Abstract
Objective
To better understand the contribution of age to the development of osteoarthritis (OA).
Methods
Surgical destabilization of the medial meniscus (DMM) was used to model OA in 12 week-old and 12 month-old male C57/BL6 mice. OA severity was evaluated histologically. RNA used for microarrays and real-time PCR was isolated from joint tissue collected from the medial side of the joint, including cartilage, meniscus, subchondral bone, and joint capsule with synovium. Computational analysis was used to identify patterns of gene expression and immunohistochemistry to evaluate tissue distribution of selected proteins.
Results
OA was more severe in older mice than young. Only 55 genes showed a similar expression with DMM-induced OA in the two age groups while 493 genes showed differential expression, the majority having increased expression in older mice. Functional categories for similarly expressed genes included extracellular matrix and cell adhesion related genes; differentially expressed genes included muscle structure and development and immunoglobulin domain genes. Comparison of expression in the sham control joints revealed an age-related decrease in matrix gene expression and an increase in immune and defense response genes. IL-33 was present in multiple joint tissue cells while CCL21 was more localized to chondrocytes and meniscal cells. Periostin was found in the extracellular matrix of cartilage and meniscus.
Conclusion
Age affects both the basal pattern of gene expression in joint tissues and the response to surgically-induced OA. Examining tissue from the joint beyond only cartilage revealed novel genes and proteins that would be important to consider in OA.
Osteoarthritis (OA) is unusual in young adults but occurs quite commonly in older adults, such that symptomatic OA affects between 10–20% of people over the age of 50 years (1). In addition to age, joint injury is a common risk factor, especially for knee OA, with a pooled odds ratio of 3.86 (95% CI 2.6–5.7) for the risk of knee OA after joint injury (2). Importantly, aging and joint injury interact. People who experienced a meniscal injury after age 30 developed radiographic evidence of OA three times faster than those who had a similar injury between 17 and 30 years of age (3). The mechanisms by which aging contributes to the development of OA and the ways in which age and joint injury interact are incompletely understood and were the subject of the present work.
Several studies have analyzed gene expression microarray data to discover the genes and pathways that are regulated at the transcriptional level during the development of OA. Studies of human OA have utilized cartilage removed at the time of joint replacement surgery, which represents end-stage disease, or from autopsies for studies of early lesions (4–7). Gene expression has also been evaluated in animal models of OA, including the rat anterior cruciate transection (ACLT) and meniscal tear models (8, 9). A limitation to these studies, is that gene expression was evaluated only in one tissue, most often the articular cartilage. It is well accepted that OA is a process that involves the joint as an organ rather than the articular cartilage alone. Microarray studies have been performed using RNA isolated from OA subchondral bone (10) and synovium (11, 12) but, like the cartilage studies, the analysis was limited to those selected tissues.
In the present study, we evaluated and compared gene expression in knee joint tissues from younger and older adult mice after the induction of OA by destabilization of the medial meniscus (DMM). The DMM model is a post-injury model described by Glasson et al (13–15) that has become popular because it involves the meniscus, which is commonly involved in human OA, and because the histological lesions within the affected joint are similar to those observed in human OA. Most commonly, young male mice (129/SvEv or C57/BL6 strains) in the age range of 8–12weeks are used in this model. Mice are considered to be skeletally mature at around 10 weeks of age, which is the age generally recommended for studies of surgically-induced OA in this species (16). However, a 10-week-old C57/BL6 mouse is the approximate equivalent of a teenaged human while a 12-month-old mouse would represent a 40 to 50 year-old human (17). Therefore, to study the effects of age on the development of post-traumatic OA, we measured OA severity histologically and analyzed gene expression by microarrays in joints from 12-week-old and 12-month-old mice which we will refer to as young and older adult mice, respectively. RNA was isolated from joint tissue that was removed from the medial (affected) side of the joint including cartilage, subchondral bone, meniscus, and the joint capsule with synovium, in order to study the joint as an organ. We identified significant histological differences in OA severity between younger and older mice as well as differences in gene expression that included genes not previously identified in OA that might play an important role in the disease process.
MATERIALS AND METHODS
Animals
Male C57/BL6 mice were purchased from Charles River Inc. (young mice) or from the National Institute on Aging aged rodent colony (older mice). All animals were housed in the same facility and provided ad libidum food and water with a 12:12hr light-dark cycle. A total of 30 animals were included in each age group (12-week-old or 12-month-old at the time of surgery). Half of the animals in each age group underwent DMM surgery and half had sham surgery, as described below. Animals were cared for following institutional animal care and use protocols and all animal studies were approved by the Wake Forest University School of Medicine Animal Care and Use Committee.
Surgical induction of OA
The DMM model of OA was established as described by Glasson et al (15) following a protocol and instructions kindly provided by Dr. Glasson. OA was induced by transection of the meniscotibial ligament. The sham controls had the same surgery except that the meniscotibial ligament was not cut. The mice were housed in cages of 5 mice per cage and allowed free activity. Mice were euthanized 8 weeks after surgery.
Histology and analysis of disease severity
Six DMM and six sham operated mice in each age group were randomly selected for histological studies. From these, four groups of joints were identified: joints that underwent DMM surgery (DMM) and their respective contralateral control joints (contralateral DMM), and joints that underwent sham surgery (sham) and their respective control joints (contralateral sham). The hindlimbs were routinely fixed in 4% paraformaldehyde, decalcified in 10% EDTA, processed, and embedded intact into paraffin for histological evaluation. The joints were sectioned in a coronal plane and serial midcoronal sections that included the femoral condyles, menisci, and tibial plateaus were cut at 4μm. Two representative sections from each joint were stained with hematoxylin and eosin (H&E) and safranin-O. Each medial and lateral tibial plateau and meniscus was graded by an evaluator, blinded to experimental group, using a recently developed murine OA grading scheme that included semiquantitative grades and quantitative measurements (18). The two semiquantitative grades consisted of an Articular Cartilage Structure (ACS) score and a Safranin-O staining score (Saf-O), both ranging in grades from 0–12, with 0 being normal and 12 indicating severe OA. The parameters evaluated quantitatively included articular cartilage (AC; thickness and area); chondrocytes (chondrocyte cell death [CCD; total area containing 2 or more necrotic chondrocytes]; total number of viable chondrocytes [#chond]; and percentage of CCD in articular cartilage [CCD%; CCD/AC area X100%]); subchondral bone (SCB; area and thickness); periarticular bone (e.g. osteophytes, OP; total area if present); and meniscus (total area and area of chondrocyte cell death, if present).
Immunohistochemistry
Serial sections immediately adjacent to those stained with H&E and safranin-O were selected from representative joints from 12 animals and were immunostained using antibodies directed against IL-33, CCL21, and periostin in order to examine protein tissue distribution. Antigen retrieval was achieved with a DAKO citrate buffer and endogenous peroxidase was blocked with 3% hydrogen peroxide. Sections were incubated with universal protein block (DAKO) and then with the appropriate primary antibody (IL-33, R&D Cat#AF3626, 1:50 dilution; CCL-21, R&D Cat#AF457, 1:100 dilution; or periostin, Invitrogen Cat#P2195, 1:200 dilution) for 30 minutes. Binding of primary antibody was detected with Biocare Goat probe for 15 minutes followed by Goat polymer for 20 minutes (IL-33 and CCL-21) or DAKO Envision+ anti-rabbit HRP Polymer for 30 minutes (periostin). All sections were developed with DAB chromagen and counterstained with Mayer’s Hematoxylin (DAKO). For negative control slides the primary antibody was substituted with negative goat (Biogenex; IL-33 andCCL-21) or rabbit (DAKO; periostin) serum.
RNA isolation and purification
Nine DMM and nine sham operated mice in each age group were used for RNA isolation. Immediately after euthanasia, the knee joints of the operative side were opened and the patella, tendons and other soft tissues around the medial side of the joint were removed. A coronal section was made through the tibial plateau and femoral condyle tissue and the medial side of the joint, including the articular cartilage, subchondral bone with any osteophytes, meniscus, and the joint capsule with synovium was collected for analysis. The tissue was placed in RNAlater® (Invitrogen) and stored overnight at 4°C. The next day the RNAlater® solution was removed and the tissue frozen at −80°C until RNA isolation. RNA was extracted from the tissue by homogenization in RNA extraction solution (UltraClean® issues and Cells RNA isolation kit from MO BIO Laboratories, Carlsbad, CA) using the Precellys 24 tissue homogenizer (Bertin Technologies purchased from MO BIO) and 1.4mm ceramic beads. The amount and quality of the RNA was determined using an Agilent 2100 Bioanalyzer. Only samples with an RIN ≥ 8 were used for analysis.
Microarrays
The Affymetrix mouse genome 430 2.0 oligonucleotide expression array chips were used. RNA samples isolated from joint tissue from three mice were pooled for analysis on each microarray chip and three separate microarray chips were run for each condition (DMM or sham) from each age group (younger or older) for a total of twelve arrays using RNA from thirty-six mice. Chip processing and image capturing was performed using Affymetrix AGCC software.
Microarray data analysis
Details of data normalization, filtering, pattern classification, and functional annotation are provided in the supplemental methods. In brief, raw microarray data was normalized using systemic variation normalization (SVN) as described (19). For each gene in each experimental replicate, the signal log ratio (SLR) was calculated as the log2 ratio of the DMM intensity to the average sham intensity. Data were then filtered according to the scheme shown in Figure 1. The SLR data associated with the filtered genes were classified into one of three groups: up, down and no change. Criteria for classification were: SLR>=0.5, up; SLR <=−0.5, down; −0.5 < SLR < 0.5, no change (NC). Once each SLR value was classified, the gene was placed into one of eight patterns (Figure 1). In order to evaluate changes in gene expression due to age, independent of surgically-induced OA, differences in expression between the young and old sham groups were also evaluated(see supplemental methods).
Figure 1.
Flowchart of filtering process. As described in the Methods, the 45,101 probe sets on the Affymetrix arrays were filtered, in a step-wise process, to identify genes that responded similarly or differently to DMM treatment between younger and older mice. Step 1 identified all genes with a signal that was significantly detected on the chip over the background. Step 2 identified significantly expressed or repressed genes in DMM joints, with respect to sham, in 2 or more replicate pools. Step 3 identified genes that either responded in a similar fashion between young and older mice, or had a significantly different response. These genes were then assigned to an expression pattern in Step 4for further analysis.
A functional analysis was performed for the genes in each pattern defined in Figure 1 and for the sham-sham comparison. The Functional Annotation Clustering tool provided by DAVID(20, 21)was used to obtain gene annotations that were significantly over-represented in each pattern as compared to the full list of genes on the Affymetrix Mouse 430 2.0 GeneChip (used as background). Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) software version 8.8 (content version 3204, 10/27/2010) was used to identify canonical pathways that were significantly over-represented. The gene list for each pattern was input into IPA and a Core Analysis was run on each.
Real time PCR
Samples of RNA from the same pools used for the microarrays were used for real-time PCR. The SYBR® Green-based method was used with optimized primer sets obtained in the RT2 qPCR Primer Assays (SABiosciences/Qiagen) with TATA box-binding protein (TBP) used to normalize relative expression as previously described(22).
Statistical analysis
Statistical analysis methods for the microarray data are provided above and in supplemental methods. Histological data from both the lateral and medial tibial plateaus was evaluated with ANOVAs (intra-animal comparisons) and repeated measures tests (inter-animal comparisons) using SPSS version 15.0 (IBM Corp, Somers, NY). The real time PCR results were analyzed using ANOVA with StatView 5.0 software (SAS, Cary, NC).
RESULTS
Development of osteoarthritic changes in young and older mice
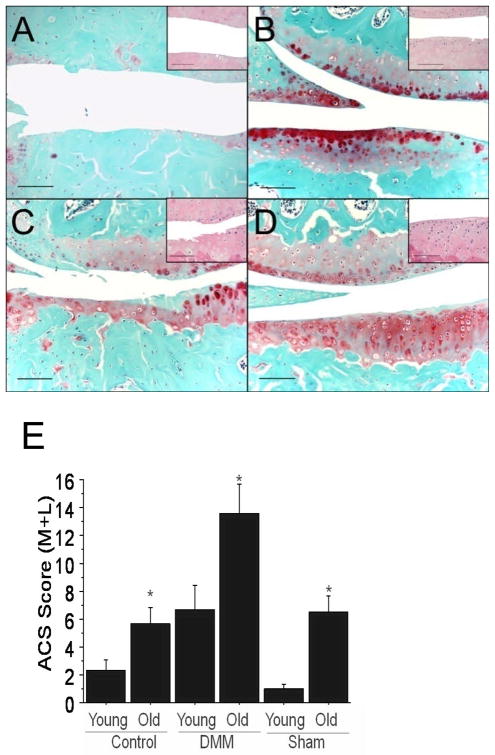
Both young and older mice exhibited typical histological features of OA in the DMM knees including cartilage surface fibrillation and clefting, medial meniscal degeneration, osteophyte formation, and subchondral bone thickening (Figure 2A–D). OA lesions were consistently more severe in the medial tibial plateau than the lateral tibial plateau and were more severe in the 12-month-old mice than in the 12-week-old mice. The average articular cartilage structure score (medial plus lateral) was about 2-fold greater in the DMM knees of the older mice than in the younger mice (Figure 2E).
Figure 2.
Histological evaluation of osteoarthritis in medial knee joint compartments of 12-week-old and 12-month-old mice. Younger and older mice underwent DMM or sham surgery and OA severity was evaluated after 8 weeks. Sections were stained with H&E or Safranin-O. A) 12-month-old DMM, B) 12-month-old sham, C) 12-week-old DMM, D) 12-week-old sham. Bar = 100 microns; insert is a higher magnification (20×) image of a central area of the medial tibial plateau of the H&E-stained section. E) Articular cartilage structure (ACS) scores were calculated on a 0–12 scale for the medial and lateral sides of the joint. Controls are the contralateral (non-operated) limb. Results are mean ± SEM presented for the medial plus lateral tibial plateaus. p < 0.05 by ANOVA.
In both the 12-week and 12-month-old DMM groups, the medial tibial plateaus had significantly higher areas and thicknesses of SCB (p<0.001 for both), higher CCD% (p=0.001 for both), lower total number of viable chondrocytes (p=0.01 for both), and larger abaxial osteophyte size (p<0.05 for both) compared with the medial tibial plateaus from the corresponding sham joints (Table S1). In addition, the medial tibial plateaus of the 12-month-old DMM joints had significantly reduced articular cartilage (p<0.001 for area & thickness) and higher ACS scores (p=0.008) than the lateral tibial plateaus (data not shown). Because the most severe lesions were present medially, only results from the medial tibial plateaus are presented in Table S1 for the surgical treatment group comparisons unless otherwise noted. In addition, no significant differences between the contralateral DMM and contralateral sham groups were identified in the younger mice. Therefore, data from these groups were combined into a single “Control” group for the young mice.
The sham-operated knees in the 12-week-old mice did not exhibit any significant changes of OA; however, the sham-operated joints in the 12-month-old mice demonstrated mild osteoarthritic-like changes, suggesting an age-related development of early spontaneous OA (Figure 2B). This was unlikely to be a result of the sham surgery since similar changes were seen in the sham contralateral (unoperated) knees of the older mice. In addition, in the older mice, the DMM contralateral joints exhibited significantly more severe OA than the sham contralateral joints, suggesting an effect of the DMM surgery on the contralateral limb in older mice. Additional histological findings are presented in the supplemental results.
Patterns of gene expression in knee joint tissue from young and older mice with surgically-induced OA
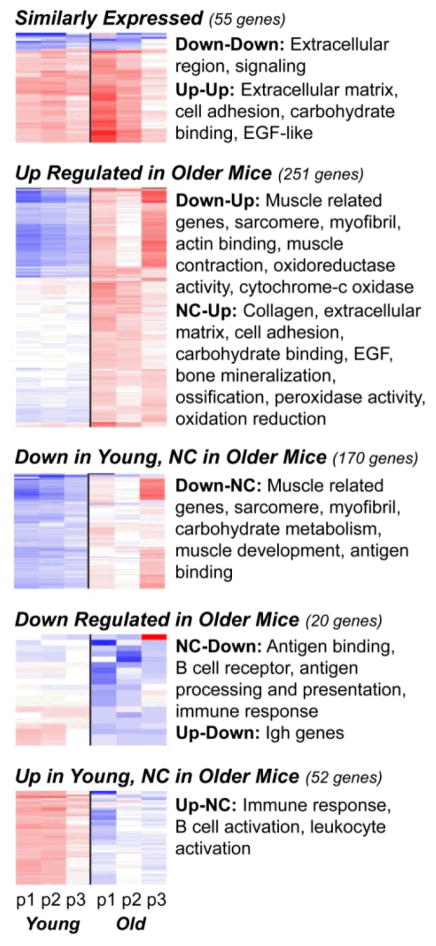
A four-step filtering and classification process (Figure 1) identified those genes that were significantly over-or under-expressed compared to sham surgery and classified them into one of eight patterns. The SLR distributions illustrate a key observation: more genes were up-regulated in the RNA from older mice, while more genes were down-regulated in the younger mice. The complete list of genes for each pattern and their DAVID annotations are provided in Supplementary File 2. The number of genes in each of the replicate pools from young and older mice that were up- or down-regulated or showed no change are provided in Figure S2 and heat maps illustrating levels of expression are shown in Figure 3 along with functional annotations significantly over-represented in each pattern identified using DAVID (20, 23).
Figure 3.
Heat maps of the genes identified in each expression pattern. Each group summarized in Step 4 of Figure 1 is represented here as a heat map with a summary of annotation terms found to be significantly over represented in each pattern by DAVID. Columns represent replicate arrays of Young mice (left 3 columns) and Older mice (right 3 columns) with each row representing a single gene. Blue=down regulation, Red=up regulation and White=no change in expression as compared to sham.
Genes regulated similarly in both 12-week and 12-month-old DMM mice relative to sham controls
55 genes were either up-regulated or down-regulated in both young and older mice. There were only eight genes in the down-down pattern. Ingenuity Pathway Analysis (IPA) identified them as belonging to metabolic pathways (citrate cycle, pyruvate, and linoleic acid metabolism), as well as the complement system (complement factor D) and metabolism of xenobiotics.
The 47 genes in the up-up pattern demonstrated significant functional annotations for extracellular matrix and cell adhesion related genes, carbohydrate binding and EGF-like proteins. Genes in this pattern included asporin, chemokine (C-C motif) ligand 21A, several collagen genes including types III, VI and XIV; dickkopf homolog 3 (DKK 3), HtrA serine peptidase 1, insulin-like growth factor 1 (IGF-1), interleukin 33, lumican, matrilin 2, matrix metalloproteinase (MMP) 2, MMP-3, periostin, and tissue inhibitor of metalloproteinase (TIMP) 1. The canonical pathways identified for these 47 genes included pathways involved in tissue remodeling including genes also classified as involved in rheumatoid arthritis (IL33, IGF1, MMP3, DKK3, MMP3, MMP2) and several signaling pathways, including IGF-1, IL17, and HIF1α. The most significant pathway identified was “hepatic fibrosis” (IGF1, TIMP1, CCL21, MMP2, COL3A1).
Genes with higher expression in older DMM mice compared to young DMM mice
The young-old patterns of NC-up, down-NC, and down-up included 421 genes for which relative expression was higher in older mice. These three patterns had similar functional annotations that included muscle, sarcomere, myofibril, and collagen-related genes. The largest gene pattern, down-NC, included 170 genes exhibiting functional groups dominated by muscle structure and development and carbohydrate metabolism. Both DAVID and IPA analysis indicated that genes involved in ion and metal homeostasis, particularly calcium signaling genes, were significantly represented. Pathway analysis also indicated that signaling genes were significantly represented, including genes involved in ILK signaling, protein kinase A signaling, actin-cytoskeleton signaling, and CDK5 signaling. Genes in this pattern also included two chemokine receptors (CCR1 and CCR2) and a chemokine ligand (CXCL10), two collagen genes (type XI and type IV), four interferon-related genes, S100b, glutaredoxin 3, and suppressor of cytokine signaling(SOCS)-6.
The NC-up (young-old) pattern was the second largest with 156 genes. DAVID annotation groups included collagen, extracellular matrix and cell adhesion, signaling (EGF), glycosaminoglycan binding, and bone mineralization. One DAVID annotation group contained genes associated with peroxidase activity and oxidative stress, suggesting perhaps a redox component to this process. Genes in this pattern included aggrecan, multiple collagen genes including types II, V, VI, XXII, XXIV, and XXVII; DDR1, matrilin-2, TRAF6, IL-13 receptor, prostaglandin-endoperoxide synthase 2, PDGF, peroxiredoxin 4, F-spondin, syndecan 1, thrombospondin 2 and 4, very low density lipoprotein receptor (VLDLR), and Wnt inhibitory factor 1.
The down-up (young-old) pattern was the third largest, containing 95 genes. As with the NC-up and down-NC patterns this gene group exhibited muscle-related annotations and calcium signaling pathways. However, this pattern also included genes such as cytochrome c oxidase genes, collagen type IX, epiphycan, hyaluronan and proteoglycan link protein 1, matrilin 3, titin, and Ucma (unique cartilage matrix-associated protein). IPA pathways included calcium signaling, actin cytoskeletal signaling, and protein kinase A signaling. A unique pathway associated with this pattern was oxidative phosphorylation.
Genes with higher expression in young DMM mice compared to older DMM mice
72 genes displayed higher expression in young mice relative to the older mice: NC-down, up-down, and up-NC (young–old). These three patterns differed from the previous patterns in that the annotations were focused on genes related to the immune response. The largest group in this pattern, up-NC, contained 52 genes which had functional annotations related to immune system development, immune regulation, and leukocyte activation. IPA analysis revealed two primary pathway groups: those associated with B-cells (including B-cell development, various types of signaling in B-cells, and altered B-cell signaling in rheumatoid arthritis) and those associated with phospholipid metabolism and signaling. Examples of genes in this category include CD19, CD5l, CD79, IL-7R, Fas apoptotic inhibitory molecule 3, peroxiredoxin 2, sphingosine kinase 1, and pre-B lymphocyte gene 1.
The two smallest patterns were the NC-down (18 genes) and up-down (2 genes) patterns. The NC-down annotations related to immunoglobulin, immune response-related processes, disulfide bond and transmembrane proteins. Genes of potential interest included the chemokine CXCL13, dual specificity phosphatase 12, phospholipase A2, group IID, radical S-adenosyl methionine domain containing 2 and immunoglobulin and histocompatibility genes. Only one gene (two probe sets) was up-regulated in young mice and down-regulated in older mice: immunoglobulin heavy chain complex.
Age-related differences in gene expression independent of surgically-induced OA
Differences in expression between the young and old sham control joints were examined and revealed significant age-related differences in gene expression. Table 1 provides a list of the ten most differentially expressed genes in the older sham adult compared to the young sham adult mice. A complete list of the 861 differentially expressed genes and their DAVID annotations can be found in Supplemental File 3. Genes expressed at higher levels in sham joints of older mice included three chemokines in the top ten: CXCL13 (B lymphocyte chemoattractant), CCL8 (MCP-2), and CCL5 (RANTES). DAVID analysis of 430 genes expressed at higher levels in older mice included annotations primarily for immune response and defense response such as response to wounding and inflammatory response. The increase in chemokine, cytokine (e.g. IL-6) and several HLA genes resulted in the top IPA canonical pathways listed as”altered T Cell and B Cell Signaling in Rheumatoid Arthritis”, “Systemic Lupus Erythematosus Signaling”, and “Graft-versus-Host Disease Signaling”.
Table 1.
List of genes with the highest and lowest signal log ratios (SLR) when comparing RNA from older sham control joints to younger sham control joints*
| Genes increased in old sham | Avg. SLR | Genes decreased in old sham | Avg. SLR |
|---|---|---|---|
| Hba-a1/2 (hemoglobin alpha, adult chain1/2) | 5.18 | Col9a1(collagen, type IX, alpha 1) | −4.14 |
| CXCL13 (B lymphocyte chemoattractant) | 2.66 | Matn3 (matrilin 3) | −3.89 |
| CCL8 (MCP-2) | 2.51 | Lect1 (leukocyte cell derived chemotaxin 1) | −2.83 |
| Pla2g2d (phospholipase A2, group IID) | 2.23 | Col9a3 | −2.82 |
| Igh-6 (immunoglobulin heavy chain 6 (heavy chain of IgM) | 2.2 | Hapln1 (hyaluronan and proteoglycan link protein 1) | −2.76 |
| Cd51 (alpha V integrin) | 1.95 | Col2a1 (collagen, type II, alpha 1) | −2.43 |
| Igh (immunoglobulin heavy chain complex) | 1.94 | Myh2 (myosin, heavy polypeptide 2, skeletal muscle, adult) | −2.43 |
| CCL5 (RANTES) | 1.92 | Epyc (epiphycan) | −2.33 |
| Ighv14–2 (immunoglobulin heavy variable V14-2) | 1.72 | Mup1(major urinary protein1) | −2.3 |
| H2-Aa (histocompatibility 2, class II antigen A, alpha) | 1.71 | Prkg2 (protein kinase, cGMP-dependent, type II) | −1.95 |
| Faim3 (Fas apoptotic inhibitory molecule 3) |
SLRs were calculated using the sham young and old microarray results as described in the methods. The eleven highest (last two genes had the same SLR) and ten lowest expressing genes in old relative to young are shown.
The 431 genes expressed at the lower levels in sham control joints of older compared to younger mice were primarily extracellular matrix genes such as type IX and type II collagen, matrillin 3, and link protein (Hapln1). DAVID analysis of this group revealed annotations for the extracellular matrix, metabolic processes, and tissue development, including cartilage development. Genes in the latter category included TGFβ2, aggrecan, SOX-9 and types II and XI collagen. Other relevant genes decreased with age were HMGB2, osteoprotegerin, and cartilage oligomeric protein (COMP). IPA canonical pathways included “calcium signaling” and “RhoA signaling’.
Real-time PCR measurement of selected genes
We selected 21 genes to measure using real-time PCR based on differences noted in the arrays and/or their potential importance in the OA process and compared expression between sham and DMM joints within an age group as well as between young and old (Table 2). Asporin (which inhibits TGF-β and has been implicated in OA) and ADAMTS5 (aggrecanase-2) had a greater increase with DMM in the old. Other genes which were increased in the DMM joints of old but not young mice included aggrecan, CXCR7, the HtrA serine peptidase 1, secreted frizzled-related protein 2, and periostin. Genes increased in DMM joints in both young and old included CCL21, IGF-1, and MMP-3 while genes increased only in young and not old were CCR7, DKK-3, and IL-33. Also of interest was that the cartilage matrix gene aggrecan was expressed at lower levels in the sham old than young while CCR7, CXCR2, insulin degrading enzyme, and IL-33 were expressed at higher levels in sham old than sham young. Type X collagen is a marker of chondrocyte hypertrophy and OA chondrocytes are thought to assume a hypertrophic phenotype but we did not see a significant difference in expression in either younger or older DMM mice nor did we find an increase in MMP-13. However, a previous study examining older C57BL/6 mice with spontaneous OA also did not find an increase in type X collagen (24) and, unlike MMP-3 and ADAMTS5, MMP-13 expression may not increase until more advanced stages of OA (4). Type III collagen RNA was increased in the DMM joints of both young and old but only reached statistical significance in the old.
Table 2.
Real-time PCR results (mean (SD)) for selected genes shown as relative expression levels normalized to expression of TATA box-binding protein (TBP)
| Gene name | Sham Young | DMM Young | Sham Older | DMM Older |
|---|---|---|---|---|
| ADAMTS5 | 5.6 (.5) | 4.8 (.1) | 8.2 (3) | 11.0 (2)# |
| Aggrecan | 1.79 (.2) | 1.9 (.3) | 0.69 (.1)** | 1.669 (.6)* |
| Asporin | 1.0 (.1) | 1.6 (.3) | 1.3 (.5) | 3.22 (1)*# |
| CCL21 | 49.7 (15.6) | 112.8 (6.2)* | 62 (3.5) | 99 (11.3)* |
| CCR7 | 0.42 (.2) | 0.77 (.1)* | 0.82 (.1)** | 0.84 (.01) |
| CXCR2 | 3.0 (.7) | 2.6 (.3) | 18.6 (.6)** | 16.7 (.2)# |
| CXCR7 | 15.4 (2.3) | 23.3 (2.7) | 22.2 (4.8) | 36.5 (8.9)*# |
| COL3 | 53.0 (5.8) | 102.1 (12.7) | 56.6 (18.1) | 195.6(91)*# |
| COL10 | 0.4 (.2) | 0.7 (.4) | 0.5 (.1) | 0.2 (.3) |
| DKK 3 | 11.3 (3.1) | 20 (6.8)* | 14.8 (2.7) | 22.5 (3.9) |
| DEL1 | 6.6 (1.4) | 5.7 (.39) | 5.4 (.35) | 7.7 (1.6) |
| Htra1 | 59.4 (13.1) | 107.6 (10.1) | 80.6 (22.1) | 216.8 (108)*# |
| IDE | 35.1 (4.8) | 43.3 (6.4) | 71.6 (9.5)** | 73.4 (13.3)# |
| IGF1 | 0.02 (.005) | 0.06 (.007)* | 0.02 (.004) | 0.04 (.006)*# |
| IL33 | 0.46 (.12) | 0.74 (.2)* | 0.76 (.01)** | 0.81 (.2) |
| MMP3 | 19.0 (5.8) | 44 (5.8)* | 24 (3.0) | 46 (8.4)* |
| MMP13 | 75.7 (9) | 82.1 (20) | 76.7 (23) | 99.7 (18) |
| Nox4 | 15 (6.6) | 12 (2.1) | 9.5 (3.7) | 11 (3.9) |
| Periostin | 6.8 (.62) | 9.8 (1.1) | 14.9 (3.8) | 25.2 (8.9)*# |
| Sfrp2 | 11.1 (1.5) | 17.3 (2) | 17.6 (6.9) | 25.7 (2.7)*# |
| TSG6 | 2.6 (.6) | 4.1 (.4) | 3.4 (.98) | 7.8 (4.8) |
DMM=destabilized medial meniscus, ADAMTS5=A Disintegrin And Metalloproteinase with Thrombospondin motif5, CCL21=chemokine (C-C motif) ligand 21, CCR7=chemokine (C-C motif) receptor 7, CXCR2= chemokine (C-X-C motif) receptor 2, CXCR7=chemokine (C-X-C motif) receptor 7, COL3=collagen type III, COL10=collagen type X, DKK3= dickkopf homolog 3, DEL1= EGF-like repeats and discoidin I-like domain, Htra1= HtrA serine peptidase 1, IDE=insulin degrading enzyme, IL33= interleukin 33, MMP3=matrix metalloproteinase-3, MMP13= matrix metalloproteinase-13, Nox4= NADPH oxidase-4, Sfrp2=secreted frizzled-related protein 2, TSG6=TNF, alpha-induced protein 6
Significantly different from age-matched sham p<0.05,
Significantly different from young sham p<0.05
Significantly different from young DMM p<0.05
Immunohistochemical analysis
We selected three genes from the list validated by real-time PCR in order to examine protein tissue distribution. The negative control sections did not exhibit any immunostaining (not shown). There was very strong cellular staining in chondrocytes in articular cartilage, meniscus, and growth plate, and in synovial cells for IL-33 (Figure 4). IL-33 was variably positive in osteocytes, osteoblasts, skeletal muscle, vascular endothelium and hemopoietic cells. CCL21 had a much more localized distribution with immunostaining in articular cartilage chondrocytes, meniscal cells and growth plate chondrocytes. Positive immunostaining was also present in vascular endothelium, very low numbers of osteocytes, growth plate matrix and (minimally) in skeletal muscle. Periostin was present in cartilage matrix (articular cartilage, meniscus, growth plate), low numbers of chondrocytes in these tissues, and in the periosteum some osteoblasts were also positive. We did not examine sufficient numbers of sections from the four groups to determine the effects of age and OA on the staining intensity but rather plan to do so as part of future studies using these and additional proteins.
Figure 4.
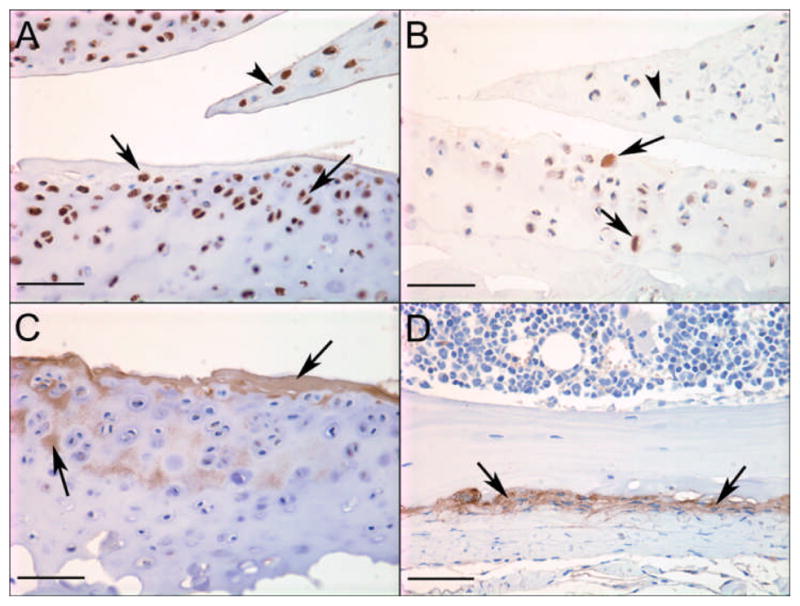
Immunohistochemical staining for selected proteins in mouse knee joints. Strong positive staining of chondrocytes in articular cartilage (arrows) and meniscus (arrowhead) for A) IL-33 in a young DMM mouse and B) CCL21 in an old sham mouse; C) positive staining for periostin in articular cartilage matrix (arrows) in a young DMM mouse and D) periosteum (arrows)in a young sham mouse; Bar = 50μm.
DISCUSSION
Similar to humans, we found that age contributes to the development of both spontaneous and injury-induced OA in mice. Mild OA-like pathology was present in the unoperated knees of the 12-month-old mice suggesting that mice at this age are in the early stages of developing naturally occurring OA, similar to what has been described in pathological studies of human knees at the equivalent age of approximately 40 years-old (25). The older mice also exhibited more severe histological OA after a joint injury that destabilized the medial meniscus, which is also consistent with studies on the development of OA after joint injury in humans (3) and supports the use of this model to study age-related differences. More severe OA lesions were also noted in the contralateral knee of older DMM mice relative to the sham operated knees which could have been due to either gait changes or systemic factors released from the DMM joint, suggesting that contralateral joints should not be used as controls in mouse models of surgically-induced OA.
Annotations of genes expressed at lower levels in the sham controls of older animals included extracellular matrix, metabolic processes and tissue development with a predominance of cartilage extracellular matrix genes including types II, IX and XI collagen, aggrecan, COMP, and link protein, as well as the transcription factor SOX-9 and the growth factor TGFβ2. These findings are consistent with studies demonstrating an age-related decline in matrix production when chondrocytes are stimulated with anabolic factors (reviewed in(26) and with a prior study showing reduced immunostaining for TGFβ2 in the articular cartilage of older mice (27). We also noted a decrease in HMGB2 consistent with a previous study in mice that showed decreased levels may contribute to chondrocyte death in the superficial zone (28). Genes expressed at higher levels in sham controls of older mice included chemokine and HLA genes and annotations for immune and defense response. CXCR2, which serves as an IL-8 receptor and has been found to play an important role in promoting cell senescence (29) was also increased in the sham joints of older mice.
In the surgically-induced OA joints, more genes were significantly up-regulated in the older mice(421) compared to younger mice (72) potentially indicating a more active disease process. Genes increased in older OA joints included extracellular matrix genes, such as aggrecan and type II collagen, that were decreased with age in the sham controls. DAVID and IPA analysis of the relatively smaller set of genes that were up-regulated in younger mice with OA (but not older) identified significant annotations for immune response genes and B cell signaling while these annotations were found in genes expressed in the older sham joints. Genes with muscle-related annotations and genes involved in calcium signaling were more often down-regulated in the younger mice with OA while these genes were upregulated in older DMM joints. Because we did not include muscle in the tissue used for RNA isolation, muscle-related genes are most likely being expressed by other tissues including cartilage as previous studies have noted chondrocyte expression of genes such as alpha-smooth muscle actin (30).
A unique aspect of the present study was that the RNA isolated for microarray analysis was extracted from the multiple tissues that make up the joint rather than from a single tissue. Although this approach might be less sensitive in detecting genes that were up-or down-regulated in a single tissue and might limit the ability to determine which particular tissue contributed to expression of a specific gene, it has the advantage of discovering genes that are more globally involved in the OA process. Despite the potential limitations, aggrecan, link protein and type II collagen, genes primarily expressed in articular cartilage and in the inner zone of the meniscus, were detected on the arrays as being up-regulated in older DMM mouse joints, even though cartilage and meniscus loss was greater in these mice than in the younger mice.
Periostin was up-regulated in older but not younger DMM joints. Periostin is a vitamin k-dependent (Gla containing) protein produced by osteoblasts and also found in the periosteum (31). We confirmed its presence in periosteum and osteoblasts and in addition found periostin was present in the cartilage matrix and in low numbers of chondrocytes. Periostin appears to play a role in diverse processes including tooth development (32), cancer metastasis (33), and tissue repair after injury, such as repair of heart tissue after a myocardial infarction (34). Likewise, secreted frizzled-related protein 2 (sFRP2), which can augment Wnt3a signaling (35) and was increased in the DMM joints from older mice, has also been shown to be involved in myocardial repair (36). The increased expression of these and other genes involved in extracellular matrix formation and tissue repair indicates an active repair response in the knee joints of the older mice.
IL-33 was found to be up-regulated in the DMM joints of the younger but not older mice although its expression was higher in older vs. younger sham joints. IL-33 is a member of the IL-1 superfamily and is thought to serve as an alarmin in a number of inflammatory diseases, including RA (reviewed in (37)) but, to our knowledge, has not been reported to be up-regulated in OA. In RA, IL-33 was found in synovial tissue, however it can also be expressed by osteoblasts, where it is thought to inhibit bone resorption (38). We found that, although fairly widely distributed in joint tissues, there was strong and consistent positive immunostaining for IL-33 in articular and growth plate chondrocytes and meniscal cells suggesting it may function in these tissues as well.
The 55 genes that were similarly expressed in DMM relative to sham knees of younger and older mice were involved in extracellular matrix remodeling and included up-regulation of genes involved in matrix degradation such as MMP-2, MMP-3, and HtrA serine peptidase 1. Although not as extensively studied as the MMPs, several serine proteases, including HtrA1, have been shown to be increased in human OA cartilage (39). HtrA1 has been implicated in degradation of fibronectin (40)and aggrecan(21) and was also found by others to be increased in the cartilage of mice 8 weeks after DMM surgery (41). In addition, HtrA1 was a prominent protease found in a proteomic analysis of human OA cartilage (42). CCL21 is a novel chemokine gene found to be increased in both young and old DMM joints which, by immunostaining, was localized to chondrocytes and meniscal cells and the growth plate matrix. CCL21 is a ligand for CCR7 which was also increased in the young DMM joints and in the old sham control joints. A recent study noted elevated levels of CCL21 in both RA and OA synovial fluid compared to normals with higher levels in RA (43).
In a recently reported cartilage microarray study that used the rat meniscal tear model of OA, Wei et al (9) integrated their findings with previous microarray studies that had reported differential gene expression in the rat ACLT model (8) and in human OA cartilage (5) and generated a list of 20 OA genes that were in common in the human study and at least one rat model. We searched for those 20 genes in our list of 548 genes that were differentially expressed in either younger or older DMM mice and in the list of 861 genes differentially regulated by age in the young and older sham joints. We found 7 genes in common in the DMM list (COL3A1, COL6A2, lumican, MMP-3, NDRG2, PCOLCE, and TIMP1), 6 were differentially regulated in the older DMM mice and 5 in the young. We also found 4 genes in the sham list differentially regulated with age in the same direction as differential regulation in the OA list (LTBP2, NDRG2, SERPINA1, and TIMP1). Given the importance of TGFβ in joint tissues, the increase in latent TGFβ binding protein 2 (LTBP2) in OA and with age may be particularly important.
We also compared our DMM gene list with a list of 150 genes noted to be differentially expressed in human OA subchondral bone reported by Hopwood et al(10)and found 4 genes in common (CCR2, crystallin alpha B, synuclein alpha, and tyrosyl-tRNA synthetase). Of these, the expression patterns matched for young but not old DMM joints for crystallin alpha B and tyrosyl-tRNA synthetase (down-regulated) and synuclein alpha (up-regulated). However, we found more genes (ten) in common in a comparison of the bone list with the sham young vs old list with eight genes having the same expression pattern (crystallin alpha B, guanine nucleotide binding protein alpha Z, glycoprotein V, lymphotoxin beta, matrix extracellular phosphoglycoprotein with ASARM motif, RAB27B, selectin P, and tubulin, beta 1). Finally, we made a similar comparison between the mouse joint microarray gene lists and a list of 260 genes found to be differentially expressed by ≥2-fold between human synovial tissue from inflamed and non-inflamed biopsy specimens taken from subjects undergoing meniscectomy for meniscal injuries (12). A total of 30 genes were present in both the synovial gene list and the mouse joint lists with 8 genes regulated in the same direction in synovium and the DMM/sham lists (Table S2). One of those was CCL21 which our immunohistochemical staining noted to be present in articular cartilage suggesting this gene is upregulated in more than one tissue in joints with meniscal damage. The comparison of genes regulated in the same direction between the sham old-young and the human synovial samples found 20 genes in common demonstrating that a significant number of genes upregulated with age in the joint are also found in inflamed synovia from meniscal injuries. This included the IL-7 receptor which we had previously shown to be expressed by human chondrocytes(44).
In summary, the analysis of gene expression in joint tissues in a meniscal injury model of OA found OA-related genes that had been previously reported in other animal models and in human OA but also revealed novel genes and pathways that could be important in the OA process. The results also demonstrated clear age-related transcriptional differences in both the sham control joints and the DMM joints. These findings demonstrate the importance of age when considering the results of animal model studies of OA. Most studies that have utilized transgenic mice to study the role of specific genes in OA have used animals at an age similar to our younger group. Genes and pathways important in the OA process may be missed if only young animals are used in such studies. Age at onset of joint injury clearly affects the way in which the cells within joint tissues respond. The older animals exhibited a very active response to joint injury, including the upregulation of matrix genes, chemokines and matrix degrading enzymes, consistent with the concept that OA is not a degenerative disease but rather a condition that activates remodelling of joint tissues. Further studies of the differences in the transcriptional response between younger and older joints should help elucidate the mechanisms underlying the contribution of age to the development of OA.
Supplementary Material
Acknowledgments
Supported by an Innovative Research Award from the Arthritis Foundation, The Wake Forest University Translational Science Institute and by the National Institute on Aging (RO1 AG16697).
The authors thank Mary Zhao for technical assistance and the microarray core facility of the Comprehensive Cancer Center of Wake Forest University for performing the microarrays. We also thank Dr. Sonya Glasson for instructions on the DMM model, Drs. Carla Scanzello and Blair Hopwood for providing microarray datasets from synovium and bone respectively, and Dr. John Williams for helpful discussions.
References
- 1.Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part II. Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18:24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Roos H, Adalberth T, Dahlberg L, Lohmander LS. Osteoarthritis of the knee after injury to the anterior cruciate ligament or meniscus: the influence of time and age. Osteoarthritis Cartilage. 1995;3:261–7. doi: 10.1016/s1063-4584(05)80017-2. [DOI] [PubMed] [Google Scholar]
- 4.Aigner T, Zien A, Gehrsitz A, Gebhard PM, McKenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum. 2001;44:2777–89. doi: 10.1002/1529-0131(200112)44:12<2777::aid-art465>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 5.Aigner T, Fundel K, Saas J, Gebhard PM, Haag J, Weiss T, et al. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–44. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 6.Fukui N, Ikeda Y, Ohnuki T, Tanaka N, Hikita A, Mitomi H, et al. Regional differences in chondrocyte metabolism in osteoarthritis: A detailed analysis by laser capture microdissection. Arthritis Rheum. 2008;58:154–63. doi: 10.1002/art.23175. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18:581–92. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Appleton CT, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum. 2007;56:1854–68. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 9.Wei T, Kulkarni NH, Zeng QQ, Helvering LM, Lin X, Lawrence F, et al. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthritis Cartilage. 2010;18:992–1000. doi: 10.1016/j.joca.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis Res Ther. 2007;9:R100. doi: 10.1186/ar2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, Matsumine A, Wakabayashi T, Hasegawa M, Sudo A, Shintani K, et al. Large-scale gene expression profiles, differentially represented in osteoarthritic synovium of the knee joint using cDNA microarray technology. Biomarkers. 2007;12:384–402. doi: 10.1080/13547500601162482. [DOI] [PubMed] [Google Scholar]
- 12.Scanzello CR, McKeon B, Swaim BH, DiCarlo E, Asomugha EU, Kanda V, et al. Synovial inflammation in patients undergoing arthroscopic meniscectomy: molecular characterization and relationship to symptoms. Arthritis Rheum. 2011;63:391–400. doi: 10.1002/art.30137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasson SS, Askew R, Sheppard B, Carito BA, Blanchet T, Ma HL, et al. Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum. 2004;50:2547–58. doi: 10.1002/art.20558. [DOI] [PubMed] [Google Scholar]
- 14.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–8. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 15.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061–9. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Poole R, Blake S, Buschmann M, Goldring S, Laverty S, Lockwood S, et al. Recommendations for the use of preclinical models in the study and treatment of osteoarthritis. Osteoarthritis Cartilage. 2010;18(Suppl 3):S10–6. doi: 10.1016/j.joca.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 17.Turnbull IR, Wlzorek JJ, Osborne D, Hotchkiss RS, Coopersmith CM, Buchman TG. Effects of age on mortality and antibiotic efficacy in cecal ligation and puncture. Shock. 2003;19:310–3. doi: 10.1097/00024382-200304000-00003. [DOI] [PubMed] [Google Scholar]
- 18.McNulty MA, Loeser RF, Davey C, Callahan MF, Ferguson CM, Carlson CS. A comprehensive histological assessment of osteoarthritis lesions in mice. Cartilage. doi: 10.1177/1947603511402665. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou JW, Paules RS, Bushel PR. Systematic variation normalization in microarray data to get gene expression comparison unbiased. J Bioinform Comput Biol. 2005;3:225–41. doi: 10.1142/s0219720005001028. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Chamberland A, Wang E, Jones AR, Collins-Racie LA, LaVallie ER, Huang Y, et al. Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. J Biol Chem. 2009;284:27352–9. doi: 10.1074/jbc.M109.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin W, Park JI, Loeser RF. Oxidative stress inhibits insulin-like growth factor-I induction of chondrocyte proteoglycan synthesis through differential regulation of phosphatidylinositol 3-Kinase-Akt and MEK-ERK MAPK signaling pathways. J Biol Chem. 2009;284:31972–81. doi: 10.1074/jbc.M109.056838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.van der Kraan PM, Stoop R, Meijers TH, Poole AR, van den Berg WB. Expression of type X collagen in young and old C57Bl/6 and Balb/c mice. Relation with articular cartilage degeneration. Osteoarthritis Cartilage. 2001;9:92–100. doi: 10.1053/joca.2000.0364. [DOI] [PubMed] [Google Scholar]
- 25.Muehleman C, Margulis A, Bae WC, Masuda K. Relationship between knee and ankle degeneration in a population of organ donors. BMC Med. 2010;8:48. doi: 10.1186/1741-7015-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis Cartilage. 2009;17:971–9. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaney Davidson EN, Scharstuhl A, Vitters EL, van der Kraan PM, van den Berg WB. Reduced transforming growth factor-beta signaling in cartilage of old mice: role in impaired repair capacity. Arthritis Res Ther. 2005;7:R1338–47. doi: 10.1186/ar1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi N, Carames B, Ronfani L, Ulmer U, Komiya S, Bianchi ME, et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci U S A. 2009;106:1181–6. doi: 10.1073/pnas.0806062106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–18. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 30.Kim AC, Spector M. Distribution of chondrocytes containing alpha-smooth muscle actin in human articular cartilage. J Orthop Res. 2000;18:749–55. doi: 10.1002/jor.1100180511. [DOI] [PubMed] [Google Scholar]
- 31.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, et al. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–49. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 32.Rios H, Koushik SV, Wang H, Wang J, Zhou HM, Lindsley A, et al. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol. 2005;25:11131–44. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruan K, Bao S, Ouyang G. The multifaceted role of periostin in tumorigenesis. Cell Molm Life Sci. 2009;66:2219–30. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–9. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 35.von Marschall Z, Fisher LW. Secreted Frizzled-related protein-2 (sFRP2) augments canonical Wnt3a-induced signaling. Biochem Biophys Res Commun. 2010;400:299–304. doi: 10.1016/j.bbrc.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirotsou M, Zhang Z, Deb A, Zhang L, Gnecchi M, Noiseux N, et al. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc Natl Acad Sci U S A. 2007;104:1643–8. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurowska-Stolarska M, Hueber A, Stolarski B, McInnes IB. Interleukin-33: a novel mediator with a role in distinct disease pathologies. J Intern Med. 2011;269:29–35. doi: 10.1111/j.1365-2796.2010.02316.x. [DOI] [PubMed] [Google Scholar]
- 38.Schulze J, Bickert T, Beil FT, Zaiss MM, Albers J, Wintges K, et al. Interleukin-33 is expressed in differentiated osteoblasts and blocks osteoclast formation from bone marrow precursor cells. J Bone Miner Res. 2011;26:704–17. doi: 10.1002/jbmr.269. [DOI] [PubMed] [Google Scholar]
- 39.Swingler TE, Waters JG, Davidson RK, Pennington CJ, Puente XS, Darrah C, et al. Degradome expression profiling in human articular cartilage. Arthritis Res Ther. 2009;11:R96. doi: 10.1186/ar2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grau S, Richards PJ, Kerr B, Hughes C, Caterson B, Williams AS, et al. The role of human HtrA1 inarthritic disease. J Biol Chem. 2006;281:6124–9. doi: 10.1074/jbc.M500361200. [DOI] [PubMed] [Google Scholar]
- 41.Polur I, Lee PL, Servais JM, Xu L, Li Y. Role of HTRA1, a serine protease, in the progression of articular cartilage degeneration. Histol Histopathol. 2010;25:599–608. doi: 10.14670/hh-25.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Liu W, Bemis A, Wang E, Qiu Y, Morris EA, et al. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum. 2007;56:3675–84. doi: 10.1002/art.22876. [DOI] [PubMed] [Google Scholar]
- 43.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM, 2nd, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum. 2011;63:914–22. doi: 10.1002/art.30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long D, Blake S, Song XY, Lark M, Loeser RF. Human articular chondrocytes produce IL-7 and respond to IL-7 with increased production of matrix metalloproteinase-13. Arthritis Res Ther. 2008;10:R23. doi: 10.1186/ar2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.