Summary
The type I interferon (IFN) response initiated by detection of nucleic acids is important for antiviral defense, but is also associated with specific autoimmune diseases. Mutations in the human 3′ repair exonuclease 1 (Trex1) gene cause Aicardi-Goutières syndrome (AGS), an IFN-associated autoimmune disease. However, the source of the type I IFN response and the precise mechanisms of disease in AGS remain unknown. Here, we demonstrate that Trex1 is an essential negative regulator of the STING-dependent antiviral response. We used an in vivo reporter of IFN activity in Trex1-deficient mice to localize the initiation of disease to non-hematopoietic cells. These IFNs drove T cell-mediated inflammation and an autoantibody response that targeted abundant, tissue-restricted autoantigens. However, B cells contributed to mortality independently of T cell-mediated tissue damage. These findings reveal a stepwise progression of autoimmune disease in Trex1-deficient mice, with implications for the treatment of AGS and related disorders.
Introduction
Innate immune detection of nucleic acids is an essential component of the host response to viral infection (Barbalat et al., 2011). In vertebrates, two families of nucleic acid receptors activate the antiviral response. Toll-like receptors (TLRs) expressed by sentinel innate immune cells survey phagocytosed material for the presence of foreign nucleic acids. In contrast, intracellular nucleic acid sensors are more broadly expressed and signal a cell-intrinsic response to viral infection. In recent years, many of these innate immune sensors have been identified and characterized in great detail. They include TLR3, TLR7, TLR8, which detect various structural features of RNA, and TLR9, which is activated by DNA. The intracellular RNA helicases RIG-I and MDA5 detect viral RNA, while intracellular DNA sensing involves AIM2, DAI, IFI16, and other, currently unknown receptors (reviewed in Barbalat et al., 2011). All of these receptors (with the exception of AIM2) activate expression of the type I interferon (IFN) family of cytokines, which act to block viral replication within infected cells and facilitate adaptive immune responses to viral antigens (Stark et al., 1998; Stetson and Medzhitov, 2006b).
The IFN response triggered by nucleic acid receptors is important for protection against infection, but it must be carefully regulated to prevent inappropriate activation by endogenous DNA and RNA. Recent studies have found that chronic activation of these antiviral sensors can cause a number of severe autoimmune diseases (Banchereau and Pascual, 2006; Theofilopoulos et al., 2005). In normal settings, this chronic activation is prevented by regulating the expression and compartmentalization of the sensors themselves (Barton and Kagan, 2009), and by the activity of RNA and DNA nucleases that metabolize the ligands for the receptors (Nagata et al., 2010). Thus, the imperfect ability of innate immune receptors to distinguish between endogenous and foreign nucleic acids is enabled in large part by the activities of accessory proteins.
Recent advances have established central roles for nucleic acid-sensing TLRs and type I IFNs in a number of severe autoimmune diseases, including systemic lupus erythematosus (SLE) and psoriasis (Barrat et al., 2005; Christensen et al., 2006; Lande et al., 2007; Leadbetter et al., 2002). In addition, chronic activation of cell-intrinsic antiviral responses can also cause autoimmunity. Specifically, we identified 3′ repair exonuclease 1 (Trex1) as an essential negative regulator of the antiviral response triggered by detection of intracellular DNA (Stetson et al., 2008). In Trex1-deficient mice, endogenous DNA substrates accumulate and trigger a lethal, type I IFN-dependent autoimmune disease. These Trex1 substrates include reverse-transcribed DNA derived from endogenous retroelements (Stetson et al., 2008), and Trex1 can also metabolize Human Immunodeficiency Virus (HIV) cDNA within infected cells (Yan et al., 2010), suggesting a key role for this pathway in anti-retroviral defense and an important contribution of nucleic acids derived from endogenous retroelements to autoimmune disease.
Interestingly, detection of intracellular DNA can activate two distinct antiviral responses in cells. One pathway, called the Interferon-Stimulatory DNA (ISD) response (Stetson and Medzhitov, 2006a), is activated by natural DNA containing all four bases and triggers type I interferon production through the adaptor protein STING (Ishikawa et al., 2009). In contrast, the synthetic DNA polymer poly-dA:dT can activate the STING-dependent pathway and can also be transcribed by cellular RNA polymerase III into a triphosphate RNA ligand that activates the RIG-I-MAVS pathway (Ablasser et al., 2009; Chiu et al., 2009; Ishii et al., 2006). Thus, while it is clear that Trex1 negatively regulates the DNA-activated antiviral response, it remains unknown whether Trex1 regulates the ISD pathway, the Pol-III pathway, or both.
Loss of function mutations in the human Trex1 gene cause Aicardi-Goutières syndrome (AGS), a rare and severe autoimmune disease that presents in infancy and mimics the features of congenital viral infection (Crow et al., 2006). AGS is characterized by elevated type I IFNs, brain inflammation, and profound psychomotor retardation, with a mortality rate approaching 35% by 15 years of age (Rice et al., 2007b). Currently, there are no effective therapies for AGS, and the precise mechanisms of disease remain incompletely defined.
Since the seminal identification of Trex1 mutations in AGS, dozens of distinct mutations within the human Trex1 open reading frame have been discovered in the context of several disease phenotypes, including chilblain lupus and SLE (de Vries et al., 2009; Lee-Kirsch et al., 2007; Namjou et al., 2011; Rice et al., 2007a). Remarkably, some of these mutations are identical to those that cause AGS. While the precise functional consequences of many of these rare, lupus-associated Trex1 mutations remain unknown, the genetic association of Trex1 mutations with SLE is the strongest of any single gene identified to date (Harley et al., 2009). Together, these studies clearly link Trex1 and the cell-intrinsic antiviral response to DNA to a number of IFN-associated human autoimmune disorders.
Trex1-deficient (Trex1−/−) mice, with a highly penetrant and severe phenotype, are an excellent model for exploring the mechanistic basis of disease, with direct relevance to human AGS and SLE. These mice are particularly amenable to genetic dissection of autoimmunity through crosses with mice deficient in key components of innate and adaptive immunity. For example, we found that Trex1−/− mice lacking interferon regulatory factor 3 (IRF3), the type I IFN receptor (Ifnar1), or RAG2 are all completely protected from autoimmune pathology and mortality (Stetson et al., 2008). However, a number of important questions remain: which DNA-activated antiviral response does Trex1 regulate? Which cells initiate the disease, where, and when? How does chronic activation of the ISD pathway lead to lymphocyte-dependent autoimmunity? And, what are the specific contributions of lymphocytes to disease?
Here, we present a detailed characterization of the development of disease in Trex1-deficient mice, from its earliest initiation to organ-specific autoimmune pathology. We establish Trex1 as a unique regulator of the STING-dependent ISD pathway. We employ an in vivo reporter of the type I IFN response to localize the initiation of disease and we determine how these IFNs drive the autoreactive lymphocyte response. We show that both T cells and B cells contribute to disease through distinct mechanisms. Together, these findings provide new insight into the progression of IFN-mediated autoimmunity, with implications for the human diseases caused by chronic activation of the ISD pathway.
Results
Trex1-deficient mice develop specific multi-organ inflammation
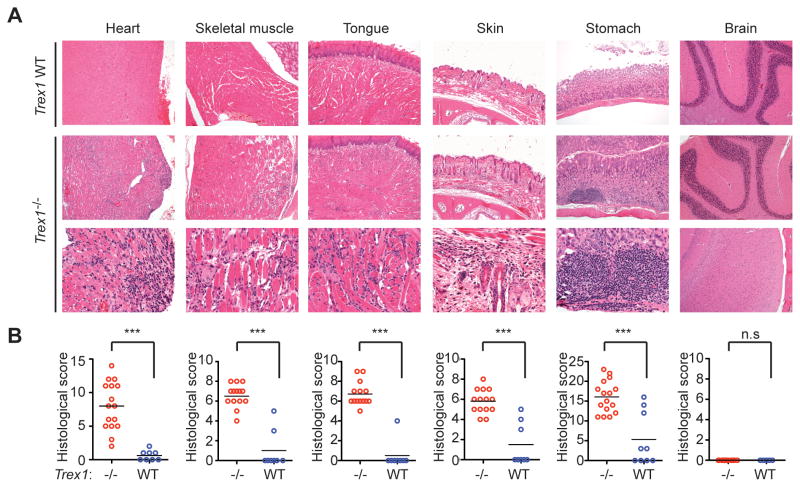
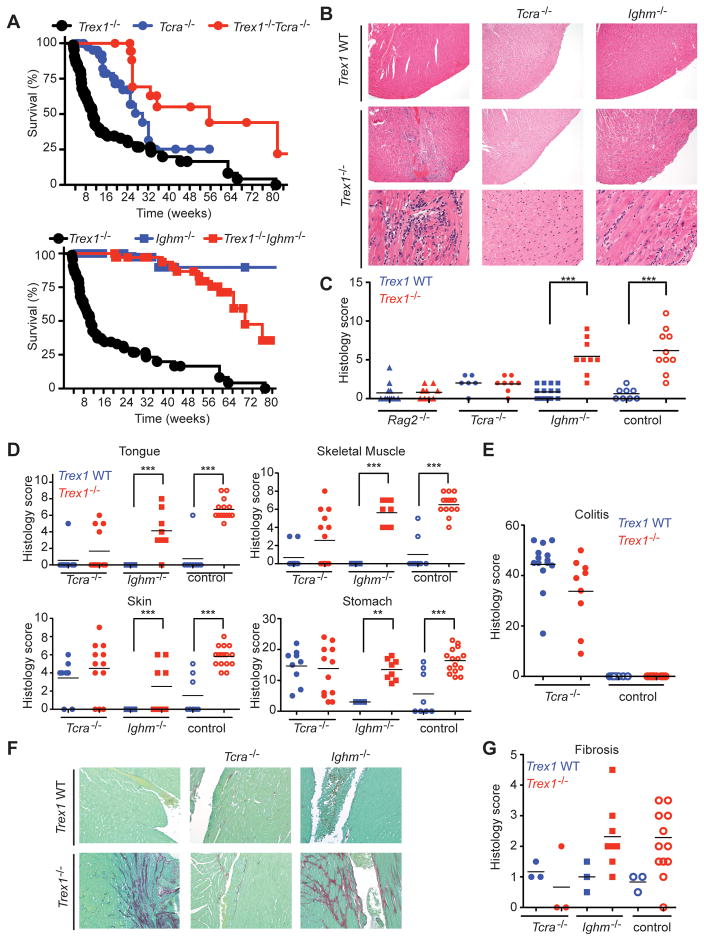
Trex1-deficient mice on a C57Bl/6 background develop a severe autoimmune disease, with a median life span of ten weeks in our colony (Morita et al., 2004; Stetson et al., 2008). Inflammatory myocarditis is evident in all Trex1−/− mice, but the extent to which other tissues are affected has not been examined. We performed a thorough histological analysis of all major tissues and organs in Trex1-deficient mice (Figure 1). In the heart, we found coalescing regions of lymphohistiocytic, degenerative and, in severe cases, fibrosing myocarditis. This myocarditis was most prominent near the endocardial surface. We also found that Trex1-deficient mice reproducibly developed profound inflammation in skeletal muscle, tongue, skin, and the glandular stomach. Similar to the heart, the skeletal muscles and tongue had lymphohistiocytic and degenerative to fibrosing myositis. In the haired skin of the muzzle, there was also mild to moderate lymphohistiocytic dermatitis, perifolliculitis and myositis. Finally, the glandular stomach had chronic lymphoid aggregates within the mucosa and moderate proliferative gastritis. Importantly, numerous organs were not affected by Trex1 deficiency, including brain, colon, small intestine, pancreas, lung, and liver (Figure 1 and unpublished data). Together, these findings reveal that the autoimmune disease in Trex1 deficient mice targets multiple, specific organs.
Figure 1. Trex1−/− mice develop specific multiorgan inflammation.
(A) Representative hematoxylin and eosin-stained (H&E) tissue sections from Trex1 WT (T1 WT, top row) and Trex1−/− mice (bottom two rows). Skeletal muscle samples were taken from the masseter. Brain sections are from the cerebellum (top panels) and the neocortex (lower panel). Original magnifications: top 2 rows 10X; bottom row 20X except for neocortex, 10X.
(B) Blinded analysis of indicated tissues from Trex1−/− (red) or WT (blue) mice. Data are represented as histological scores from individual animals and the mean (horizontal line) of experimental groups. Statistical analysis was performed using a two-tailed, unpaired Student’s t test. n.s. = not statistically significant (P > 0.05) *** = P ≤ 0.0005.
Trex1 is a specific negative regulator of STING-dependent signaling
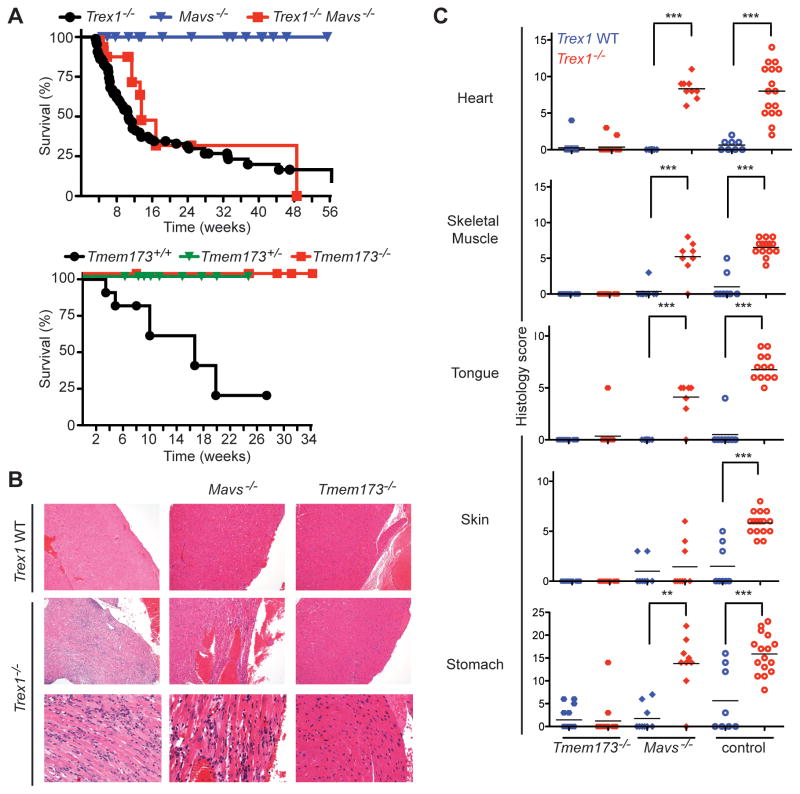
The multi-organ autoimmune disease in Trex1-deficient mice requires IRF3 and type I IFNs (Stetson et al., 2008), and extranuclear DNA accumulates in Trex1-deficient cells (Stetson et al., 2008; Yang et al., 2007). This accumulated DNA could conceivably activate two distinct antiviral responses: the ISD-STING pathway (Ishikawa et al., 2009), or the Pol-III-RIG-I-MAVS pathway (Ablasser et al., 2009; Chiu et al., 2009). To determine which of these two DNA-activated antiviral responses is regulated by Trex1, we crossed Trex1−/− mice to Mavs−/− mice and to Tmem173−/− (STING-deficient) mice. We found that Trex1−/− Mavs−/− mice developed identical autoimmune disease and succumbed with identical kinetics when compared to Trex1−/− mice (Figure 2A–2C). In contrast, Trex1−/−Tmem173−/− mice were completely rescued from mortality and autoimmune tissue destruction (Figure 2A–2C). Trex1−/− mice that were heterozygous for Tmem173 were also protected from mortality (Figure 2A), even more so than Trex1−/−Irf3+/− mice (Stetson et al., 2008). These two crosses formally establish Trex1 as a specific and essential negative regulator of the STING-dependent ISD pathway and suggest a prominent role for STING in AGS and related human diseases. Moreover, we uncover a dramatic phenotypic consequence of STING haploinsufficiency – protection from autoimmune disease – that may offer an evolutionary explanation for the recently reported existence of hypomorphic TMEM173 alleles in the human population (Jin et al., 2011).
Figure 2. Trex1 is a specific negative regulator of STING-dependent signaling.
(A) Survival curves of Trex1−/− mice, Mavs−/− mice, and Trex1−/−Mavs−/− mice. The Mavs−/− mice were generated on a pure C57Bl/6 background, so the plain Trex1−/− controls include mice generated from other contemporary crosses within this background. The Tmem173−/− mice were on a mixed C57Bl/6:129 background, and plain Trex1−/− mice generated from intercrossing Trex1+/−Tmem173+/+ mice are shown as controls.
(B) Representative H&E-stained heart tissue sections from mice of the indicated genotypes. The original magnification in the top row is 10X and in the bottom two rows is 20X.
(C) Blinded analysis of the indicated tissues of Trex1−/− mice crossed to Tmem173−/− and Mavs−/− mice. Plain Trex1−/− mice and controls are the same as in figure 1B and are presented for direct comparison to the other genotypes. For these and all other histological analyses presented below, statistical analysis was performed using a two-tailed, one-way ANOVA with Tukey’s multiple comparison post-test. * = p < 0.05, ** p ≤ 0.005, *** = P ≤ 0.0005.
Tracking the origins of type I interferon-mediated disease in vivo
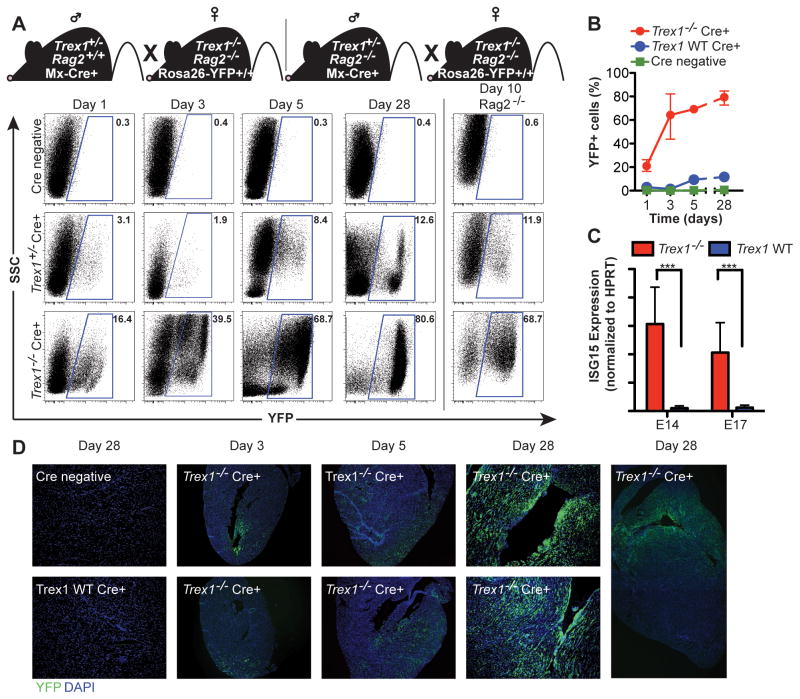
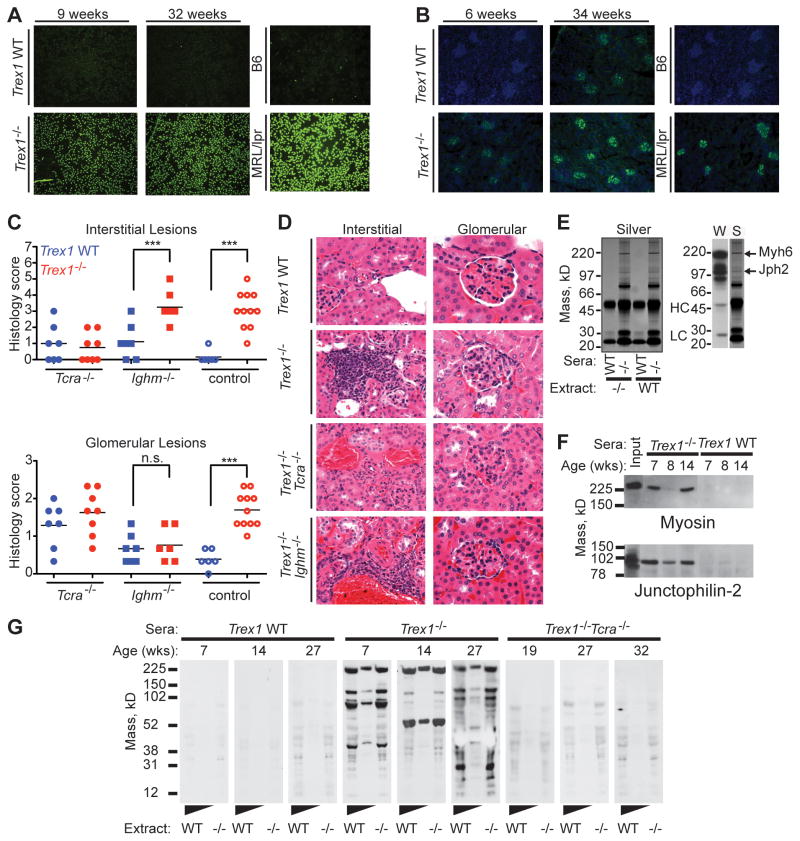
AGS in humans is strongly associated with an elevated type I IFN response (Lebon et al., 1988), and Trex1-deficient mice lacking the type I IFN receptor (Ifnar1) are completely protected from tissue damage and mortality (Stetson et al., 2008). Given the absolute requirement for STING-dependent type I IFNs in this mouse model, we performed a simple genetic cross to track the IFN response in vivo during disease initiation and progression. We bred Trex1+/− mice carrying an Mx1-Cre transgene (Kuhn et al., 1995) to Trex1−/−Rag2−/− mice homozygous for the Cre-activated Rosa26-YFP reporter allele (Srinivas et al., 2001), thus generating Trex1+/− controls and Trex1−/− mice with an in vivo reporter of IFN activity (Figure 3A). Specifically, IFN signaling activates expression of the IFN-inducible Mx-Cre transgene, which then excises the LoxP-flanked “stop cassette” in the Rosa26-YFP reporter allele, thus turning any IFN-responsive cell brightly and permanently YFP+. We examined peripheral blood from Trex1−/− and control reporter mice and found that one day after birth, 20% of circulating leukocytes were YFP+ in Trex1-deficient reporter mice, compared with only 3% YFP+ cells in control mice (Figure 3A, 3B). By day 3, the frequency of YFP+ cells in Trex1−/− reporter mice increased to over 60%, and then further increased to a maximum of almost 80% by four weeks of age. We observed a similar early emergence and accumulation of YFP+ cells in Trex1−/− reporter mice on a Rag2−/− background, which demonstrated that the IFN response was independent of lymphocytes (Figure 3A, right column). We confirmed these findings by measuring expression of ISG15, an interferon-inducible gene, in whole embryos of plain Trex1−/− mice and controls. We found that at E14 and E17, ISG15 expression was elevated in Trex1-deficient mice compared to controls (Figure 3C). Thus, the IFN response that drives disease in Trex1-deficient mice develops in utero and precedes lymphocyte-dependent inflammation and autoimmune tissue damage.
Figure 3. Origins of the type I IFN response in Trex1-deficient mice.
(A) Representative flow cytometry plots of YFP expression in peripheral blood of Trex1−/−Rag2+/− Rosa26-YFPR/WT Mx-Cre+ or Trex1−/−Rag2−/− Rosa26-YFPR/WT Mx-Cre+ and control mice of indicated ages.
(B) Percentage of YFP+ cells from Trex1−/−Rag2+/− Rosa26-YFPR/WT Mx-Cre+ and control mice of indicated ages. Data are represented as means of percentages (n ≤ 7) and range.
(C) Trex1−/− and control embryos of the indicated gestational age were analyzed by quantitative RT-PCR for expression of ISG15. Data are from 3–6 littermates per genotype and are presented as the ratio of ISG15 to HPRT. ***p ≤ 0.0005.
(D) Longitudinal analysis of YFP expression in heart tissue sections from Trex1−/−Rag2+/− Rosa26-YFPR/WT Mx-Cre+ and control mice of the indicated ages. Sections were stained with anti-GFP rabbit polyclonal antibody followed by tyramide amplification (green fluorescence), and counterstained with DAPI (blue).
Based on the detection of an early systemic type I IFN response in circulating leukocytes in Trex1-deficient reporter mice, we examined heart tissue by immunofluorescence microscopy to track the in situ emergence of this response within an organ that is strongly affected by the autoimmune disease. By day 3 after birth, we reproducibly detected (in 3 out of 4 mice examined) a localized focus of YFP+ cells near the endocardial surface of the apex of the heart in Trex1-deficient reporter mice (Figure 3D). The distribution of YFP+ cells became more widespread by day 5, but remained concentrated near the endocardial surface (Figure 3D). By day 28, we observed a dramatic expansion of reporter fluorescence with the most robust signal along the entire endocardial surface of the heart, which correlated strongly with the site of the most extensive inflammation (Figures 3D and 1A). These in situ data, while qualitative, reveal a number of important insights into the origins of disease in Trex1-deficient mice. First, the initial emergence of a detectable IFN response in the heart occurs shortly after birth in a geographically restricted subset of cardiac cells. Second, this IFN response spreads rapidly but remains localized near the endocardial surface, suggesting a process that drives the IFN response specifically in these cells but not other, nearby cells. Finally, the spatial overlap between the early IFN reporter signal and the later inflammatory infiltrate suggests a causal relationship between the site of the initial, tissue-restricted IFN response and the site of subsequent autoimmune inflammation within the target tissue.
Nonhematopoietic cells initiate autoimmune disease in Trex1-deficient mice
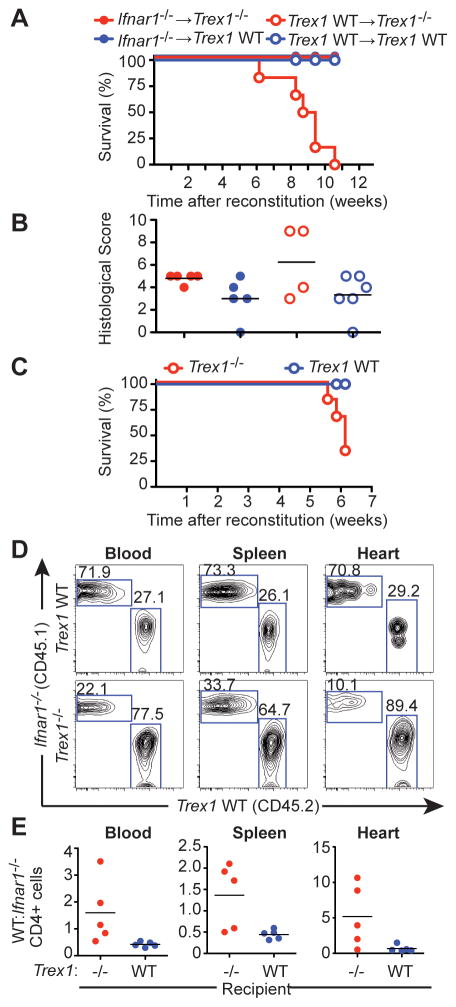
The early emergence of a type I IFN response within the endocardial region of Trex1-deficient hearts suggested that a tissue-restricted IFN response might initiate the disease process. To test this possibility, we performed a series of bone marrow chimeras to establish tissue-specific requirements for Trex1-deficiency in the progression of autoimmune inflammation. We used Trex1+/− or Trex1−/− mice on a Rag2−/− background as recipients; Trex1−/−Rag2−/− mice are completely rescued from autoimmune pathology and mortality but still initiate a type I IFN response (Figure 3A and Stetson et al., 2008), thus allowing us to examine the effects of hematopoietic reconstitution without potentially confounding our results with preexisting inflammation in the mutant recipients. We reconstituted irradiated Rag2−/− controls or Trex1−/−Rag2−/− mice with either wild-type (WT) bone marrow or Ifnar1−/− bone marrow. All of the Trex1−/−Rag2−/− mice that received WT bone marrow exhibited dramatic morbidity and mortality beginning six weeks after reconstitution (Figure 4A). In contrast, all of the Trex1−/−Rag2−/− mice reconstituted with Ifnar1−/− bone marrow remained alive and healthy. Rag2−/− control recipients survived after reconstitution with either WT or Ifnar1−/− bone marrow. Histological analysis of hearts using the same criteria described in Figure 1 revealed a radiation-induced heart inflammation that was present in all mice, regardless of the genotype of the recipient or the source of the bone marrow (Figure 4B). Within the background of this radiation-induced inflammation, we observed a slight but statistically insignificant increase in inflammation in the Trex1−/−Rag2−/− mice reconstituted with WT bone marrow (Figure 4B).
Figure 4. Non-hematopoietic cells initiate disease in Trex1-deficient mice.
(A) Survival curves of female Trex1−/−Rag2−/− or Rag2−/− mice reconstituted with either Ifnar1−/− or WT bone marrow. Graph shows results from one out of two independent experiments, each with 5 mice per group.
(B) Histological scores of hearts from Trex1−/− or WT female mice reconstituted with either Ifnar1−/− or WT bone marrow.
(C) Percent survival of Trex1−/−Rag2−/− or Rag2−/− female mice (n=5 per group) reconstituted with a ~2:1 ratio of Ifnar1−/− (CD45.1):WT (CD45.2) bone marrow. Graph is representative of one out of two independent experiments.
(D) Representative flow cytometry plots of CD45.1 (Ifnar1−/−) and CD45.2 (WT) expression by CD4+ cells in the indicated tissues of mixed bone marrow recipients.
(E) The ratio of WT:Ifnar1−/− CD4 T cells was calculated for the indicated organs. We obtained identical results when examining CD8 T cells and B cells.
We next created mixed bone marrow chimeras in which we reconstituted Rag2−/− mice or Trex1−/−Rag2−/− mice with a ~2:1 mixture of Ifnar1−/− (CD45.1):WT (CD45.2) bone marrow. Trex1−/−Rag2−/− mice that received the mixed bone marrow succumbed to disease with similar kinetics to those that received only WT bone marrow (Figure 4C), whereas the Rag2−/− recipients remained healthy. We examined the ratio of WT:Ifnar1−/− CD4 T cells in the blood, spleens, and hearts of a cohort of recipient mice euthanized six weeks post-reconstitution. The Rag2−/− control recipients maintained a ratio of WT:Ifnar1−/− cells in all three tissues that was identical to the input ratio. In contrast, we observed a strong bias towards WT CD4 T cells in the Trex1−/−Rag2−/− recipients that was most dramatic in the heart tissue (Figure 4D, 4E). This strong WT bias was also present in CD8 T cells and in B cells, again most prominently among the cells recovered from the heart (data not shown). Taken together with the in situ analysis of the IFN response in the reporter mice, these data define tissue-specific requirements for Trex1 deficiency and type I IFNs in the initiation and progression of disease. First, Trex1 deficiency in non-hematopoietic cells is sufficient to drive disease, even in the presence of a WT hematopoietic system. Second, Trex1 deficiency in hematopoietic cells, specifically lymphocytes, is not required for the autoimmune response. Third, type I IFN receptor signaling in hematopoietic cells is required to sense the IFNs produced by the initiating cells, and this signaling favors the expansion and/or recruitment of autoreactive lymphocytes to the target organ. This IFN-dependent lymphocyte expansion is identical to what has been reported for T cell responses to viral infection (Kolumam et al., 2005), suggesting that similar mechanisms mediate both protective and autoimmune lymphocyte responses triggered by innate immune detection of nucleic acids.
T cells and B cells drive inflammation and mortality in Trex1-deficient mice
The IFN-dependent appearance of lymphocytes in the hearts of Trex1-deficient recipients, together with our previous observation that lymphocytes are required for inflammation and mortality (Stetson et al., 2008), led us to explore the individual contributions of αβ T cells and B cells to the autoimmune inflammation in Trex1−/− mice. We generated Trex1−/−Tcra−/− mice and Trex1−/−Ighm−/− mice and performed an extensive survival and histological analysis of these mice and controls, compared with plain Trex1−/− mice. Tcra-deficient mice develop an inflammatory bowel disease that is caused by a population of TCRββ+ lymphocytes that respond to environmental Helicobacter antigens (Takahashi et al., 1999). Indeed, we found that the plain Tcra−/− mice in our colony suffered from increased mortality associated with severe colitis (Figure 5A, 5E). Remarkably, Trex1−/−Tcra−/− mice were rescued from mortality compared with plain Trex1−/− mice, and even compared with plain Tcra−/− mice (Figure 5A). Thus, αβ T cells are essential for the mortality caused by Trex1 deficiency.
Figure 5. Contributions of αβ T cells and B cells to autoimmunity in Trex1−/− mice.
(A) Survival curves for mice of indicated Trex1/Tcra (top panel) and Trex1/Ighm genotypes (bottom panel). Median lifespans of each Trex1−/− genotype are as follows. Trex1−/−: 10 weeks; Trex1−/−Tcra−/−: 56 weeks (p < 0.0001); Trex1−/−Ighm−/−: 73 weeks (p < 0.0001). Statistical analysis was performed using a Log-rank (Mantel-Cox) test. All mice were on a C57BL/6 background.
(B) Representative H&E-stained sections of heart apex from mice of the indicated genotypes.
(C) Blinded analysis of the endocardial region in Trex1−/− (red) or Trex1 WT (blue) mice of indicated Rag2, Tcra Ighm genotype, scored as in Figure 1B. *** = P ≤ 0.0005.
(D) Blinded analysis of tissues from Trex1−/− (red) or Trex1 WT (blue) mice of indicated Tcra Ighm genotype. * = p < 0.05, ** p ≤ 0.005, *** = P ≤ 0.0005.
(E) Colon sections from Trex1−/− (red) or Trex1 WT (blue) mice of indicated Tcra genotype were graded for inflammation, mucosal changes, dysplasia and extent of pathology.
(F) Examples of picrosirius red/fast green-stained sections of left ventricle (endocardial region) to highlight collagen (fibrosis = red staining).
(G) Endocardial and epicardial regions of the heart from Trex1−/− (red) or Trex1 WT (blue) mice of indicated Tcra Ighm genotype were graded for fibrosis. The average of the two scores was considered the total fibrosis score for each animal.
We found that Trex1−/−Ighm−/− mice that lack B cells were dramatically rescued from mortality, with a median lifespan that was over seven times longer than plain Trex1−/− mice (73 weeks versus 10 weeks; Figure 5A). However, after one year, survival of Trex1−/−Ighm−/− mice declined steadily compared to Ighm−/− littermate controls, suggesting that while B cells clearly play a central role in the rapid mortality caused by Trex1 deficiency, Trex1-deficient mice lacking B cells are not completely protected from disease.
We performed a thorough histological analysis of the affected tissues of Trex1−/− mice, Trex1−/−Tcra−/− mice, and Trex1−/−Ighm−/− mice, along with Trex1 WT controls from each cross. Remarkably, we found that αβ T cells were absolutely required for inflammation and tissue damage associated with Trex1 deficiency, but B cells were largely dispensable for this inflammation. Representative heart tissue sections in Figure 5B and histological scores in Figure 5C revealed that Trex1−/−Tcra−/− mice were completely protected from inflammation, but Trex1−/−Ighm−/− mice developed extensive heart inflammation that was statistically indistinguishable from that which developed in plain Trex1−/− mice. In the other organs affected by Trex1 deficiency, we found that Trex1−/−Tcra−/− mice were indistinguishable from plain Tcra−/− littermates, although both Tcra−/− and Trex1−/−Tcra−/− mice had elevated inflammation in skeletal muscle, skin, and stomach (Figure 5D). Importantly, examination of multiple tissues allowed us to separate the effects of Tcra deficiency from the effects of Trex1 deficiency, most notably in the tongue and heart, where we found no evidence of any inflammation in Trex1−/−Tcra−/− mice (Figure 5D, 5E). Thus, we conclude that the increased mortality in Trex1−/−Tcra−/− mice is due to the inflammatory bowel disease caused by Tcra deficiency, not the autoimmune disease caused by Trex1 deficiency. In contrast, every tissue that exhibited autoimmune pathology in plain Trex1−/− mice showed similar inflammatory infiltrates in Trex1−/−Ighm−/− mice (Figure 5D), demonstrating that B cells are dispensable for autoimmune inflammation in this model.
We next tested whether differential development of fibrotic damage could explain the dramatic rescue of Trex1−/−Ighm−/− mice from mortality without amelioration of inflammation. We stained heart tissue sections with picrosirius red, which detects collagen, and found that both Trex1−/− mice and Trex1−/−Ighm−/− mice developed fibrosis that was most extensive near the endocardial surface, but Trex1−/−Tcra−/− mice were completely protected from fibrosis (Figure 5F, 5G). Together, these data demonstrate that the inflammation and fibrosis in Trex1−/− mice absolutely requires αβ T cells but is largely independent of B cells. However, B cells dramatically accelerate mortality in Trex1−/− mice, suggesting an uncoupling of T cell-dependent inflammation from B cell-dependent processes that may drive end-stage tissue damage.
T cell-dependent autoantibody specificities in Trex1-deficient mice
We previously showed that Trex1−/− mice develop a type I IFN-dependent autoantibody response that targets heart tissue (Stetson et al., 2008). Based on the contribution of B cells to mortality in Trex1-deficient mice described above and the strong genetic association in humans between rare Trex1 mutations and SLE, we characterized the specificity of this autoantibody response. We found that Trex1−/− mice developed anti-nuclear autoantibodies (ANA), progressive IgG deposition in kidney glomeruli, and kidney inflammation, three hallmark features of lupus (Figure 6A–6C and S1). However, we did not find significantly elevated levels of anti-chromatin IgG or anti-dsDNA autoantibodies in Trex1−/− mice (Figure S1), suggesting that dsDNA reactivity was not the source of the ANA signal.
Figure 6. T cell-dependent autoantibody specificities in Trex1−/− mice.
(A) Antinuclear antibodies in Trex1 WT (top panel) or Trex1−/− (bottom panel) sera from age-matched mice. Serum from a MRL-lpr/lpr mouse served as a positive control.
(B) Kidney sections from Trex1 WT (top panel) or Trex1−/− (bottom panel) were stained with anti-mouse IgG (green) and DAPI (blue).
(C) Interstitial kidney inflammation scores (top) and glomerulonephritis scores (bottom) for mice of the indicated genotypes. Each glomerular data point represents the average score of three individual glomeruli per animal. *** = P ≤ 0.0005.
(D) Representative images of interstitial inflammation (left column) and individual glomeruli (right column) from mice of the indicated genotypes. Note that the cuboidal parietal epithelium of Bowman’s capsule is a normal finding in male mice. All images are shown at 600X magnification.
(E) Silver stain of heart extract immunoprecipitations, using sera from WT or Trex1−/− mice. Immunoglobulin heavy chain (HC) and light chain (LC) are indicated. The right panel shows a Western blot (W) of heart extract blotted with Trex1−/− sera compared to the silver stain (S) of the proteins immunoprecipitated with the same sera.
(F) Co-immunoprecipitation of heart extracts with sera from Trex1−/− or WT mice, immunoblotted with the indicated antibodies.
(G) Heart extracts from Rag2−/− (neat and 1:5 diluted) and Trex1−/−Rag2−/− mice blotted with sera from Trex1 WT, Trex1−/− or Trex1−/−Tcra−/− mice of indicated ages, and detected with HRP-conjugated anti-mouse IgG.
We investigated the contributions of T and B cells to these lupus-like features by performing a comparative analysis of kidney pathology in Trex1−/−Tcra−/− and Trex1−/−Ighm−/− mice. Similar to all other affected tissues in Trex1−/− mice, we found extensive interstitial inflammation in Trex1−/− kidneys that was most prominent in perivascular areas (Figure 6C, 6D). This interstitial kidney inflammation required T cells but not B cells (Figure 6C). We also observed mild membranoproliferative glomerulonephritis in Trex1-deficient kidneys, including hypersegmentation, expanded mesangial matrix, and increased cellularity with immune cell infiltrates (Figure 6C, 6D). Interestingly, and consistent with a causal role for immune complexes in glomerular damage, the glomeruli of Trex1−/−Ighm−/− mice were devoid of this inflammation (Figure 6C, 6D). Thus, Trex1-deficient mice develop some of the cardinal features of lupus, which provides mechanistic support for the strong genetic association between TREX1 mutations and SLE in humans (Lee-Kirsch et al., 2007; Namjou et al., 2011). Moreover, we genetically separate interstitial kidney inflammation from glomerulonephritis by showing that the former requires T cells and the latter requires B cells.
We next characterized the targets of the tissue-specific autoantibody response by immunoprecipitating autoantigens from heart extracts with sera from Trex1−/− mice. We reproducibly recovered a number of proteins using Trex1−/− sera but not control sera, two of which were identical in mass to the strongest autoantibody signals observed by western blotting of heart extracts with Trex1−/− sera (Figure 6E). We identified these autoantigens by mass spectrometry as cardiac myosin (Myh6) and junctophilin-2 (Jph2; Figure 6E). We confirmed the mass spectrometry data with western blot of heart immunoprecipitates using antisera specific for Myh6 and Jph2 and found that we immunoprecipitated Myh6 and Jph2 with multiple serum samples from Trex1−/− mice but not from WT littermate controls (Figure 6F). Thus, Trex1-deficient mice reproducibly develop autoantibodies to two abundant, heart-specific, cytosolic proteins.
We tested the contribution of αβ T cells to the heart-specific autoantibody response in Trex1−/− mice. Sera from Trex1−/− mice showed strong IgG autoreactivity by western blot with extracts from both Rag2−/− and Trex1−/−Rag2−/− hearts. In contrast, these autoantibodies were absent in Trex1−/−Tcra−/− mice, demonstrating that the autoantibody response in Trex1−/− mice is T cell-dependent (Figure 6G).
Discussion
Type I IFNs are associated with a number of autoimmune diseases in humans, including AGS and SLE. We describe here a detailed characterization of the origins and progression of the type I interferon-mediated autoimmune disease in a mouse model of AGS. We show that STING (Tmem173) is essential for disease in Trex1-deficient mice, thus formally establishing a role for the STING-dependent ISD pathway in autoimmune diseases associated with Trex1 mutations. We identify the earliest sites of IFN production and show that these IFNs act on hematopoietic cells to enable the autoimmune response. We delineate the relative contributions of T and B cells to disease and unexpectedly reveal a prominent role for B cells that is independent of T cell-mediated inflammation and fibrotic damage. We identify lupus-like features as well as tissue-specific autoantibody responses in Trex1−/− mice. Together, these findings provide an integrated picture of the development of autoimmune disease caused by Trex1 deficiency and a framework for understanding its progression, with important implications for the development of therapies for human AGS, which is currently untreatable and incurable.
We propose a model - based on the six genetic crosses that ameliorate disease and our in vivo tracking of the type I IFN response in Trex1−/− mice - that links chronic activation of the ISD pathway to lymphocyte-dependent autoimmune attack of specific target tissues (Figure S2). First, a geographically restricted subset of nonhematopoietic cells in the target tissue accumulates sufficient endogenous DNA substrates to trigger a STING- and IRF3-dependent type I IFN response. This response initiates in utero and is reliably detectable in specific tissues within a day after birth, prior to the emergence of mature lymphocytes in mice. Next, the type I IFNs produced by the initiating cells signal to hematopoietic cells and are required to drive a T cell-dependent inflammation and autoantibody response that results in a rapidly progressing autoimmune destruction of the target tissue. Genetic deletion of Tmem173, Irf3, Ifnar1, Rag2, Tcra, or Ighm rescues the mice from mortality at distinct points along this continuum. This model suggests a non-hematopoietic origin of the dysregulated innate immune response that ultimately leads to autoimmunity, and reveals how therapies targeted to specific points in this pathway might intercept disease progression. For example, efforts to block the accumulation of Trex1 substrates or inhibit the ISD pathway would act at the earliest phase of the autoimmune disease. In contrast, lymphocyte depletion would potentially and temporarily ameliorate inflammation and tissue damage, but the dysregulated innate immune response would remain.
The application of an in vivo reporter of the IFN response in Trex1-deficient mice reveals the initiation of this autoimmune disease with unprecedented temporal and spatial resolution. Perhaps most remarkably, we show that a relatively small number of cells near the endocardial surface generates the earliest detectable IFN response in the heart, raising the important question of what is unique about these cells compared to other cells in the same region. We speculate that these cells are the first to reach a threshold level of accumulated Trex1 DNA substrates that triggers the ISD pathway, and, more broadly, that the specific target organs affected by Trex1 deficiency reflect specific cell types that trigger the early IFN response within these tissues. We are currently exploring why certain cells initiate early IFN production in the target organs of Trex1−/− mice, and the ability to isolate and sort these cells based on the fluorescent reporter of type I IFN signaling will allow for a direct comparison of these cells to their uninvolved neighbors within the same tissue.
We show that both T cells and B cells contribute to the autoimmune disease in Trex1-deficient mice, but through distinct and genetically separable mechanisms. T cells are necessary and sufficient for the tissue-specific autoimmune inflammation and fibrosis, whereas B cells are not required for either of these key disease features. However, the presence of B cells is associated with a dramatic acceleration of morbidity and mortality in Trex1-deficient mice. We found that membranoproliferative glomerulonephritis in Trex1-deficient mice uniquely requires B cells, but we find it unlikely that this relatively mild glomerular damage explains the dramatic difference in mortality between plain Trex1−/− mice and Trex1−/−Ighm−/− mice. Importantly, our finding that B cells are dispensable for nearly all of the autoimmune inflammation in Trex1-deficient mice may offer a partial explanation for the disappointing results of some of the B cell-targeted therapies that are currently being trialed in SLE patients (Looney, 2010), especially given the strong genetic association of rare TREX1 mutations with this disease. We suggest that some of the inflammatory processes in a subset of SLE patients might progress in the absence of B cells, particularly in the context of established disease.
How might B cells contribute to disease in Trex1-deficient mice? Current models suggest at least three possibilities, all tied to the antigen specificity of the autoreactive B cells: antigen presentation to T cells, pathogenic autoantibodies, and production of proinflammatory cytokines. Since Trex1−/−Ighm−/− mice still develop extensive T cell-dependent inflammation and fibrosis, B cells cannot be the major antigen presenting cells in this disease. Notably, our findings in Trex1−/− mice differ from those in the MRL-lpr/lpr model of murine lupus, in which B cells are prominent antigen presenting cells and are required for tissue inflammation (Chan and Shlomchik, 1998). Thus, whereas B cells are clearly important in both model systems, the mechanisms by which they contribute to disease may be different and determined by the nature of the genetic predisposition to autoimmunity. We propose that the various (and still poorly understood) genetic contributions to human SLE will also reveal distinct mechanistic contributions of B cells, suggesting that a pharmacogenomics approach to SLE would yield more effective therapies to target these different disease mechanisms.
We identified T cell-dependent autoantibodies that target abundant, tissue-specific autoantigens, suggesting that these autoantibodies might contribute to the rapidly progressing autoimmune pathology in Trex1-deficient mice. To further explore this possibility, we injected pooled sera from Trex1−/− mice into WT recipients twice in two days and sacrificed the recipients five days after the second injection to look for signs of a type II hypersensitivity response. However, we did not find any evidence for inflammation in WT mice injected with Trex1−/− sera, suggesting that at least in this acute experimental setting, the autoantibodies do not mediate direct toxicity in an otherwise healthy recipient (data not shown). It remains possible, however, that the autoantibodies directly contribute to disease in the context of chronic, T cell-dependent inflammation. Interestingly, the major heart autoantigens in Trex1−/− mice are cytosolic proteins, which suggests that exposure of these autoantigens during inflammation-associated cell death in the target tissue might be required to reveal the epitopes detected by the autoreactive B cells. Finally, whether B cells produce relevant proinflammatory cytokines in Trex1-deficient mice is currently unknown, but can be explored in detail now that the major heart autoantigen specificities have been identified. Thus, while the precise mechanisms by which B cells contribute to disease will require further study, our findings suggest that B cells may play an important role in AGS and warrant a more thorough search for autoantibody specificities in the human disease.
In summary, we describe the initiation and stepwise development of IFN-dependent autoimmunity in a mouse model of specific human autoimmune diseases. These findings provide detailed insight into how dysregulated activation of cell-intrinsic antiviral sensors leads to autoimmune pathology, and suggest new avenues for therapeutic intervention to intercept the disease process.
Experimental Procedures
Mice
Rag2−/−, Tcra−/−, and Ighm−/− mice were purchased from Jackson Laboratories. Rosa26-YFP and Mx-Cre mice were generously provided by Dr. Alexander Rudensky and Dr. Klaus Rajewsky, respectively. Ifnar1−/− CD45.2 mice were kindly provided by Dr. Murali-Krishna Kaja. Trex1−/− mice were generously provided by Dr. Deborah Barnes and Dr. Tomas Lindahl. Tmem173−/− mice were generated as described (Ishikawa et al., 2009), and backcrossed three times to C57Bl/6 before intercrossing with Trex1−/− mice. Mavs−/− mice on a C57Bl/6 background were generated by M. Loo, M. Gale Jr, and colleagues (manuscript in preparation). All mice were maintained and used in a Specific Pathogen Free facility at the University of Washington in accordance with the guidelines of the UW Institutional Animal Care and Use Committee.
Pathology
Tissues were fixed in 10% neutral buffered formalin, paraffin embedded, cut into 5μm sections and stained with hematoxylin and eosin or picrosirius red. All tissues were coded to remove genotype identification and were evaluated for evidence of inflammation, fibrosis and in some cases mucosal lesions. Numerical scores were assigned based on degree of severity (0=normal to 5 = most severe) and extent of pathology (E1 = % of tissue affected in any manner, E2 = % of tissue affected in most severe manner). Unless otherwise indicated, the sum of individual scores was used to obtain a total tissue histological score. Please see Table S1 for more specific information about the scoring criteria for each tissue.
Bone marrow chimeras and flow cytometry
Rag2−/− or Trex1−/−Rag2−/− female mice were irradiated with 800 rads from a cesium source and reconstituted 24 hours later with 2 × 106 bone marrow cells. Mice were pretreated with and maintained on antibiotics (neomycin sulfate + polymixin B) for three weeks post reconstitution. After 6–9 weeks mice were harvested for histology and flow cytometry analysis. Spleens and hearts were treated with collagenase/DNase prior to surface antibody staining with anti-CD4 (GK1.5, BD Biosciences), anti-CD45.1 (A20, eBiosciences) and anti-CD45.2 (104, eBiosciences). Samples were acquired on a FACSCanto (BD Biosciences) and analyzed using FlowJo software (TreeStar).
Immunofluorescence and ANA/ELISA assays
For visualization of YFP reporter fluorescence, hearts were fixed in 4% paraformaldehyde overnight, cryoprotected in PBS with 30% sucrose, and frozen in Tissue-Tek OCT embedding compound (Electron Microscopy Sciences). 5 μm tissue sections were cut on a Leica CM1900 cryostat (Leica Microsystems) and stained with polyclonal rabbit anti-GFP (Abcam) followed by biotinylated donkey anti-rabbit F(ab′)2 (Jackson ImmunoResearch). YFP signal was further amplified using a FITC-TSA detection kit (Perkin Elmer) according to manufacturer’s instructions, and nuclei were counterstained with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI; Invitrogen). For visualization of immune complex deposition in the kidneys, 5 μm sections of fresh frozen tissue were fixed in 100% acetone at −20°C for 10 minutes, and then stained with anti-mouse IgG Alexa-Fluor 488 (Invitrogen) and DAPI. Digital images were acquired using a fluorescence microscope equipped with a monochrome digital CCD camera (Orca-ER, Hamamatsu Photonics) and converted to red-blue-green images using Adobe Photoshop CS3 software (Adobe Systems). Detection of antinuclear antibodies was performed with 1:200 diluted sera using an ANA (HEp-2) antigen substrate slide kit (MBL-BION) according to manufacturer’s instructions. Anti-dsDNA and anti-chromatin ELISAs were performed as previously described (Martin et al., 2007).
Quantitative RT-PCR
Embryos were harvested into RNAlater (Qiagen) and homogenized the next day using a TissueRuptor (Qiagen) followed by RNA extraction using an RNeasy kit (Qiagen) according to manufacturer’s instructions. Quantitative PCR was performed on a CFX 96 Real-Time System instrument (Biorad). The primers used were: ISG15 Sense - 5′ GGT GTC CGT GAC TAA CTC CAT 3′; ISG15 Antisense - 5′ TGG AAA GGG TAA GAC CGT CCT 3′; HPRT Sense - 5′ GTT GGA TAC AGG CCAGAC TTT GTT G 3′; HPRT Antisense – 5′ GAG GGT AGG CTG GCC TAT AGG CT 3′.
Western Blots/Co-Immunoprecipitation
Heart extracts were prepared and blotted as described previously (Stetson et al., 2008). For co-immunoprecipitations, heart extracts were pre-cleared with Protein G Dynabeads (Invitrogen) and incubated overnight with 1:200 diluted sera from Trex1−/− or control mice. Immunoprecipitated samples were then captured by Protein G Dynabeads, washed extensively and uncoupled from the beads by boiling. Samples were separated by SDS-PAGE, transferred to PVDF membranes, and incubated with anti-myosin heavy chain or anti-junctophillin-2 (Santa Cruz Biotechnology), followed by HRP-conjugated secondary antibodies.
Supplementary Material
Acknowledgments
We are grateful to Deborah Barnes and Tomas Lindahl for providing Trex1-deficient mice; to Alexander Rudensky for Rosa26-YFP reporter mice; to Klaus Rajewsky for Mx-Cre mice; to Murali-Krishna Kaja for CD45.1 congenic Ifnar1−/− mice; to Martin Prlic and Nu Zhang for help with bone marrow chimeras; to Dick Winant and the Stanford PAN facility for mass spec analysis; to Brian Johnson for expert histology preparation; to Lee Reinhardt for help with immunofluorescence; and to Yanick Crow and the Stetson lab for helpful discussions. DBS is a scholar of the Rita Allen Foundation. This work was supported by grants from the NIH (AI084914-DBS; AR48796-KBE), the European Union (FP7/2007–2013) grant agreement number 241779 to DBS (NIMBL: http://www.NIMBL.eu), and the Lupus Research Institute (DBS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan O, Shlomchik MJ. A new role for B cells in systemic autoimmunity: B cells promote spontaneous T cell activation in MRL-lpr/lpr mice. J Immunol. 1998;160:51–59. [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- de Vries B, Steup-Beekman GM, Haan J, Bollen EL, Luyendijk J, Frants RR, Terwindt GM, van Buchem MA, Huizinga TW, van den Maagdenberg AM, Ferrari MD. TREX1 gene variant in neuropsychiatric systemic lupus erythematosus. Ann Rheum Dis. 2009;69:1886–1887. doi: 10.1136/ard.2009.114157. [DOI] [PubMed] [Google Scholar]
- Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lebon P, Badoual J, Ponsot G, Goutieres F, Hemeury-Cukier F, Aicardi J. Intrathecal synthesis of interferon-alpha in infants with progressive familial encephalopathy. J Neurol Sci. 1988;84:201–208. doi: 10.1016/0022-510x(88)90125-6. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Looney RJ. B cell-targeted therapies for systemic lupus erythematosus: an update on clinical trial data. Drugs. 2010;70:529–540. doi: 10.2165/11535420-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Martin DA, Zhang K, Kenkel J, Hughes G, Clark E, Davidson A, Elkon KB. Autoimmunity stimulated by adoptively transferred dendritic cells is initiated by both alphabeta and gammadelta T cells but does not require MyD88 signaling. J Immunol. 2007;179:5819–5828. doi: 10.4049/jimmunol.179.9.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Namjou B, Kothari PH, Kelly JA, Glenn SB, Ojwang JO, Adler A, Alarcon-Riquelme ME, Gallant CJ, Boackle SA, Criswell LA, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–279. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O’Hara A, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet. 2007a;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007b;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006a;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006b;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Takahashi I, Iijima H, Katashima R, Itakura M, Kiyono H. Clonal expansion of CD4+ TCRbetabeta+ T cells in TCR alpha-chain- deficient mice by gut-derived antigens. J Immunol. 1999;162:1843–1850. [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 Exonuclease Degrades ssDNA to Prevent Chronic Checkpoint Activation and Autoimmune Disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.