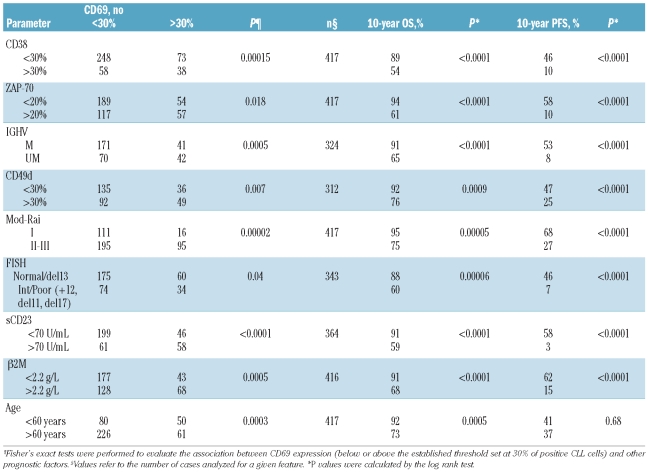
Expression of CD69 revealed variable patterns of florescence intensity, corresponding to different percentages of positive CLL cells (Figure 1A–C). On the basis of a 30% threshold value, 111 patients (26.6%) were CD69 positive and 306 (73.4%) were CD69 negative. Bi-variant plots for CD69 versus CD38 or ZAP70 or CD49d or IGHV status (Figure 2A–D) showed four subpopulations expressing concordant and discordant profiles. Table 1 shows the association of CD69 with biological markers (CD38, CD49d, ZAP-70, IGHV gene mutational status), markers of tumor burden (β2M, sCD23 and mod-Rai), chromosomal aberrations, and age. All these factors were demonstrated to be significant prognosticators for OS and/or PFS in our CLL series (Table 1). Significant correlations were found between CD69 and some markers of tumor burden (sCD23, P<0.0001; mod-Rai, P=0.00002; β2M, P=0.0005, Table 1). Moreover, the presence of multiple (3 or more) intrathoracic/abdominal lymphoadenopathies (>3 cm in diameter) and/or splenomegaly was significantly correlated with CD69 over 30% (59 of 111, P=0.00005). CD69 over 30% was observed in 35 of 80 patients with lymphocyte doubling time (LDT) of less than 12 months (P=0.0002).
Figure 1.
Characteristics of CD69 expression. (A) Forward and side scatter of a representative PB sample is shown with a region (R1) drawn around the lymphocytes. Lymphocytes were R1 gated and then B-CLL (CD5+CD19+) cells (the R2 region in plot (B) were selected according to their phenotype. (C) Expression of mouse IgG1 isotypic controls after lymphocyte (R1) gating. (D–F) Flow cytometric dot-plots of CD19-APC versus CD69-PE expression on PB samples of 3 representative CLL cases. Reported percentage of CD69-expressing cells are calculated on the gated CD5+CD19+ CLL population (R2). In each case, mouse IgG1 isotypic antibody is used as control for CD69 positivity.
Figure 2.
Bi-variant plots for CD69 and CD38 or ZAP-70 or CD49d or IGHV. y axis reports values for CD69 and x axis values for (A) CD38, (B) ZAP-70, (C) CD49d and (D) IGHV mutations, measured as percentage of positive cells (CD69, CD38, ZAP-70, CD49d) or percentage mutations (IGHV). Number of cases expressing concordant and discordant profiles are reported within each quadrant of the plots.
Table 1.
Distribution of prognostic factors in CLL according to CD69 expression.
Relevance of CD69 as biological prognostic marker
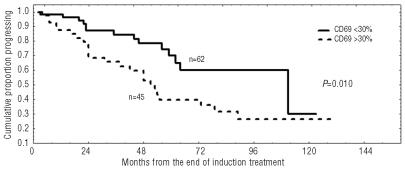
A significant correlation was found between treatment and CD69 expression. In particular, 82 of 111 (74%) CD69 positive patients received chemotherapy, while only 123 of 306 (40%) CD69 negative patients were treated (P<0.0001). It is worthy of note that CD69 negative cases showed a significantly longer response duration (60% vs. 40% at six years, P=0.010) in a series of 107 patients achieving a complete or partial response after an induction treatment with fludarabine plus rituximab (Figure 3).28
Figure 3.
Duration of response after fludarabine and rituximab in relation to CD69 expression. Progression free survival after treatment was significantly longer within CLL subgroup showing CD69 lower than 30% (60% vs. 40% at six years; P=0.010).
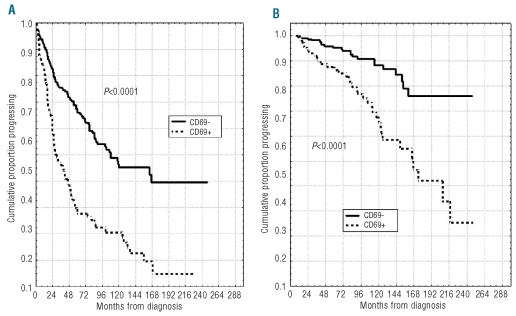
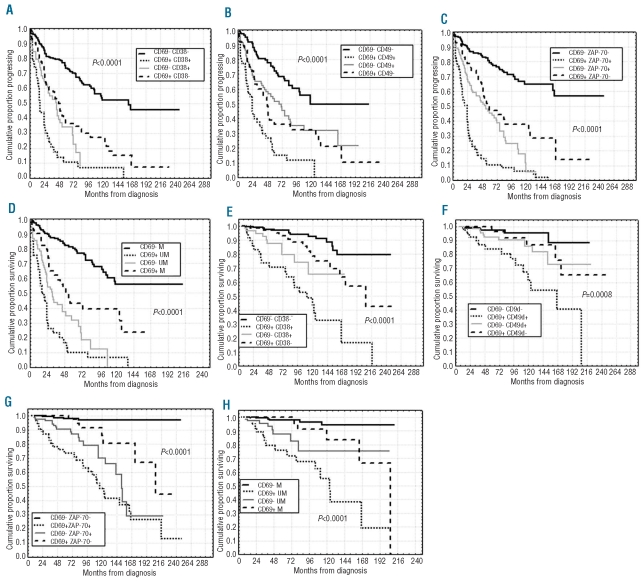
Our cohort of cases had a median PFS of 3.8 years and 39% PFS at ten years. A significantly shorter PFS was observed in CD69 positive cases (20% vs. 45% at ten years, P<0.0001; Figure 4A) compared with the CD69 negative subgroup. Likewise, a shorter OS was found in CD69 positive patients (58% vs. 85% at 12 years; P<0.0001, Figure 4B). Similar results were observed when investigating the impact on PFS and OS of the three immunophenotypic markers CD38, CD49d and ZAP-70 or of IGHV gene mutations (Table 1). A more refined prognostic assessment of PFS and OS was obtained by combining CD69 expression with that of CD38, CD49d and ZAP-70 (Figure 5A–C and E–G). For all these variables, CD69 expression had true additive properties. In particular, the simultaneous positivity or negativity for CD69 and CD38 or CD49d or ZAP-70 identified 2 subsets of patients: the former with the worst prognosis and the latter with the best prognosis with regard to both PFS (7% vs. 52% at ten years, P<0.0001; 12% vs. 49% at ten years, P<0.0001 and 6% vs. 65% at ten years, P<0.0001) and OS (44% vs. 91% at ten years, P<0.0001; 61% vs. 96%, P=0.0008 and 49% vs. 97%, P<0.0001). In all combinations, patients who did not fit these results showed intermediate outcomes (Figure 5A–C and E–G). Similar behavior was observed by combining CD69 and IGHV mutational status in additional bivariate analyses, the shortest PFS and OS time intervals being found for CD69+/UM IGHV patients (7% vs. 56% at ten years and 55% vs. 94% at ten years, respectively, P<0.0001; Figure 5D and H).
Figure 4.
Progression-free survival (PFS) and overall survival (OS) curves based on CD69 expression. Kaplan-Meier plot comparing (A) PFS and (B) OS based on the detection of greater than 30% (CD69+) or lower than 30% CD69+ B cells (CD69−). CD69− patients both experienced a longer PFS and OS (P<0.0001).
Figure 5.
PFS and OS curves in relation to combined CD69 and CD38 expression or CD69 and CD49d or CD69 and ZAP-70 or CD69 and IGHV mutational status. PFS and OS were significantly longer (A, E) within the CD69-CD38- subgroup, or (B, F) within CD69-CD49d- patients or (C, G) within CD69-ZAP-70 negative cases. (D, H) Equally, CD69-IGHV mutated (M) patients experienced longer PFS and OS. (A–H) Conversely, double positive (CD69+CD38+ or CD69+CD49d+ or CD69+ZAP−70+) or CD69+IGHV unmutated patients demonstrated worse PFS and OS. Discordant patients showed intermediate outcomes.
CD69 and tumor burden markers
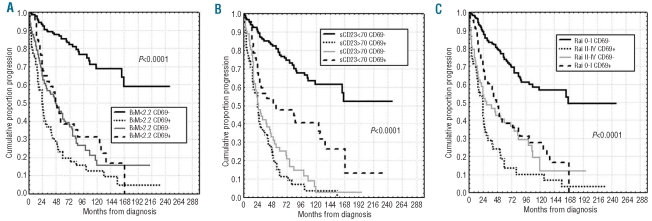
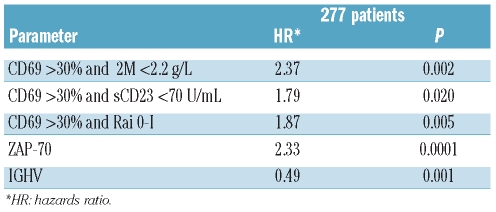
Interestingly, CD69 expression levels were found to be necessary to correctly prognosticate PFS duration in the context of favorable prognosis patients with regard to tumor burden markers. Indeed, only cases in which a low tumor burden (β2M <2.2.g/L or sCD23 <70 U/mL or 0-I Rai stage) was associated with CD69 below 30% had significantly longer PFS. Conversely, expression of CD69 over the threshold of 30% even in the β2Mlow, sCD23low, 0-I Rai categories identified patients with a poor prognosis, whose PFS duration was similar to those characterized by an unfavorable profile of tumor burden markers (i.e. β2Mhigh, sCD23high or II–IV Rai) (Figure 6A–C).
Figure 6.
PFS of CD69 in combination with markers of tumor burden. Kaplan-Meier curves obtained by associating CD69 expression to 3 tumor burden markers, i.e. (A) β2M, (B) sCD23 serum levels and (C) Rai stage risk groups. For each Kaplan-Meier analysis, 4 groups were compared: patients lacking both negative prognosticators, patients with presence of both negative prognosticators, patients with presence of either one unfavorable prognosticator.
Multivariate analyses
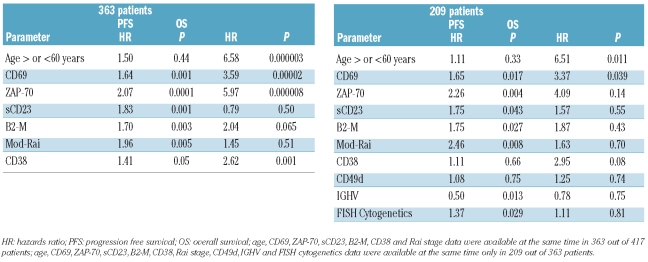
CD69, CD38 ZAP-70, β2M, sCD23 and mod-Rai were included in a Cox’s proportional hazard regression model to test their strength as independent prognostic factors for PFS. In a cohort of 363 of 417 patients, in which all the above mentioned prognostic factors were available at the same time, CD69 was an independent prognosticator, along with ZAP-70 and the 3 markers of tumor burden (Table 2). On the other hand, if we included CD49d, the IGHV mutational status and FISH cytogenetics, available only in 209 of 363 patients at the same time, the multivariate model again selected this covariate as an independent prognosticator (P=0.017) (Table 2). Because of the differences in the prognostic relevance of CD69 expression in β2Mlow versus β2Mhigh CLL, or in CLL with sCD23low versus sCD23high or in 0-I Rai versus II–IV Rai CLL, as seen by the trend of PFS curves (Figure 6A–C), the possibility of interactions between CD69 and these clinical variables was considered and assessed by bivariate Cox’s proportional hazards models. In fact, CD69 over 30% in CLL with β2Mlow or sCD23low or 0-I Rai stages was associated with a hazard ratio (HR) for progressive disease of 2.88 or 2.74 or 2.79, respectively. On the other hand, no additional prognostic information was provided when CD69 over 30% or below 30% was tested in the context of CLL expressing β2Mhigh (HR=1.34) or sCD23high (HR=1.27) or II–IV Rai stages (HR=1.21).
Table 2.
Multivariate Cox’s regression analysis of PFS and OS.
To consider interactions between CD69 and β2M or CD69 and sCD23, and to evaluate their independent prognostic value for PFS, these combinations were incorporated in a further multivariate analysis along with all the other biological markers (CD38, ZAP-70 and IGHV mutational status) tested in our study. CLL expressing a CD69high/b2Mlow or a CD69high/sCD23low phenotype or a CD69high/Rai 0-I were associated with an increased independent risk of progressive disease (HR=2.13, HR=1.82 and HR=1.87, respectively) compared to CLL expressing CD69low/b2Mlow or CD69low/sCD23low or CD69low/Rai 0-I phenotypes (Table 3). Conversely, CD69 values failed to provide additional prognostic information in the β2Mhigh or sCD23high or Rai II–IV categories.
Table 3.
Multivariate Cox’s regression analyses of PFS with interactions between prognostic factors.
The value of CD69 as independent prognosticator for OS was checked by multivariate Cox’s proportional hazards analysis applied to models including the prognosticators proven to be significant in univariate analyses (Table 1). In our cohort of 363 of 417 patients, multivariate analysis selected CD69 (P=0.00002), ZAP-70 (P=0.000008), CD38 (P=0.001), age (P=0.000003) as significant independent prognosticators (Table 2). In a further model, adjusting also for the IGHV status, FISH cytogenetics and CD49d, available at the same time in a smaller number of patients, (209 of 363 cases; Table 2) again CD69 was an independent prognostic factor for OS (P=0.039), along with age (P=0.011).
Discussion
The aim of our study was to validate in a large cohort of patients the clinical impact of CD69 as independent prognostic factor for CLL in relationship with other biological prognosticators (CD38, ZAP-70, IGHV mutations, cytogenetic abnormalities) or with tumor burden indicators (Rai clinical stages, β2M, sCD23).8–12
Expression of CD69 was performed by a direct 3-color immunofluorescence and this combination provided a simple flow cytometric method that could be introduced into a routine immunophenotyping panel in a clinical diagnostic laboratory. However, a standardized procedure which avoids some of the difficulties encountered with ZAP-70, and to a lesser extent with CD38, has still not been established. The optimal cut off for CD69 expression yielding the best separation of CLL patients into 2 subsets with significantly different prognosis was fixed at 30% of positive cells by applying the most commonly used methods.9,13,32,34 Since our proposal is to use CD69 for prognostic purposes, we confirmed its inter-laboratory reproducibility and the stability over time of its expression.21,35 We demonstrated that CD69 expression was significantly correlated with CD38, CD49d and ZAP-70 and IGHV mutational status. CD69 that is up-regulated rapidly with cellular activation resembling B cells at an earlier state of activation, might be able to transduce BCR–mediated signals better with the help of simultaneous ZAP-70 expression,36,37 and may reflect ongoing in vivo stimulation, thereby explaining the more aggressive disease course observed in these patients. Such increased intracellular signaling could influence the survival or proliferation of CLL cells, leading to a tendency toward disease progression. It is worthy of note that micro-array analysis revealed an increased expression of CD69 gene in UM CLL upon BCR cross-linking.38 In fact, IgM stimulation of BCR up-regulates several genes associated with the cell cycle allowing cell growth and expansion, thus ultimately contributing to the unfavorable clinical course of these CLL patients. In addition, CD69, tested on CLL cells by flow cytometry, was found to be up-regulated on cells in the tissue microenvironment, both in bone marrow (BM) and lymph nodes (LN). Interestingly, stronger upregulation of CD69 was also found in LN-resident compared with BM-resident CLL cells and LN-derived cells showed an increase in BCR signature genes compared with those from the BM and the PB.39 Moreover, because CD69 is involved in retaining lymphocytes at the site of stimulation,40–42 the levels of this molecule on circulating B-CLL cells might be less than those in the solid tissue (BM and LN) and, therefore, might indicate that B-CLL cells expressing CD69 are recent emigrants from such sites.43 Tumor proliferation, quantified by the expression of the E2F and c-MYC target genes and verified with Ki67 staining by flow cytometry was highest in the LN and was correlated with clinical disease progression.39 Thus, CD69 appear to offer an easy to perform and reliable marker both of progression and poor outcome in CLL.
From a clinical point of view, CD69 overexpression was significantly correlated with more advanced Rai stages, with large intrathoracic/abdominal lymphoadenopathies, splenomegaly and a shorter LDT, all characteristics of an active and aggressive disease. Significant differences in treatment histories were found between CD69 positive and CD69 negative patients because CD69 positive cases underwent treatment more frequently and presented a shorter response duration (P<0.0001 and P=0.010).
We demonstrated that CD69 protein expression was an independent risk factor for PFS and OS in our large series of CLL patients. Furthermore, also higher CD38 or ZAP-70 or CD49d expression and sCD23 levels were significantly associated with shorter PFS and OS in this large cohort of patients, confirming our previous results.21 Interestingly, combined analysis of CD69 with CD38 or CD49d or ZAP-70 demonstrated that CD69 expression had true additive properties, allowing us to identify CLL subsets (CD69-CD38-; CD69-CD49d-; CD69-ZAP-70-; CD69-M IGHV) presenting a very good outcome with regard to PFS and OS. Conversely, double positive (CD69+CD38+, CD69+CD49d+, CD69+ZAP-70+) subsets and CD69 positive UM IGHV patients showed the worst outcome. Discordant cases showed an intermediate clinical course (Figure 5). Moreover, CD69 was also necessary to correctly prognosticate the active/progressive disease status within selected subsets of patients.9 In particular, CD69 levels were found to be necessary to correctly classify groups of patients in the context of CLL with β2Mlow or sCD23low or 0-I Rai stage (Figure 6A-C). In particular, the slow disease progression of β2Mlow or of sCD23low or of 0-I Rai stage CLL holds true only if associated with low CD69 expression levels. Notably, no statistical differences in terms of PFS were observed by comparing β2Mlow/CD69high or sCD23low/CD69high or 0-I Rai stages/CD69high with cases with β2Mhigh, scCD23high or II-IV Rai stage, irrespective of CD69 expression (Figure 6A-C). Therefore, CD69 appear to be crucial to refine prognosis in CLL patients with a low tumor burden.
As shown by multivariate analysis, CD69 had an independent prognostic value with regard both to PFS and OS (Table 2), along with ZAP-70 and the 3 markers of tumor burden. Additional multivariate analysis, including also CD49d, FISH cytogenetics and IGHV mutational status, confirmed CD69 as an independent prognosticator (Table 2).
Datasets compiled by the prospective collection of well-characterized CLL patients are now surely warranted in order to confirm the clinical significance of CD69 in CLL.
Footnotes
Funding: supported in part by MURST, Programmi di Ricerca di Interesse Nazionale, 2007; Ministero della Salute (Ricerca Finalizzata I.R.C.C.S., “Alleanza Contro il Cancro” and Rete Nazionale Bio-Informatica Oncologica/RN-BIO; “Giovani Ricercatori” project, grant GR-2008-1138053), Rome; Fondazione Internazionale di Ricerca in Medicina Sperimentale, Turin, Italy; Associazione Italiana Contro le Leucemie, Linfomi e Mielomi (AIL), Venezia Section, Pramaggiore (VE) Group; the Associazione Italiana Ricerca Cancro (AIRC, Investigator Grant IG-8701 to VG; MFAG-10327 to RB); “5×1000 program” of the Centro di Riferimento Oncologico, Aviano, Italy; Ricerca Scientifica Applicata, Regione Friuli Venezia Giulia, Trieste (“Linfonet” Project).
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Zwiebel JA, Cheson BD. Chronic lymphocytic leukemia: Staging and prognostic factors. Semin Oncol. 1998;25(1):42–59. [PubMed] [Google Scholar]
- 2.Binet JL, Auquier A, Dighiero G, Chastang C, Piguet H, Goasguen J, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48(1):198–206. doi: 10.1002/1097-0142(19810701)48:1<198::aid-cncr2820480131>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 3.Rai K. A critical analysis of staging in CLL. Chronic Lymphocytic Leukemia. In: Gale R, Rai K, editors. Recent Progress and Future Directions. New York, NY: Liss; 1987. p. 253. [Google Scholar]
- 4.Montserrat E, Sanchez-Bisono J, Vinolas N, Rozman C. Lymphocyte doubling time in chronic lymphocytic leukaemia: analysis of its prognostic significance. Br J Haematol. 1986;62(3):567–75. doi: 10.1111/j.1365-2141.1986.tb02969.x. [DOI] [PubMed] [Google Scholar]
- 5.Keating MJ, Lerner S, Kantarjian H, Freireich EJ, O'Brien S. The serum β2-microglobulin level is more powerful than stage in predicting response and survival in chronic lymphocytic leukemia. Blood. 1995;86 Abstract 606. [Google Scholar]
- 6.Sarfati M, Chevret S, Chastang C, Biron G, Stryckmans P, Delespesse G, et al. Prognostic importance of serum soluble CD23 level in chronic lymphocytic leukemia. Blood. 1996;88(11):4259–64. [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Del Poeta G, Maurillo L, Venditti A, Buccisano F, Epiceno AM, Capelli G, et al. Clinical significance of CD38 expression in chronic lymphocytic leukemia. Blood. 2001;98(9):2633–9. doi: 10.1182/blood.v98.9.2633. [DOI] [PubMed] [Google Scholar]
- 9.Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111(2):865–73. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 10.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. IgV gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. [PubMed] [Google Scholar]
- 11.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. [PubMed] [Google Scholar]
- 12.Degan M, Bomben R, Dal Bo M, Zucchetto A, Nanni P, Rupolo M, et al. Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antige-driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery. Br J Haematol. 2004;126(1):29–42. doi: 10.1111/j.1365-2141.2004.04985.x. [DOI] [PubMed] [Google Scholar]
- 13.Krober A, Seller T, Benner A, Bullinger L, Brückle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100(4):1410–6. [PubMed] [Google Scholar]
- 14.Bomben R, Dal Bo M, Capello D, Benedetti D, Marconi D, Zucchetto A, et al. Comprehensive characterization of IGHV3-21-expressing B-cell chronic lymphocytic leukemia: an Italian multicenter study. Blood. 2007;109(7):2989–98. doi: 10.1182/blood-2006-10-051110. [DOI] [PubMed] [Google Scholar]
- 15.Capello D, Guarini A, Berra E, Mauro FR, Rossi D, Ghia E, et al. Evidence of biased immunoglobulin variable gene usage in highly stable B-cell chronic lymphocytic leukemia. Leukemia. 2004;18(12):1941–7. doi: 10.1038/sj.leu.2403537. [DOI] [PubMed] [Google Scholar]
- 16.Hamblin TJ, Orchard JA, Gardiner A, Oscier DG, Davis Z, Stevenson FK. Immunoglobulin V genes and CD38 expression in CLL. Blood. 2000;95(7):2455–7. [PubMed] [Google Scholar]
- 17.Ibrahim S, Keating M, Do KA, O'Brien S, Huh YO, Jilani I, et al. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood. 2001;98(1):181–6. doi: 10.1182/blood.v98.1.181. [DOI] [PubMed] [Google Scholar]
- 18.Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348(18):1764–75. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 19.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;35(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 20.Orchard JA, Ibbotson RE, Davis Z, Wiestner A, Rosenwald A, Thomas PW, et al. ZAP-70 expression and prognosis in chronic lymphocytic leukaemia. Lancet. 2004;363(9403):105–11. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- 21.Del Principe MI, Del Poeta G, Buccisano F, Maurillo L, Venditti A, Zucchetto A, et al. Clinical significance of ZAP-70 protein expession in B-cell chronic lymphocytic leukemia. Blood. 2006;108(3):853–61. doi: 10.1182/blood-2005-12-4986. [DOI] [PubMed] [Google Scholar]
- 22.Zucchetto A, Bomben R, Dal Bo M, Nanni P, Bulian P, Rossi FM, et al. ZAP-70 expression in B-cell chronic lymphocytic leukemia: evaluation by external (isotypic) or internal (T/NK cells) controls and correlation with IgV(H) mutations. Cytometry B Clin Cytom. 2006;70(4):284–92. doi: 10.1002/cyto.b.20127. [DOI] [PubMed] [Google Scholar]
- 23.Damle RN, Ghiotto F, Valetto A, Albesiano E, Fais F, Yan XJ, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002;99(11):4087–93. doi: 10.1182/blood.v99.11.4087. [DOI] [PubMed] [Google Scholar]
- 24.Testi R, D’Ambrosio D, De Maria R, Santoni A. The CD69 receptor: a multipurpose cell-surface trigger for hematopoietic cells. Immunol Today. 1994;15(10):479–83. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 25.Marzio R, Manuel J, Betz-Corradin S. CD69 and regulation of the immune function. Immunopharmacol Immunotoxicol. 1999;21(3):365–82. doi: 10.3109/08923979909007126. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler SF, Ramsdell F, Alderson MR. The activation antigen CD69. Stem Cells. 1994;12(5):456–65. doi: 10.1002/stem.5530120502. [DOI] [PubMed] [Google Scholar]
- 27.D’Arena G, Musto P, Nunziata G, Cascavilla N, Savino L, Pistolese G. CD69 expression in B-cell chronic lymphocytic leukemia: a new prognostic marker? Haematologica. 2001;86(9):995–6. [PubMed] [Google Scholar]
- 28.Del Poeta G, Del Principe MI, Consalvo MA, Maurillo L, Buccisano F, Venditti A, et al. The addition of rituximab to fludarabine improves clinical outcome in untreated patients with ZAP-70-negative chronic lymphocytic leukemia. Cancer. 2005;104(12):2743–52. doi: 10.1002/cncr.21535. [DOI] [PubMed] [Google Scholar]
- 29.Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376(9747):1164–74. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 30.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Comp Statist Data Analysis. 2003;43:121–37. [Google Scholar]
- 31.Del Principe MI, Del Poeta G, Venditti A, Buccisano F, Maurillo L, Marini R, et al. Clinical significance of soluble p53 protein in B-cell chronic lymphocytic leukemia. Haematologica. 2004;89(12):1468–75. [PubMed] [Google Scholar]
- 32.Bomben R, Dal Bo M, Zucchetto A, Zaina E, Nanni P, Sonego P, et al. Mutational status of IgV(H) genes in B-cell chronic lymphocytic leukemia and prognosis: percent mutations or antigen-driven selection? Leukemia. 2005;19(8):1490–2. doi: 10.1038/sj.leu.2403830. [DOI] [PubMed] [Google Scholar]
- 33.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O'Brien S, Rai KR. National Cancer Institute Sponsored Working Group Guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87(12):4990–7. [PubMed] [Google Scholar]
- 34.Zucchetto A, Bomben R, Dal Bo M, Sonego P, Nanni P, Rupolo M, et al. A scoring system based on the expression of six surface molecules allows the identification of three prognostic risk groups in B-cell chronic lymphocytic leukemia. J Cell Physiol. 2006;207(2):354–63. doi: 10.1002/jcp.20570. [DOI] [PubMed] [Google Scholar]
- 35.D’Arena G, Nunziata G, Coppola G, Vigliotti ML, Tartarone A, Carpinelli N, et al. CD38 expression does not change in B-cell chronic lymphocytic leukemia. Blood. 2002;100(8):3052–3. doi: 10.1182/blood-2002-07-2249. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Widhopf G, Huynh L, Rassenti L, Rai KR, Weiss A, Kipps TJ. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100(13):4609–14. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Apgar J, Huynh L, Dicker F, Giago-McGahan T, Rassenti L, et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 2005;105(5):2036–41. doi: 10.1182/blood-2004-05-1715. [DOI] [PubMed] [Google Scholar]
- 38.Guarini A, Chiaretti S, Tavolaro S, Maggio R, Peragine N, Citarella F, et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood. 2008;112(3):782–92. doi: 10.1182/blood-2007-12-127688. [DOI] [PubMed] [Google Scholar]
- 39.Herishanu Y, Perez-Galan P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-{kappa} B activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563–74. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol. 1996;3(3):301–4. doi: 10.1128/cdli.3.3.301-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440(7083):540–4. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 42.Kishimoto TK, Jutila MA, Butcher EC. Identification of a human peripheral lymph node homing receptor: a rapidly down-regulated adhesion molecule. Proc Natl Acad Sci USA. 1990;87(6):2244–8. doi: 10.1073/pnas.87.6.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damle RN, Temburni S, Calissano C, Yancopoulos S, Banapour T, Sison C, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007;110(9):3352–9. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]