Abstract
Background
In only months-to-years a primary cancer can progress to an advanced phenotype that is metastatic and resistant to clinical treatments. As early as the 1900s, it was discovered that the progression of a cancer to the advanced phenotype is often associated with a shift in the metabolic profile of the disease from a state of respiration to anaerobic fermentation – a phenomenon denoted as the Warburg Effect.
Scope of Review
Reports in the literature strongly suggest that the Warburg Effect is generated as a response to a loss in the integrity of the sequence and/or copy number of the mitochondrial genome content within a cancer. Multiple studies regarding the progression of cancer indicate that mutation, and/or, a flux in the copy number, of the mitochondrial genome content can support the early development of a cancer, until; the mutational load and/or the reduction-to-depletion of the copy number of the mitochondrial genome content induces the progression of the disease to an advanced phenotype.
General Significance
Collectively, evidence has revealed that the human cell has incorporated the mitochondrial genome content into a cellular mechanism that, when pathologically actuated, can de (un)differentiate a cancer from the parental tissue of origin into an autonomous disease that disrupts the hierarchical structure-and-function of the human body.
Keywords: Mitochondria, cancer, mitochondrial DNA, mutation, depletion
Introduction
Respiration in a human cell is primarily localized to the mitochondria that reside in the cytosol of the eukaryote. The mitochondria harbor the only extra-nuclear genome in the form of a compact (~ 16,569 base pair) sequence of double-stranded DNA [1, 2]. The two strands are differentiated by the nucleotide content with the guanine rich strand referred to as the heavy strand, and the cytosine rich strand referred to as the light strand. The heavy strand encodes 28 genes, and the light strand encodes 9 genes for a total of 37 genes. Of the 37 genes, 13 are for proteins (polypeptides), 22 are for transfer RNA (tRNA) and two are for the small and large subunits of ribosomal RNA (rRNA) [1, 2]. It is important to note that the remaining polypeptides that are critical for the structure and the function of mitochondria are encoded in the nuclear genome. In a somatic cell the nuclear genome is represented as two copies of a linear sequence that is partitioned into 23 chromosomes; on the other hand, the mitochondrial genome is a circular construct that is harbored at a copy number in the thousands within the mitochondrial matrixes of a single cell [3].
This review will begin with a general discussion of the mitochondrial genome, and the potential repercussions that are generated when a human inherits a variant (haplotype) of the genome. The review will, also, address the development and significance of somatic mutations in cancer, in addition to, the cellular effects that are produced as a response to a heteroplasmic to homoplasmic shift in the representation of a mutant sequence within the mitochondrial genome content. With respect to the copy number of the mitochondrial genome, the review will discuss the reports that strongly suggest that the early development of cancer is characterized with a flux in the copy number of the mitochondrial genome, until; a reduction-to-depletion of the copy number of the mitochondrial genome content is noted in the progression of the disease to an advanced phenotype. The review will then describe mitochondrial-genomic knock-out and mitochondrial genomic knock-in models which have confirmed that a loss in the integrity of the mitochondrial genome content of a cancer is transduced into the de (un)differentiation of the cells from the parental tissue of origin into an autonomous disease of enhanced metastasis, and resistance to the apoptotic effects of clinical therapeutics. In conclusion will be discussed the potential for the utilization of the mitochondrial genome content as a biomarkers in clinical approaches to the maintenance and treatment of cancer and other degenerative diseases.
Maintenance of the Mitochondrial Genome
The transcription and translation of the 13 mitochondrial-encoded genes is mediated by an essential set of 22 tRNAs and 2 rRNAs (12S and 16S rRNAs) that are, also, encoded within the mitochondrial genome [1, 2]. All 13 of the mitochondrial-encoded polypeptides are components of the mitochondrial respiratory chain (MRC), including: 7 of the at least 46 polypeptides in complex I, NADH dehydrogenase (ND1, 2, 3, 4L, 4, 5, 6); 1 of 11 polypeptides in complex III, cytochrome bc1 (cytb); 3 of 13 polypeptides of complex IV, cytochrome c oxidase (COI, II, III); and, 2 of 16 polypeptides of complex V, F1F0 ATP synthetase (ATP6 and 8) [1, 2]. The remaining polypeptides of the MRC are nuclear-encoded genes; thus, the MRC is the only cellular structure to be encoded from the nuclear genome and the mitochondrial genome.
Within the mitochondrial genome is a non-coding displacement (D)-loop (~ 1,122 base pairs) that harbors the main promoter for the transcription of the heavy strand and the light strand of the genome. Additional components of the D-loop include the origin of replication of the heavy strand, mitochondrial transcription factor (mtTFA) binding sites, and conserved sequence blocks (CSB I, II, III) [4]. The collective properties of the D-loop integrate nuclear-encoded processes into the maintenance, replication and transcription of the mitochondrial genome. Since the mitochondrial genome is expressed as polycistronic transcripts [5], it is important to note that the entire repertoire of mitochondrial-encoded polypeptides can be potentially jeopardized with a mutation to the D-loop [6].
A reduction-to-depletion of the mitochondrial transcriptome can, also, be a consequence of mutation(s) to the nuclear genome [7–9]. Nuclear-encoded proteins that are essential for the replication of the mitochondrial genome include mitochondrial DNA polymerase γ (POLG), DNA helicase twinkle, and single-stranded DNA-binding protein (mtSSB). For the repair of the mitochondrial genome the mammalian mitochondria imports mechanisms, such as: elimination of mutagenic 8-oxodeoxyguanosine triphosphate (8-oxodGTP) [10], short patch base excision repair (SP-BER) [11, 12], long-patch base excision repair (LP-BER) [13, 14], mismatch repair [15, 16], and nonhomologous end-joining [17, 18]. The nuclear-encoded maintenance of the mitochondrial genome, also, includes sanitation of the mitochondrial dNTP pool [19, 20], and the selective destruction of damaged sequences of the mitochondrial genome [21, 22].
The complexity of the cellular mechanisms that maintain the integrity of the mitochondrial genome is further evident with the degree of protection that can be generated from a single molecular component. For example, p53 is an enzyme that facilitates multiple facets of genomic maintenance and repair [23–25]. The mitochondrial genome content (sequence and copy number) is predisposed to somatic mutations in cells deficient of p53 [26, 27]. New mechanisms for the maintenance of the mitochondrial genome continues to be discovered [28, 29] and warrants a re-evaluation on the impact that a loss of integrity within the mitochondrial genome content generates in human health.
Mitochondrial Genome Haplotypes
The mitochondrial genome content is exclusively inherited from the oocyte in embryogenesis; thus, variations in the mitochondrial genome of a mother can be passed on to prodigy as restriction fragment length polymorphisms (RFLPs). The isolation of subpopulations of humans over time, amongst diverse geographical locations on Earth, has generated a variety of polymorphisms within the mitochondrial genome. A polymorphism that has become established in a population is denoted as a haplotype. Within a human cell can be a combination of polymorphic sequences of the mitochondrial genome that are collectively classified as a haplogroup. The mitochondrial genomic haplogroups reflect major branch points in the mitochondrial phylogenetic tree and are named alphabetically (A to Z) in the order of discovery [30, 31]. African descendents are reported to have the highest diversity of haplogroups in the world, including L1, L2, and L3 [32]. European and Asian decedents are often found to harbor the distinct set of haplogroups H, I, J, K, T, U, V, W, X and A, B, C, D, F, G respectively [33, 34].
Maternal propagation of the mitochondrial genome can generate inherited diseases with the passage of mutations that shut down the electron transport (complex I–IV) and/or the ATP synthesis (complex V) of mitochondrial respiration [2]. In respect to the development of cancers, the inheritance of the mitochondrial genome has been determined to be a biomarker of carcinogenesis within an array of tissue, including; breast [35–37], prostate [38], thyroid [39], oral [40], endometrial [41, 42], colorectal [43], and renal tissues [38, 44]. Reports have suggested that the inheritance of haplogroup U in European-descendents is associated with an increased risk of prostate and renal cancer in North American populations [38], however; other reports indicate that the risk of prostate cancer is not associated with the haplogroups of European [45–47] and Asians descent [48]. For example, the Carolina Breast Cancer Study has indicated that haplogroup U is a risk factor for breast cancer in African-American and North Indian women, but not European descendants [35, 36, 49]. The collective evidence suggests that the background of the nuclear-genome and/or environment of a cancer could modify the impact that its mitochondrial haplotype has in the development of the disease.
The biological significance of the inheritance of the mitochondrial genome has been verified with cybrid models that are generated from the complete trade of the mitochondrial genome contents between cells. Briefly, cybrid models can be constructed with the transfer of a mitochondrial genome content of an enucleated (donor) cell to a recipient cell that has been depleted of the mitochondrial genome. Cell lines depleted of the mitochondrial genome are denoted as rho (ρ0) cells [50]. Mitochondrial genome contents of interest can, also, be evaluated with cybrid models that are generated from the fusion of ρ0 cells with platelets or synaptosomes, i.e., the remnant cytoplasm of a megakaryocyte or a nerve terminal, respectively [51, 52].
Kulawiec et al. has reported that when a ρ0 cell was fused with platelets that contain a G10398A missense (ND3) polymorphism, from an African-American patient with breast cancer, the activity of NADH dehydrogenase was diminished, the generation of oxidative stress was amplified, and tumor growth was enhanced in comparison to a control that was constructed with the cell-fusion of the ρ0 cells to platelets that were collected from healthy patients [51]. The experimental evidence supports observational analysis that has associated the G10398A polymorphism with an increased risk of cancer in Indian, African, Polish, and European descendants [35–37, 49, 53]. In general, the collective reports of haplotypes/groups in cancer development have revealed that inherited variants of the mitochondrial genome are key initiators in cancer development, and modifiers of carcinogenesis. Moreover, it has been postulated that specific mitochondrial haplotypes are differentially associated with etiological processes of carcinogenesis, e.g., HBV-hepatocellular carcinoma (HCC) versus alcohol-HCC [54].
Somatic Mitochondrial Mutations
Statistics from the American Cancer Society indicate that cancers, such as, breast and prostate cancer, are triggered to metastasize with the ageing of a human [55]. A direct cause-consequence relationship has been associated with the ageing of humans and the accumulation of somatic mutations in the mitochondrial genome [56, 57]. The mitochondrial genome is predisposed to mutation as a consequence of the sheer number of replications of the genome in the life cycle of a cell, in addition to, the oxidative stress that is generated in the near vicinity of the mitochondrial genome content as a byproduct of mitochondrial respiration [58, 59]. Mutations in the mitochondrial genome that are characterized with oxidative stress (e.g., purine G-A and T-C transitions) are often noted within cancers [60–62].
When mutation in the mitochondrial genome impairs the transport of electrons, or, the synthesis of ATP synthesis, within the MRC the dysfunction in mitochondrial respiration can further enhance the cellular level of oxidative stress [51, 63]. Interestingly, cancer progression that is initiated with a reduction-to-depletion of the mitochondrial genome content is not always associated with a significant flux in super oxide, the source of reactive oxygen species (ROS) [64]. The evidence suggests that a loss in the integrity of the mitochondrial genome (sequence and/or copy number) can progress a cancer to an advanced phenotype by way of ROS-dependent and -independent events. For example, a persistent source of mitochondrial genomic damage can, also, be generated in response to mutant mitochondrial-encoded tRNA genes that form secondary structures that resemble the mitochondrial origins of replication [65, 66].
The detection of temporal changes in a mitochondrial genome content with respect to the key developmental stages of a cancer has improved with advances in the ability to sequence the entire genome within a single-cancerous cell, adjacent benign cell, and circulating lymphocytes from a patient [3, 54, 67–69]. Evidence indicates that somatic mutations accumulate in the mitochondrial genome, in part, as a result of a life time of exposure to environmental factors such as ultraviolet light [70], bacteria, viruses [71], and tobacco [72, 73]. For example, lung tissues from smokers are generally acknowledged to have a mitochondrial genome content with an increased density of mutations, at a higher copy number [74], than is observed in tissues of non-smokers [72, 73, 75–77]. The environmental toxins can preferentially generate oxidative damage and/or form adducts in the mitochondrial genome over the nuclear genome [50, 78–82].
The presence of a tumor may, in return, exert a field effect that leads to mutations in ‘bystander cells’ that are adjacent or distant to the tumor. The evidence indicates that cells directly exposed to the mutagens can generate cell-to-cell signals that, ultimately, generate in the anatomically distant ‘bystander’ cells a similar profile of acute or chronic genomic damage noted in the mitochondria of cells directly exposed to the mutagen [83–90]. The bystander effect appears to be a general phenomena that is inducible by radiation [88, 91], thermal injury [92, 93], and chemo-therapeutics/toxicities [94, 95]. Gorman et al. has demonstrated in an ex vivo system of human colorectal cancer that conditioned medium from an irradiated tumor can generate a rapid and transient increase in the frequency of mutant sequences of the mitochondrial genome in bystander tissue, as a secondary effect to dysfunction of mitochondrial respiration [88, 91].
It is postulated that the D-loop of the mitochondrial genome is a mutational hotspot that bears the burden of mutations in the ageing and carcinogenesis of human cells, often before the morphological indications of malignant transformation are visible [60, 66, 96–99]. For example, Tan et al. reported that tissues from smokers accumulate mutations more than those from nonsmokers and those tissues from nonsmokers accumulate mutations predominately in the D-loop, whereas mutations of smokers were observed in the D-loop and coding regions [73]. The postulate has been further supported by the evaluation of prostate needle biopsies from patients suspected of having prostate cancer. Patients that were found to be histologically and symptomatically benign contained somatic mutations that were confined to the D-loop. In patients with adenocarcinoma, especially of an invasiveness phenotype, mutations were randomly distributed across the mitochondrial-encoded genes [66, 99]. The collective evidence strongly suggest that aging and carcinogenesis introduces mutations into the D-loop of the mitochondrial genome of a cancer, which can be followed by a subsequent burst of mutations throughout the genome as the disease progresses to an advanced phenotype [98, 100–102].
Cancer Development
The ability to evaluate if a loss in the integrity of the mitochondrial genome (sequence and/or copy number) generates or enhances the progression of a cancer is subject to the scrutiny of reverse causation. Since eukaryote cells are prone to rapid death after the acute inhibition of mitochondrial respiration, the issue of reverse causation cannot be resolved by pharmacological analysis alone. As an alternative, the mitochondrial genome content from patients or well characterized cell lines can be evaluated by cybrid models [103–108]. For example, cancers have been observed to progress to an advanced disease after obtaining a mitochondrial genome content from platelets of an African-American breast cancer patient with a polymorphic ND3 gene (G10398A) [51], or, cytoplasm generated from a MDA-MB-435 breast cancer cell line with a mutant tRNA gene (Leu(CUN)) [109]. The characterization of cybrid models strongly suggests that variants of the mitochondrial genome are modifiers in carcinogenesis, in addition to, primary initiators for the progression of cancer to an advanced disease [63, 106, 107].
Specific variants of the mitochondrial genome can differentially affect the function of the MRC; thus, it is critical in the study of cancer development that the biological significance of mitochondrial-encoded genes be evaluated on a gene-by-gene basis. Structural limitations hinder the single transfer of a mitochondrial-encoded gene into the mitochondria; and, unfortunately, the non-universal code of the mitochondrial-encoded genes limits the use of conventional expression vectors for the expression of mitochondrial-encoded polypeptides. The direct translation of mitochondrial-encoded sequences in the cytosol results in multiple amino acid alterations and/or premature truncation. With advances in long-range gene synthesis, mutant mitochondrial-encoded genes have been isolated from primary cancer, and, subsequently, converted into nuclear (universal) codons that can be expressed from vectors in the cytosol. The vectors also allow for the addition of mitochondrial targeting sequences to the gene products. As a proper control, a wild-type of a specific mitochondrial-encoded gene can be converted to the universal-code that is, then, concurrently transfected with a variant of the gene. Such studies have shown that variants of ND2 (G4831A or A4605G) can enhance anchorage-dependent and anchorage-independent growth, in addition to, glycolysis in HeLa and head and neck squamous cancer [63]. Mutant ATP6 (T8993G or T9176C) has been observed to increase tumor growth and diminish the apoptotic sensitivity of HeLa cells [105]; while, a variation of the D-loop from colorectal cancer cells (SW480) is reported to enhance tumorigenicity [110]. The collective evidence further emphasizes the need for knowledge regarding how variants of the mitochondrial genome are biologically significant in the development of cancer.
Heteroplasmy to Homoplasmy
A human cell has multiple copies of the mitochondrial genome, and, thus, a single cell can contain a variety of sequences of the genome. The coexistence of two or more sequences of the mitochondrial genome with slightly different nucleotide compositions in a single cell or tissue is defined as heteroplasmy [31]. In the early attempts of mitochondrial genomic analysis, experimental techniques were limited to the lysates of bulk tissues; and, heteroplasmic variations were noted to be quite rare in the mitochondria of healthy individuals. As quantitative-sequencing techniques improved to utilize the mitochondrial genomic content of single-cells isolated from tissues, the sensitivity and resolution in the detection of variants of the mitochondrial genome was increased. Moreover, the advent of resequencing arrays allowed for evaluations of large quantities of DNA in a single run at low cost. The advances have revealed that heteroplasmy is observed and varies markedly in the mitochondrial genome content of a broad array of tissues within an individual [63, 96, 111–116].
It has been reported that in the progression of cancer (s) to an advanced disease a mitochondrial genome sequence can shift from a heteroplasmic-to-homoplasmic representation [66, 117]. In a study of the carcinogenesis of normal colorectal mucosa, oncogenesis has been associated with the deletion of specific mutations in the cancer cells; while, a significant number of mutants are observed to become homoplasmic [66, 96]. The heteroplasmic-to-homoplasmic shift of a variant sequence in the mitochondrial genome content of a cell can, also, be influenced by environmental factors as observed in buccal cells exposed to cigarette smoke [73].
Specific variants of the mitochondrial genome are known to be prone to undergo a heteroplasmic-to-homoplasmic shift. For example, Turner et al. has observed in a cybrid model that the introduction of the A3234G mutation (tRNALeu(UUR)) is subsequently followed by the mitotic segregation and homoplasmic representation of the mutation [118]. Multiple reports have demonstrated the biased segregation of additional variants of the mitochondrial genome, in vitro and in vivo [119–127]. If a variant of the mitochondrial genome produces a selective cellular advantage, over the thousands of generations required for tumorigenesis, the variant could become homoplasmic within a tissue as the oncogenic cell overtakes the tissue by clonal growth [128]. Alternatively, computer models suggest that during the unbiased states of replication and genomic sorting in the propagation of a cell, a variant sequence within the mitochondrial genome content can become homoplasmic [129]. Thus, warrants the need for the continued investigation to determine how the development of cancer is impacted as a response to shifts in the proportions of variant sequences within the mitochondrial genome content of a cell.
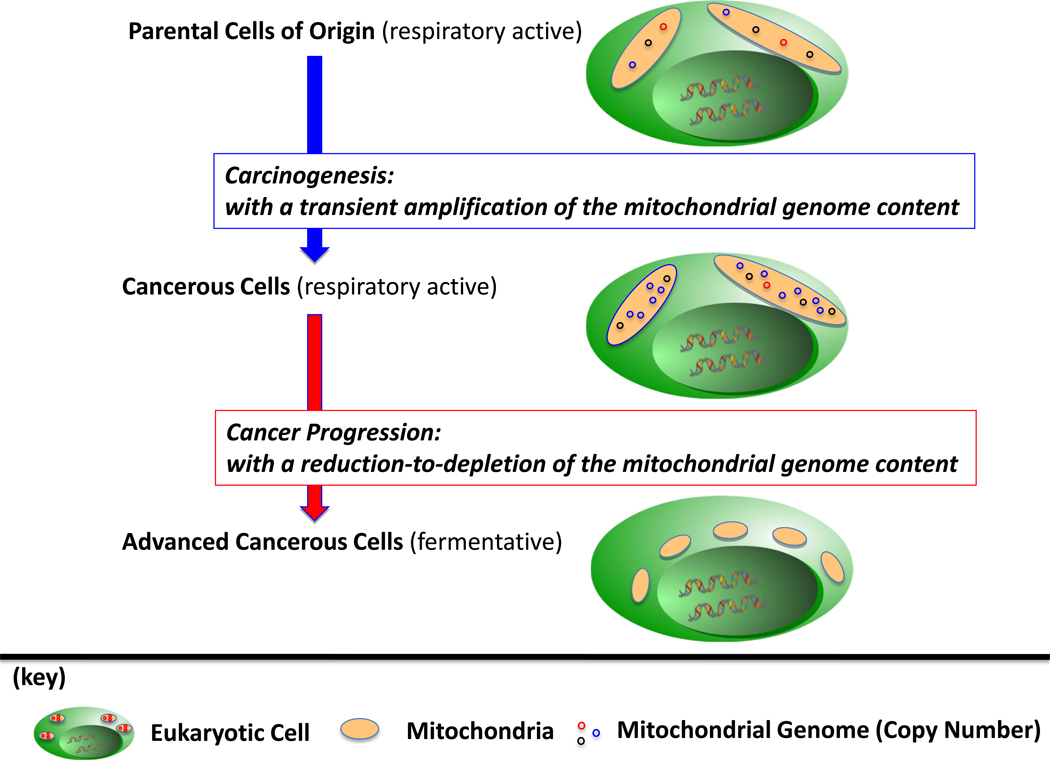
Copy Number, Amplification to Depletion (Figure 1)
Figure 1. The Mitochondrial Genome Content in Cancer Development.
In general, cells of human origin are respiratory-competent with mitochondria that harbor a mixture (haplotype) of sequences of the mitochondrial genome. Evidence indicates that in carcinogenesis the copy number of the mitochondrial genome is amplified, potentially as a consequence to the generation, and/or heteroplasmic-to-homoplasmic shift, of a mutant sequence of the mitochondrial genome; until, the progression of the cancer to an advanced (fermentative) phenotype is observed with a reduction-to-depletion of the mitochondrial genome content.
The mitochondrial genome content is reported to vary within the subpopulations of oncogenic cells collected from the same cancerous tumor. For example, Mizumachi et al. reported that tumors of prostate cancers are composed of cells that contain an amplified copy number of the mitochondrial genome content, in addition to, cells with a diminished-to-depleted copy number of the genome [3]. Mizumachi et al. also observed in primary prostate cancer cell lines a higher distributional variance in the copy number of the mitochondrial genome content than was reported in immortalized adjunct normal cells collected from the same tumor. It has been postulated that the copy number of the mitochondrial genome is amplified as a compensatory response to oxidative stress [58, 130]. Unfortunately, an amplified copy number of the mitochondrial genome in a cancer could be advantages for tumorigenesis, e.g., if a subsequent enhancement of mitochondrial-respiration enhances hypoxia-inducing signals and/or ROS-generated signals [131–134]. For example, in head and neck cancer it has been reported that the amplification of the mitochondrial genome content enhances the resistance of cells to docetaxel, in part, as a response to an increase in the activity of Fo-ATPase [3].
While the copy-number of the mitochondrial genome of a cell is noted to be amplified after the carcinogenesis of lung tissue [135, 136], the progression of the cancer to an advanced disease is often associated with a depletion of the mitochondrial genome and low oxidative stress [94]. In general, the depletion of the mitochondrial genome content is a common characteristic in the progression of a variety of cancers [102], as observed in ovarian [137], gastric [138], hepatocellular [6, 139, 140], renal cell [44, 141–143], prostate [144–147], and breast cancers [147–150].
Multiple reports have shown that specific variants of the mitochondrial genome, particularly the hypervariability of the D-loop, are deleterious to the copy-number of the genome [6, 61, 149]. A complete depletion of the mitochondrial genome and/or the polycistronic transcriptome has been documented in cells following the heteroplasmic-to-homopasmic shift of a mitochondrial genomic sequence(s) that harbors mutation mapping to a tRNA, rRNA or the D-loop [123, 151–153]. For example, Turner et al. has demonstrated that a heteroplasmic-to-homoplasmic shift of the A3234G mutation (tRNALeu(UUR)) is frequently followed by a depletion of the mitochondrial genome [118].
From the perspective of nuclear-encoded genes, reports have shown that mutants of the primary machinery for the replication of the mitochondrial genome (e.g., DNA polymerase γ (POLG) and DNA helicase twinkle) can also perpetuate a dysregulation and depletion in the copy number of the genome [9, 154, 155]. For example, Singh et al. noted that the majority of breast cancers harbor a nuclear genome with mutant POLG genes, and mitochondria that are depleted of the mitochondria genome; moreover, the report demonstrated that the expression of vector-encoded mutant POLG in breast cancer cell lines depleted the mitochondrial genome content, and signaled the progression of the cancers to enhanced states of invasiveness [8]. From another perspective, Pursell et al. has reported that the activity of POLG can be diminished if the level of ROS in a cell leads to a minute shift of the total cellular dGTP pool to 8-oxo-dGTP [156].
Damage to nuclear-encoded proteins (e.g., p53, Ras and p66shc) that perturb mitochondrial homeostasis or inter organelle signaling within a cell are likewise to contribute instability to the copy number of the mitochondrial genome [157–159]. Mutations in nuclear genes involved in deoxynucleotide metabolism (e.g., a p53-regulated subunit (p53R2) of ribonucleotide reductase, RNR) generate diseases that are characterized by a severe reduction in the mitochondrial genome content [25, 160–162]. Models of p53 null mice and p53 knockdown human primary fibroblasts have also been observed to have mitochondria that are depleted of the mitochondrial genome content [27]. It has been proposed that p53 associates with the mitochondrial genome through physical interactions within the genome [163], in addition to, modifying the functions of POLG [164] and mtTFA [165]. The repertoire of nuclear-encoded genes involved in the homeostasis of the mitochondrial genome continues to expand; recent evidence strongly implicates that the actomyosin cytoskeleton, in addition to, autocrine and paracrine signaling pathways (e.g., insulin-like growth factor, IGF-1, by way of mitochondrial pyrimidine nucleotide carrier 1, PNC1) are involved in the maintenance of the mammalian mitochondrial genome [28, 166].
Exogenous toxins such as alcohol, benzene, and herpes simplex virus are also linked to a reduction-to-depletion in the copy number of the mitochondrial genome [167–170]. Exogenous toxins such as benzene, and herpes simplex virus are also linked to a reduction-to-depletion in the copy number of the mitochondrial genome, as reported in buccal, parotid and lung tissues [169, 170]. Interestingly, in response to carcinogens, the mitochondrial genome content of a cell is often noted to be transiently amplified, and, then, depleted in a dose-dependent manner [58, 73–77, 130, 135]. The endogenous formation of toxins, such as, abnormalities in endocrine (estrogen and androgen) signaling pathways that develop in the progression of cancers, has also been associated with a reduction in the copy number of the mitochondrial genome [171–173].
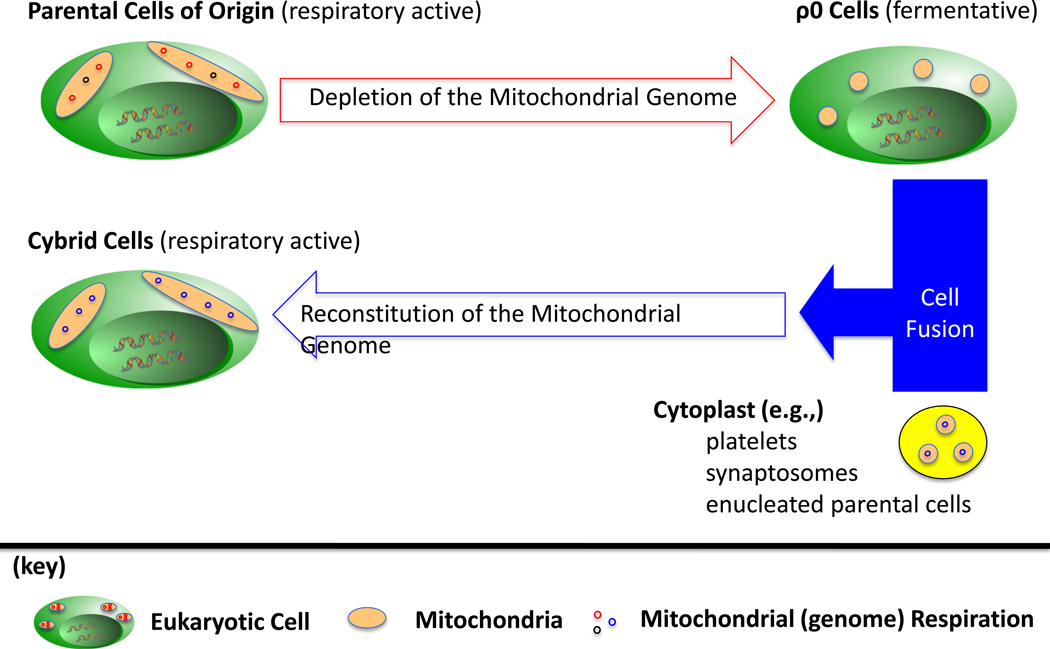
Mitochondrial Genomic Knock-Out (ρ0) and Mitochondrial Genomic Knock-In (Cybrid) Models (Figure 2)
Figure 2. Mitochondrial Genome Knock-In (ρ0) and Mitochondrial Genome Knock-Out (Cybrid) Models.
In general, cells of human origin are respiratory-competent with mitochondria that harbor the mitochondrial genome. Viable cells (ρ0) can be generated with the depletion of the mitochondrial genome. As a control, the mitochondrial genome content can be reconstituted to ρ0 cells with the fusion of ρ0 to cytoplast that act as membrane bound donors of mitochondria, and, thus, the mitochondrial genome.
The mitochondria as a whole cannot be excluded from cells, as the mitochondria serve multiple roles for the eukaryote, including ATP synthesis, redox regulation, thermogenesis, calcium regulation, and production of secondary messengers [2]. From the perspective of the mitochondrial genome, the generation of viable (ρ0) cell lines after the depletion of the genome from cancer cell lines reveals that a loss in the integrity of the mitochondrial genome content (sequence and/or copy number) progresses the disease to an advanced phenotype [145–147, 150, 174]. It should be noted that the in vitro cultures of ρ0 cells are auxotrophic for pyruvate and uridine [50, 175]. The observations can be attributed to the fact that uridine synthesis requires mitochondrial respiration, and that the ability to sustain a persistent flux in glycolysis is dependent on the conversion of pyruvate to lactate as a means to oxidize NADH to NAD+. In terms of the in vivo growth of ρ0 cells, multiple reports have demonstrated the ability to generate xenograft tumors of ρ0 cells in immune-deficient mice [176, 177].
Within human cells that are competent for mitochondrial respiration the mitochondria form an elaborate tubular network of mitochondrial respiration that maintains the mitochondrial membrane potential. The mitochondria of the ρ0 cells appear as punctate spots that localize around the nucleus [178]. Interestingly, the mitochondria of (respiratory-incompetent) ρ0 cells can still obtain nuclear encoded proteins [179, 180], a process that is generally acknowledged to be mediated by the mitochondrial membrane potential [181]. Evidence indicates that ρ0 cells generate the mitochondrial membrane potential by the remnant components of F1Fo ATP synthase; it has been postulated that the mitochondrial membrane potential is generated, in part, as a response to an import of ‘glycolytic’ ATP by way of ATP synthase, that is coupled, to the (reverse) ATP/ADP translocase activity of ANT [179, 182]. Reports also suggest that the mitochondrial membrane potential is maintained with additional electrogenic pumps, e.g., the mitochondrial chloride intracellular channel (mtCLIC) [179, 183].
It is possible that in the formation of ρ0 cells a mutation can be incidentally introduced into the nuclear genome that, ultimately, mask the cellular signaling pathways and phenotypes that are generated with the depletion of the mitochondrial genome content. As a proper control to a ρ0 cell line, a mitochondrial genome content can be reconstituted to ρ0 cells with the creation of cybrid cell line, i.e., the fusion of a ρ0 cell to cytoplasmic vessels that harbor donor mitochondria from platelets, synaptosomes or enucleated parental cells of the ρ0 [51, 52, 146]. The shared nuclear background in a ρ0/cybrid model allows for the verification that a cellular event or phenotype which is observed in the ρ0 and not the cybrid is a direct result of the depletion of the mitochondrial genome, and not the product of an incidental mutation to the nuclear genome.
A Cellular and Pathological Signaling Pathway
The ρ0 cells are characterized with a reduced sensitivity to death-stimuli in comparison to parental cells. For example, Higuchi et al. reported that the depletion of the mitochondrial genome content in ML-1a cells generated a resistant to TNF-induced apoptosis, even though TNF-induced cellular proliferation and differentiation could still be observed in theρ0 cells [174]. The report also noted that the mitochondrial apoptotic machinery in the ρ0 cells was not completely obliterated, based on evidence that the viability of the culture of the cells was decreased in the presence of staurosporine-induced apoptosis [174]. Suzuki et al. would later reveal that the ρ0 cells generate a resistance to TNF-induced apoptosis by the constitutive activation of Akt [184]. A flux in the Akt signal is a key mechanism that can enhance the resistance of cancer to apoptotic-stimuli [51, 145, 185].
Additional reports of the molecular pathways within ρ0 cells have demonstrated that Akt is one of a diverse array of signals (e.g., Raf/MAPK, PI3K/Akt, JNK, AP1, and NF-κB) that are concurrently and reversibly activated with flux in the copy number of the mitochondrial genome content within a cell [64, 147, 184]. For example, Naito et al. noted that the depletion the mitochondrial genome in breast cancer and prostate cancer generated the constitutive activation of the Raf/MAPK signaling cascade, in addition to, the enhanced expression of transforming growth factor beta (TGFβ) 1 and type I TGFβ receptor (TGFβRI) [147]. Naito et al. also observed the ρ0 cells of the cancers to be of a mesenchymal-like and invasive phenotype, in comparison to the epithelial-like phenotype of the parental cells. In a corresponding report, Xie et al. noted in ρ0 cells that the depletion of the mitochondrial genome generated the hypermethylation of promoter regions (i.e, DNA methyltransferase 1), and, thus, the silencing of epithelial biomarkers (E-cadherin) and a variety of tumor suppressor genes [186]. It is also heavily reported that the depletion of the mitochondrial genome content in ρ0 cells elevates the transcription of genes relevant to metal homeostasis, initiation of EMT, glucuronidation pathways, autophagy of defective mitochondria, and elimination of lipophilic molecules via peroxisomal lipid metabolic pathways [187–189]. Furthermore, the silencing of systemic (hormone) signaling pathways that maintain the viability, growth, and differentiation of prostate cancer and breast cancer has also been noted in ρ0 cells [144, 146–149]. The overall phenotype of the ρ0 cells strongly indicates that a depletion of the copy number of the mitochondrial genome in cancer is integrated into a mitochondrial-generated progression signal that generates the progression of the disease to an advanced phenotype that is highly invasive-to-metastatic, and resistance to clinical (apoptotic) stimuli [145–147, 150, 174].
An Intracellular and Extracellular Biomarker
With the high frequency of variant sequences of the mitochondrial genome in cancer, at a copy number that is often more numerous than any mutant nuclear-encoded gene, the mitochondrial genome is potentially a convenient biomarker for cancer development. Reports have shown that the copy number of the mitochondrial genome is a biomarker associated with the risk of carcinogenesis in tissues such as the lung [135, 136]. Furthermore, mitochondrial genomic analysis can potentially enhance the histopathological evaluation of tumors and could be beneficial in the classification of malignant, benign, and proximal to malignant lesions [190]. Evidence also suggests that the characterization of the mitochondrial genome content (sequence and copy number) within cells could aid the identification of the etiology behind the carcinogenesis of the tissue [54], the designation of effective therapeutic regimens [3, 108, 146, 150], and the prognosis for a patient [191].
Outside of the cell, fractions of the mitochondria genome of cancers have been isolated from extracellular fluids such as saliva [192], blood and plasma [97, 193], urine [97, 112], pancreatic juice [112], breast nipple aspirate fluid [194], and cerebral spinal fluid [195]. It is important to note that that the measurement of circulating fragments of the mitochondrial genome can be masked by paraneoplastic characteristics; for example, enhanced protease activity can cause a decrease in the nucleic acid binding capacity on the surface of blood cells (erythrocytes and leukocytes) [193, 196–198], nuclear complexes [199], and/or apoptotic bodies [200].
Accumulation of fragments of the mitochondrial genome (DNA, mtDNA) or transcriptome (RNA, mtRNA) in extra-cellular fluids is presumably generated by, a active release of the molecules from circulating cancer cells [201–203], in addition to, a passive release in response to apoptosis or necrosis of cancer and bystander tissue [204–207]. The quantification of circulating mtDNA has been postulated to be a prognostic marker of cancers that is of an increased sensitivity and specificity over circulating nuclear DNA [191]. It has been reported that a significant increase of mtDNA is observed in the saliva of patients with head and neck cancer, and, the cerebral spinal fluid of patients with medulloblastomas, in comparison to patients with no cancer [192, 195]. The circulating mtDNA levels were also observed to decrease after radiotherapy or hormone therapy. A similar observation has been characterized for prostate cancer. Patients that die from prostate cancer within 2 years of initial presentation are reported to maintain higher levels of mtDNA in circulation, in comparison to patients with prostate cancer who survived for longer periods [191, 208]. A concurrent elevation of circulating mtDNA and PSA has also been characterized with the early biochemical recurrence of prostate cancer after radial prostectomy [191]. The collective observations suggest that intracellular and extracellular levels of the mitochondrial genome can be in direct relationship with the cancer cell burden, and, thus, provide a biomarker for the disease-free and the overall survival status of a patent diagnosed with cancer.
Revival of the Mitochondrial Genome
It is generally acknowledged that the endosymbiotic relationship of the mitochondrial genome and nuclear genome in the eukaryote enables the human cell to form tissue structures that can support the complex functions of the human body. An appreciation of the significance of the mitochondrial genome in cancer development was initially delayed with, the late discovery of the mitochondrial genome in comparison to the nuclear genome, in addition to, a time lapse in the adaptation of nuclear genomic analysis to the unique characteristics (sequence and copy number) of the mitochondrial genome content of a cell. Meanwhile, the identification of oncogenes and tumor suppressors revealed a broad spectrum of cellular signaling pathways that can, switch the cellular proteome from a metabolic state of mitochondrial respiration towards glycolytic fermentation, and, concurrently, de(un)differentiate a cancer into an advanced disease [209–211]. Near half of a century has passed since the discovery of oncogenes and tumor suppressors and cancer progression still remains a deadly event in human society [55, 212]; thus, a return is warranted to the classical observations of the Warburg Effect that hinted to the postulate that the mitochondrial genome is more than a compensatory mechanism to genomic insults [213, 214].
This review presented multiple lines of evidence which demonstrate that a loss in the integrity of the mitochondrial genome can lead to the de(un)differentiation of a variety of cancers from the parental tissue of origin, into autonomous (fermentative) diseases that disrupt the organization of tissue structure and function [146, 147, 150]. Interestingly, the depletion of the mitochondrial genome from cancer generates a de (un)differentiated cell that harbors mitochondria of similar morphology and genealogy as observed in undifferentiated embryonic stem cells (ESCs) and pluripotent blastomeres (pre-implantation embryos) [178, 215]; the small and immature mitochondria are localized around the nucleus, and have a low copy number of the mitochondria genome and diminished MRC activity. From the alternative perspective, mitochondria in respiratory-competent cancers resemble eukaryotes that have differentiated to a specific cell fate [178, 215]; the mitochondria are observed to form dense and wide networks with enriched mitochondrial genome contents and enhanced MRC activity.
The collective evidence suggests that the early (fermentative) stages of embryogenesis are maintained by a mitochondrial-generated progression signal that is generated in response to a low copy number of the mitochondrial genome. Moreover, the evidence strongly indicates that the mitochondrial-generated progression signal is pathologically ‘reawakened’ in the later stages of human life when the copy number of the genome is reduced within an adult (respiratory competent) cell. To this date no study has elicited a mechanism that links the mitochondrial genome content to a mitochondrial-generated progression signal of embryogenesis, or the progression of cancer. It is plausible that a loss in the integrity of the mitochondrial genome is a common pathological event that drives the progression of a broad spectrum of human diseases that are attributed to respiratory-incompetent mitochondria, such as the clearly defined ‘mtDNA depletion syndrome’ [216–219]. Future studies are warranted to define the mechanism(s) that transduces the integrity of the mitochondrial genome content into a mitochondrial-generated progression signal; such a discovery could reveal cellular processes that could be targeted in the attempt to stall and/or reverse the progression of cancer and other degenerative diseases, to enable a realistic fight as we await the cure.
Highlights.
This review describes change in mitochondrial genome in cancer.
Evidence shows mitochondrial genome is predisposed to mutation in carcinogenesis.
Then a burst of mutations occurred associated with advanced malignant phenotype.
Acknowledgement
This study was supported by State of Arkansas Tobacco Settlement, NIH grants CA100846 and DOD PCRP Prostate Cancer Training Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. A mitochondrial paradigm of metabolic anddegenerative diseases aging, cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizumachi T, Muskhelishvili L, Naito A, Furusawa J, Fan CY, Siegel ER, Kadlubar FF, Kumar U, Higuchi M. Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate. 2008;68:408–417. doi: 10.1002/pros.20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koehler CM, Bauer MF. Mitochondrial function and biogenesis. Berlin; New York: Springer; 2004. [Google Scholar]
- 5.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs--like members of all families similar but different Biochimica et Biophysica Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res. 2004;547:71–78. doi: 10.1016/j.mrfmmm.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Loon B, Hubscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc Natl Acad Sci U S A. 2009;106:18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topping RP, Wilkinson JC, Scarpinato KD. Mismatch repair protein deficiency compromises cisplatin-induced apoptotic signaling. J Biol Chem. 2009;284:14029–14039. doi: 10.1074/jbc.M809303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa J, Tsuchimoto D, Oka S, Ohno M, Furuichi M, Sakumi K, Nakabeppu Y. Oxidation of mitochondrial deoxynucleotide pools by exposure to sodium nitroprusside induces cell death. DNA Repair (Amst) 2008;7:418–430. doi: 10.1016/j.dnarep.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Nakabeppu Y, Oka S, Sheng Z, Tsuchimoto D, Sakumi K. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat Res. 2010;703:51–58. doi: 10.1016/j.mrgentox.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim I, Lemasters JJ. Mitophagy Selectively Degrades Individual Damaged Mitochondria After Photoirradiation Antioxid Redox Signal. 2011 doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci U S A. 2010;107:12828–12833. doi: 10.1073/pnas.0910885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura N, Nakamura Y, Miyamoto Y, Miyamoto T, Kabu K, Yoshida M, Futamura M, Ichinose S, Arakawa H. Mieap a p53-inducible protein controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One. 2011;6:16060. doi: 10.1371/journal.pone.0016060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B encoding p53-controlled ribonucleotide reductase (p53R2)causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 26.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, Harbour ME, Fearnley IM, Crouch RJ, Conti MA, Adelstein RS, Walker JE, Holt IJ. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 31.MITOMAP. A Human Mitochondrial Database. 2009 [Google Scholar]
- 32.Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19:255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- 33.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 34.Pasternak JJ. An introduction to human molecular genetics : mechanisms of inherited diseases. 2nd ed. Hoboken, N.J: Wiley-Liss; 2005. [Google Scholar]
- 35.Mims MP, Hayes TG, Zheng S, Leal SM, Frolov A, Ittmann MM, Wheeler TM, Prchal JT. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2006;66:1880. doi: 10.1158/0008-5472.CAN-05-3774. author reply 1880–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 37.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 38.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, Lim SD, Fernandez-Golarz C, Lyles RH, Brown MD, Marshall FF, Petros JA. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol. 2006;175:468–472. doi: 10.1016/S0022-5347(05)00163-1. discussion 472-463. [DOI] [PubMed] [Google Scholar]
- 39.Yeh JJ, Lunetta KL, van Orsouw NJ, Moore FD, Jr, Mutter GL, Vijg J, Dahia PL, Eng C. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000;19:2060–2066. doi: 10.1038/sj.onc.1203537. [DOI] [PubMed] [Google Scholar]
- 40.Datta S, Majumder M, Biswas NK, Sikdar N, Roy B. Increased risk of oral cancer in relation to common Indian mitochondrial polymorphisms and Autosomal GSTP1 locus. Cancer. 2007;110:1991–1999. doi: 10.1002/cncr.23016. [DOI] [PubMed] [Google Scholar]
- 41.Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY, Wong LC, Nagley P, Ngan HY. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Hum Mutat. 2003;22:173–174. doi: 10.1002/humu.10244. [DOI] [PubMed] [Google Scholar]
- 42.Czarnecka AM, Klemba A, Semczuk A, Plak K, Marzec B, Krawczyk T, Kofler B, Golik P, Bartnik E. Common mitochondrial polymorphisms as risk factor for endometrial cancer. Int Arch Med. 2009;2:33. doi: 10.1186/1755-7682-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aikhionbare FO, Khan M, Carey D, Okoli J, Go R. Is cumulative frequency of mitochondrial DNA variants a biomarker for colorectal tumor progression? Mol Cancer. 2004;3:30. doi: 10.1186/1476-4598-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller EE, Eder W, Mayr JA, Paulweber B, Sperl W, Horninger W, Klocker H, Kofler B. Mitochondrial haplogroups and control region polymorphisms are not associated with prostate cancer in Middle European Caucasians. PLoS One. 2009;4:6370. doi: 10.1371/journal.pone.0006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1455–1459. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, McDonnell SK, Hebbring SJ, Cunningham JM, St Sauver J, Cerhan JR, Isaya G, Schaid DJ, Thibodeau SN. Polymorphisms in mitochondrial genes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3558–3566. doi: 10.1158/1055-9965.EPI-08-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim W, Yoo TK, Shin DJ, Rho HW, Jin HJ, Kim ET, Bae YS. Mitochondrial DNA haplogroup analysis reveals no association between the common genetic lineages and prostate cancer in the Korean population. PLoS One. 2008;3:2211. doi: 10.1371/journal.pone.0002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 50.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 51.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009;54:647–654. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isobe K, Kishino S, Inoue K, Takai D, Hirawake H, Kita K, Miyabayashi S, Hayashi JI. Identification of inheritance modes of mitochondrial diseases by introduction of pure nuclei from mtDNA-less HeLa cells to patient-derived fibroblasts. J Biol Chem. 1997;272:12606–12610. doi: 10.1074/jbc.272.19.12606. [DOI] [PubMed] [Google Scholar]
- 53.Czarnecka AM, Krawczyk T, Zdrozny M, Lubinski J, Arnold RS, Kukwa W, Scinska A, Golik P, Bartnik E, Petros JA. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res Treat. 2010;121:511–518. doi: 10.1007/s10549-009-0358-5. [DOI] [PubMed] [Google Scholar]
- 54.Zhang R, Zhang F, Wang C, Wang S, Shiao YH, Guo Z. Identification of sequence polymorphism in the D-Loop region of mitochondrial DNA as a risk factor for hepatocellular carcinoma with distinct etiology. J Exp Clin Cancer Res. 2010;29:130. doi: 10.1186/1756-9966-29-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.American Cancer Society. Cancer facts & figures. Atlanta, GA: The Society; 2011. p. v. [Google Scholar]
- 56.Jessie BC, Sun CQ, Irons HR, Marshall FF, Wallace DC, Petros JA. Accumulation of mitochondrial DNA deletions in the malignant prostate of patients of different ages. Exp Gerontol. 2001;37:169–174. doi: 10.1016/s0531-5565(01)00153-x. [DOI] [PubMed] [Google Scholar]
- 57.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348(Pt 2):425–432. [PMC free article] [PubMed] [Google Scholar]
- 59.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 60.Yu M, Shi Y, Zhang F, Zhou Y, Yang Y, Wei X, Zhang L, Niu R. Sequence variations of mitochondrial DNA D-loop region are highly frequent events in familial breast cancer. J Biomed Sci. 2008;15:535–543. doi: 10.1007/s11373-007-9229-4. [DOI] [PubMed] [Google Scholar]
- 61.Liou CW, Lin TK, Chen JB, Tiao MM, Weng SW, Chen SD, Chuang YC, Chuang JH, Wang PW. Association between a common mitochondrial DNA D-loop polycytosine variant and alteration of mitochondrial copy number in human peripheral blood cells. J Med Genet. 2010;47:723–728. doi: 10.1136/jmg.2010.077552. [DOI] [PubMed] [Google Scholar]
- 62.Abu-Amero KK, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene. 2005;24:1455–1460. doi: 10.1038/sj.onc.1208292. [DOI] [PubMed] [Google Scholar]
- 63.Zhou S, Kachhap S, Sun W, Wu G, Chuang A, Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, Westra WH, Maitra A, Glazer C, Carducci M, Sidransky D, McFate T, Verma A, Califano JA. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci U S A. 2007;104:7540–7545. doi: 10.1073/pnas.0610818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higuchi M, Manna SK, Sasaki R, Aggarwal BB. Regulation of the activation of nuclear factor kappaB by mitochondrial respiratory function: evidence for the reactive oxygen species-dependent and -independent pathways. Antioxid Redox Signal. 2002;4:945–955. doi: 10.1089/152308602762197489. [DOI] [PubMed] [Google Scholar]
- 65.Seligmann H. Mitochondrial tRNAs as light strand replication origins: similarity between anticodon loops and the loop of the light strand replication origin predicts initiation of DNA replication. Biosystems. 2010;99:85–93. doi: 10.1016/j.biosystems.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 66.Kloss-Brandstatter A, Schafer G, Erhart G, Huttenhofer A, Coassin S, Seifarth C, Summerer M, Bektic J, Klocker H, Kronenberg F. Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am J Hum Genet. 2010;87:802–812. doi: 10.1016/j.ajhg.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao YG, Ogasawara Y, Kajigaya S, Molldrem JJ, Falcao RP, Pintao MC, McCoy JP, Jr, Rizzatti EG, Young NS. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007;109:756–762. doi: 10.1182/blood-2006-01-011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salas A, Yao YG, Macaulay V, Vega A, Carracedo A, Bandelt HJ. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandelt HJ, Lahermo P, Richards M, Macaulay V. Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med. 2001;115:64–69. doi: 10.1007/s004140100228. [DOI] [PubMed] [Google Scholar]
- 70.Birch-Machin MA, Swalwell H. How mitochondria record the effects of UV exposure and oxidative stress using human skin as a model tissue. Mutagenesis. 2010;25:101–107. doi: 10.1093/mutage/gep061. [DOI] [PubMed] [Google Scholar]
- 71.Machado AM, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection generates genetic instability in gastric cells. Biochim Biophys Acta. 2010;1806:58–65. doi: 10.1016/j.bbcan.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 72.Prior SL, Griffiths AP, Baxter JM, Baxter PW, Hodder SC, Silvester KC, Lewis PD. Mitochondrial DNA mutations in oral squamous cell carcinoma. Carcinogenesis. 2006;27:945–950. doi: 10.1093/carcin/bgi326. [DOI] [PubMed] [Google Scholar]
- 73.Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, Orden RA, Chen J, Goldman R, Shields PG. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29:1170–1177. doi: 10.1093/carcin/bgn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masayesva BG, Mambo E, Taylor RJ, Goloubeva OG, Zhou S, Cohen Y, Minhas K, Koch W, Sciubba J, Alberg AJ, Sidransky D, Califano J. Mitochondrial DNA content increase in response to cigarette smoking. Cancer Epidemiol Biomarkers Prev. 2006;15:19–24. doi: 10.1158/1055-9965.EPI-05-0210. [DOI] [PubMed] [Google Scholar]
- 75.Fahn HJ, Wang LS, Kao SH, Chang SC, Huang MH, Wei YH. Smoking-associated mitochondrial DNA mutations and lipid peroxidation in human lung tissues. Am J Respir Cell Mol Biol. 1998;19:901–909. doi: 10.1165/ajrcmb.19.6.3130. [DOI] [PubMed] [Google Scholar]
- 76.Lee HC, Lu CY, Fahn HJ, Wei YH. Aging- and smoking-associated alteration in the relative content of mitochondrial DNA in human lung. FEBS Lett. 1998;441:292–296. doi: 10.1016/s0014-5793(98)01564-6. [DOI] [PubMed] [Google Scholar]
- 77.Lewis PD, Fradley SR, Griffiths AP, Baxter PW, Parry JM. Mitochondrial DNA mutations in the parotid gland of cigarette smokers and non-smokers. Mutat Res. 2002;518:47–54. doi: 10.1016/s1383-5718(02)00066-9. [DOI] [PubMed] [Google Scholar]
- 78.Almeida AM, Bertoncini CR, Borecky J, Souza-Pinto NC, Vercesi AE. Mitochondrial DNA damage associated with lipid peroxidation of the mitochondrial membrane induced by Fe2+-citrate. An Acad Bras Cienc. 2006;78:505–514. doi: 10.1590/s0001-37652006000300010. [DOI] [PubMed] [Google Scholar]
- 79.Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 80.Allen JA, Coombs MM. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980;287:244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- 81.Cakir Y, Yang Z, Knight CA, Pompilius M, Westbrook D, Bailey SM, Pinkerton KE, Ballinger SW. Effect of alcohol and tobacco smoke on mtDNA damage and atherogenesis. Free Radic Biol Med. 2007;43:1279–1288. doi: 10.1016/j.freeradbiomed.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 82.Ballinger SW, Bouder TG, Davis GS, Judice SA, Nicklas JA, Albertini RJ. Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 1996;56:5692–5697. [PubMed] [Google Scholar]
- 83.Montironi R, Diamanti L, Pomante R, Thompson D, Bartels PH. Subtle changes in benign tissue adjacent to prostate neoplasia detected with a Bayesian belief network. J Pathol. 1997;182:442–449. doi: 10.1002/(SICI)1096-9896(199708)182:4<442::AID-PATH866>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 84.Mairinger T, Mikuz G, Gschwendtner A. Nuclear chromatin texture analysis of nonmalignant tissue can detect adjacent prostatic adenocarcinoma. Prostate. 1999;41:12–19. doi: 10.1002/(sici)1097-0045(19990915)41:1<12::aid-pros3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 85.Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 87.Mothersill C, Seymour C. Radiation-induced bystander effects: past history and future directions. Radiat Res. 2001;155:759–767. doi: 10.1667/0033-7587(2001)155[0759:ribeph]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 88.Gorman S, Fox E, O’Donoghue D, Sheahan K, Hyland J, Mulcahy H, Loeb LA, O’Sullivan J. Mitochondrial mutagenesis induced by tumor-specific radiation bystander effects. J Mol Med. 2010;88:701–708. doi: 10.1007/s00109-010-0616-3. [DOI] [PubMed] [Google Scholar]
- 89.Prise KM, Belyakov OV, Folkard M, Michael BD. Studies of bystander effects in human fibroblasts using a charged particle microbeam. Int J Radiat Biol. 1998;74:793–798. doi: 10.1080/095530098141087. [DOI] [PubMed] [Google Scholar]
- 90.Shao C, Folkard M, Michael BD, Prise KM. Targeted cytoplasmic irradiation induces bystander responses. Proc Natl Acad Sci U S A. 2004;101:13495–13500. doi: 10.1073/pnas.0404930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorman S, Tosetto M, Lyng F, Howe O, Sheahan K, O’Donoghue D, Hyland J, Mulcahy H, O’Sullivan J. Radiation and chemotherapy bystander effects induce early genomic instability events: telomere shortening and bridge formation coupled with mitochondrial dysfunction. Mutat Res. 2009;669:131–138. doi: 10.1016/j.mrfmmm.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 92.Purschke M, Laubach HJ, Anderson RR, Manstein D. Thermal injury causes DNA damage and lethality inunheated surrounding cells: active thermal bystander effect. J Invest Dermatol. 2010;130:86–92. doi: 10.1038/jid.2009.205. [DOI] [PubMed] [Google Scholar]
- 93.Dabrowska A, Gos M, Janik P. “Bystander effect” induced by photodynamically or heat-injured ovarian carcinoma cells (OVP10) in vitro. Med Sci Monit. 2005;11:BR316–BR324. [PubMed] [Google Scholar]
- 94.Lin CS, Wang LS, Tsai CM, Wei YH. Low copy number and low oxidative damage of mitochondrial DNA are associated with tumor progression in lung cancer tissues after neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg. 2008;7:954–958. doi: 10.1510/icvts.2008.177006. [DOI] [PubMed] [Google Scholar]
- 95.Mumford JL, He XZ, Chapman RS, Cao SR, Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC, et al. Lung cancer indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi: 10.1126/science.3798109. [DOI] [PubMed] [Google Scholar]
- 96.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr., Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195–5198. doi: 10.1038/sj.onc.1204646. [DOI] [PubMed] [Google Scholar]
- 98.Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62:6470–6474. [PubMed] [Google Scholar]
- 99.Parr RL, Dakubo GD, Crandall KA, Maki J, Reguly B, Aguirre A, Wittock R, Robinson K, Alexander JS, Birch-Machin MA, Abdel-Malak M, Froberg MK, Diamandis EP, Thayer RE. Somatic mitochondrial DNA mutations in prostate cancer and normal appearing adjacent glands in comparison to age-matched prostate samples without malignant histology. J Mol Diagn. 2006;8:312–319. doi: 10.2353/jmoldx.2006.050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, Kirkwood TB, Turnbull DM. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res. 2005;3:14–20. [PubMed] [Google Scholar]
- 102.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, Chi CW, Tam TN, Wei YH. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 103.Arnold RS, Sun CQ, Richards JC, Grigoriev G, Coleman IM, Nelson PS, Hsieh CL, Lee JK, Xu Z, Rogatko A, Osunkoya AO, Zayzafoon M, Chung L, Petros JA. Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment. Prostate. 2009;69:1–11. doi: 10.1002/pros.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- 106.Ishikawa K, Koshikawa N, Takenaga K, Nakada K, Hayashi J. Reversible regulation of metastasis by ROS-generating mtDNA mutations. Mitochondrion. 2008;8:339–344. doi: 10.1016/j.mito.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 107.Ishikawa K, Hashizume O, Koshikawa N, Fukuda S, Nakada K, Takenaga K, Hayashi J. Enhanced glycolysis induced by mtDNA mutations does not regulate metastasis. FEBS Lett. 2008;582:3525–3530. doi: 10.1016/j.febslet.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 108.Mizutani S, Miyato Y, Shidara Y, Asoh S, Tokunaga A, Tajiri T, Ohta S. Mutations in the mitochondrial genome confer resistance of cancer cells to anticancer drugs. Cancer Sci. 2009;100:1680–1687. doi: 10.1111/j.1349-7006.2009.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8:1378–1385. doi: 10.4161/cbt.8.14.8751. [DOI] [PubMed] [Google Scholar]
- 110.Li Y, Weibing S, Liu H, Hongli J, Zhuosheng L, Yadong W, Bing X, Daiming F. Mitochondrial DNA from colorectal cancer cells promotes the malignant phenotype of NIH3T3 cells. Cell Biol Int. 2008;32:979–983. doi: 10.1016/j.cellbi.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 111.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 112.Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Van Trappen PO, Cullup T, Troke R, Swann D, Shepherd JH, Jacobs IJ, Gayther SA, Mein CA. Somatic mitochondrial DNA mutations in primary and metastatic ovarian cancer. Gynecol Oncol. 2007;104:129–133. doi: 10.1016/j.ygyno.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y, Xue WC, Liu VW, Ngan HY. Detection of mosaic pattern of mitochondrial DNA alterations in different populations of cells from the same endometrial tumor. Mitochondrion. 2007;7:171–175. doi: 10.1016/j.mito.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 115.Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Irwin JA, Saunier JL, Niederstatter H, Strouss KM, Sturk KA, Diegoli TM, Brandstatter A, Parson W, Parsons TJ. Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples. J Mol Evol. 2009;68:516–527. doi: 10.1007/s00239-009-9227-4. [DOI] [PubMed] [Google Scholar]
- 117.Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, Wei YH. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg. 2010;139:189–197. doi: 10.1016/j.jtcvs.2009.04.007. e184. [DOI] [PubMed] [Google Scholar]
- 118.Turner CJ, Granycome C, Hurst R, Pohler E, Juhola MK, Juhola MI, Jacobs HT, Sutherland L, Holt IJ. Systematic segregation to mutant mitochondrial DNA and accompanying loss of mitochondrial DNA in human NT2 teratocarcinoma Cybrids. Genetics. 2005;170:1879–1885. doi: 10.1534/genetics.105.043653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci U S A. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bourgeron T, Chretien D, Rotig A, Munnich A, Rustin P. Fate and expression of the deleted mitochondrial DNA differ between human heteroplasmic skin fibroblast and Epstein-Barr virus-transformed lymphocyte cultures. J Biol Chem. 1993;268:19369–19376. [PubMed] [Google Scholar]
- 121.Dunbar DR, Moonie PA, Jacobs HT, Holt IJ. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci U S A. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 123.Vergani L, Rossi R, Brierley CH, Hanna M, Holt IJ. Introduction of heteroplasmic mitochondrial DNA (mtDNA) from a patient with NARP into two human rho degrees cell lines is associated either with selection and maintenance of NARP mutant mtDNA or failure to maintain mtDNA. Hum Mol Genet. 1999;8:1751–1755. doi: 10.1093/hmg/8.9.1751. [DOI] [PubMed] [Google Scholar]
- 124.Blok RB, Gook DA, Thorburn DR, Dahl HH. Skewed segregation of the mtDNA nt 8993 (G) mutation in human oocytes. Am J Hum Genet. 1997;60:1495–1501. doi: 10.1086/515453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weber K, Wilson JN, Taylor L, Brierley E, Johnson MA, Turnbull DM, Bindoff LA. A new mtDNA mutation showing accumulation with time and restriction to skeletal muscle. Am J Hum Genet. 1997;60:373–380. [PMC free article] [PubMed] [Google Scholar]
- 126.Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A-->G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet. 2001;68:238–240. doi: 10.1086/316930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Battersby BJ, Loredo-Osti JC, Shoubridge EA. Nuclear genetic control of mitochondrial DNA segregation. Nat Genet. 2003;33:183–186. doi: 10.1038/ng1073. [DOI] [PubMed] [Google Scholar]
- 128.Kurtz A, Lueth M, Kluwe L, Zhang T, Foster R, Mautner VF, Hartmann M, Tan DJ, Martuza RL, Friedrich RE, Driever PH, Wong LJ. Somatic mitochondrial DNA mutations in neurofibromatosis type 1-associated tumors. Mol Cancer Res. 2004;2:433–441. [PubMed] [Google Scholar]
- 129.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 130.Lee CF, Liu CY, Hsieh RH, Wei YH. Oxidative stress-induced depolymerization of microtubules and alteration of mitochondrial mass in human cells. Ann N Y Acad Sci. 2005;1042:246–254. doi: 10.1196/annals.1338.027. [DOI] [PubMed] [Google Scholar]
- 131.Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Telang S, Lane AN, Nelson KK, Arumugam S, Chesney J. The oncoprotein H-RasV12 increases mitochondrial metabolism. Mol Cancer. 2007;6:77. doi: 10.1186/1476-4598-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Groof AJ, te Lindert MM, van Dommelen MM, Wu M, Willemse M, Smift AL, Winer M, Oerlemans F, Pluk H, Fransen JA, Wieringa B. Increased OXPHOS activity precedes rise in glycolytic rate in H-RasV12/E1A transformed fibroblasts that develop a Warburg phenotype. Mol Cancer. 2009;8:54. doi: 10.1186/1476-4598-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hosgood HD, 3rd, Liu CS, Rothman N, Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL, Albanes D, Lan Q. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis. 2010;31:847–849. doi: 10.1093/carcin/bgq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bonner MR, Shen M, Liu CS, Divita M, He X, Lan Q. Mitochondrial DNA content lung cancer risk in Xuan Wei. China Lung Cancer. 2009;63:331–334. doi: 10.1016/j.lungcan.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95:1087–1091. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 139.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, Wei YH, Liu TY, Chi CW. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90:2390–2396. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N, Nakao A. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol. 2006;32:303–307. doi: 10.1016/j.ejso.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 141.Selvanayagam P, Rajaraman S. Detection of mitochondrial genome depletion by a novel cDNA in renal cell carcinoma. Lab Invest. 1996;74:592–599. [PubMed] [Google Scholar]
- 142.Meierhofer D, Mayr JA, Foetschl U, Berger A, Fink K, Schmeller N, Hacker GW, Hauser-Kronberger C, Kofler B, Sperl W. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25:1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- 143.Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 144.Moro L, Arbini AA, Marra E, Greco M. Mitochondrial DNA depletion reduces PARP-1 levels and promotes progression of the neoplastic phenotype in prostate carcinoma. Cell Oncol. 2008;30:307–322. doi: 10.3233/CLO-2008-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moro L, Arbini AA, Yao JL, di Sant’Agnese PA, Marra E, Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death Differ. 2009;16:571–583. doi: 10.1038/cdd.2008.178. [DOI] [PubMed] [Google Scholar]
- 146.Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y, Shirakawa T, Sawyer JR, Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene. 2006;25:1437–1445. doi: 10.1038/sj.onc.1209190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT, Kelly T, Higuchi M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci. 2008;99:1584–1588. doi: 10.1111/j.1349-7006.2008.00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 149.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 150.Naito A, Carcel-Trullols J, Xie CH, Evans TT, Mizumachi T, Higuchi M. Induction of acquired resistance to antiestrogen by reversible mitochondrial DNA depletion in breast cancer cell line. Int J Cancer. 2008;122:1506–1511. doi: 10.1002/ijc.23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chomyn A, Meola G, Bresolin N, Lai ST, Scarlato G, Attardi G. In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Mol Cell Biol. 1991;11:2236–2244. doi: 10.1128/mcb.11.4.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with tha tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy encephalopathy lactic acidosis and strokelike episodes. Mol Cell Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5' -> 3' DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 155.Correia RL, Oba-Shinjo SM, Uno M, Huang N, Marie SK. Mitochondrial DNA depletion it correlation with TFAM TFB1M TFB2M and POLG in human diffusely infiltrating astrocytomas. Mitochondrion. 2011;11:48–53. doi: 10.1016/j.mito.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 156.Pursell ZF, McDonald JT, Mathews CK, Kunkel TA. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity. Nucleic Acids Res. 2008;36:2174–2181. doi: 10.1093/nar/gkn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen D, Yu Z, Zhu Z, Lopez CD. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res. 2006;66:3485–3494. doi: 10.1158/0008-5472.CAN-05-4103. [DOI] [PubMed] [Google Scholar]
- 158.Trinei M, Berniakovich I, Pelicci PG, Giorgio M. Mitochondrial DNA copy number is regulated by cellular proliferation: a role for Ras and p66(Shc) Biochim Biophys Acta. 2006;1757:624–630. doi: 10.1016/j.bbabio.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 159.Donahue RJ, Razmara M, Hoek JB, Knudsen TB. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. FASEB J. 2001;15:635–644. doi: 10.1096/fj.00-0262com. [DOI] [PubMed] [Google Scholar]
- 160.Saada A. Deoxyribonucleotides and disorders of mitochondrial DNA integrity. DNA Cell Biol. 2004;23:797–806. doi: 10.1089/dna.2004.23.797. [DOI] [PubMed] [Google Scholar]
- 161.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 162.Pontarin G, Ferraro P, Hakansson P, Thelander L, Reichard P, Bianchi V. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem. 2007;282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 163.Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS Lett. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 164.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 166.Favre C, Zhdanov A, Leahy M, Papkovsky D, O’Connor R. Mitochondrial pyrimidine nucleotide carrier (PNC1) regulates mitochondrial biogenesis and the invasive phenotype of cancer cells. Oncogene. 2010;29:3964–3976. doi: 10.1038/onc.2010.146. [DOI] [PubMed] [Google Scholar]
- 167.Larosche I, Choumar A, Fromenty B, Letteron P, Abbey-Toby A, Van Remmen H, Epstein CJ, Richardson A, Feldmann G, Pessayre D, Mansouri A. Prolonged ethanol administration depletes mitochondrial DNA in MnSOD-overexpressing transgenic micebut not in their wild type littermates. Toxicol Appl Pharmacol. 2009;234:326–338. doi: 10.1016/j.taap.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 168.Mansouri A, Demeilliers C, Amsellem S, Pessayre D, Fromenty B. Acute ethanol administration oxidatively damages depletes mitochondrial dna in mouse liver brain eart and skeletal muscles: protective effects of antioxidants. J Pharmacol Exp Ther. 2001;298:737–743. [PubMed] [Google Scholar]
- 169.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shen M, Zhang L, Bonner MR, Liu CS, Li G, Vermeulen R, Dosemeci M, Yin S, Lan Q. Association between mitochondrial DNA copy number, blood cell counts, and occupational benzene exposure. Environ Mol Mutagen. 2008;49:453–457. doi: 10.1002/em.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yager JD. Endogenous estrogens as carcinogens through metabolic activation. J Natl Cancer Inst Monogr. 2000:67–73. doi: 10.1093/oxfordjournals.jncimonographs.a024245. [DOI] [PubMed] [Google Scholar]
- 172.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 173.Chen JQ, Brown TR, Yager JD. Mechanisms of hormone carcinogenesis: evolution of views role of mitochondria. Adv Exp Med Biol. 2008;630:1–18. [PubMed] [Google Scholar]
- 174.Higuchi M, Aggarwal BB, Yeh ET. Activation of CPP32-like protease in tumor necrosis factor-induced apoptosis is dependent on mitochondrial function. J Clin Invest. 1997;99:1751–1758. doi: 10.1172/JCI119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 176.Morais R, Zinkewich-Peotti K, Parent M, Wang H, Babai F, Zollinger M. Tumor-forming ability in athymic nude mice of human cell lines devoid of mitochondrial DNA. Cancer Res. 1994;54:3889–3896. [PubMed] [Google Scholar]
- 177.Magda D, Lecane P, Prescott J, Thiemann P, Ma X, Dranchak PK, Toleno DM, Ramaswamy K, Siegmund KD, Hacia JG. mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC Genomics. 2008;9:521. doi: 10.1186/1471-2164-9-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gilkerson RW, Margineantu DH, Capaldi RA, Selker JM. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 2000;474:1–4. doi: 10.1016/s0014-5793(00)01527-1. [DOI] [PubMed] [Google Scholar]
- 179.Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J Biol Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- 180.Nijtmans LG, Spelbrink JN, Van Galen MJ, Zwaan M, Klement P, van den Bogert c. Expression and fate of the nuclearly encoded subunits of cytochrome-c oxidase in cultured human cells depleted of mitochondrial gene products. Biochim Biophys Acta. 1995;1265:117–126. doi: 10.1016/0167-4889(94)00203-q. [DOI] [PubMed] [Google Scholar]
- 181.Gasser SM, Daum G, Schatz G. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J Biol Chem. 1982;257:13034–13041. [PubMed] [Google Scholar]
- 182.Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- 183.Arnould T, Mercy L, Houbion A, Vankoningsloo S, Renard P, Pascal T, Ninane N, Demazy C, Raes M. mtCLIC is up-regulated and maintains a mitochondrial membrane potential in mtDNA-depleted L929 cells. FASEB J. 2003;17:2145–2147. doi: 10.1096/fj.03-0075fje. [DOI] [PubMed] [Google Scholar]
- 184.Suzuki S, Naito A, Asano T, Evans TT, Reddy SA, Higuchi M. Constitutive activation of AKT pathway inhibits TNF-induced apoptosis in mitochondrial DNA-deficient human myelogenous leukemia ML-1a. Cancer Lett. 2008;268:31–37. doi: 10.1016/j.canlet.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, Zhang W, Plunkett W, Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA, Fan CY, Higuchi M. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun. 2007;364:656–661. doi: 10.1016/j.bbrc.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets transcription regulation and induced resistance to apoptosis. Gene. 2005;354:132–139. doi: 10.1016/j.gene.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Biswas G, Anandatheerthavarada HK, Avadhani NG. Mechanism of mitochondrial stress-induced resistance to apoptosis in mitochondrial DNA-depleted C2C12 myocytes. Cell Death Differ. 2005;12:266–278. doi: 10.1038/sj.cdd.4401553. [DOI] [PubMed] [Google Scholar]
- 189.Kwong JQ, Henning MS, Starkov AA, Manfredi G. The mitochondrial respiratory chain is a modulator of apoptosis. J Cell Biol. 2007;179:1163–1177. doi: 10.1083/jcb.200704059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Maki J, Robinson K, Reguly B, Alexander J, Wittock R, Aguirre A, Diamandis EP, Escott N, Skehan A, Prowse O, Thayer RE, Froberg MK, Wilson MJ, Maragh S, Jakupciak JP, Wagner PD, Srivastava S, Dakubo GD, Parr RL. Mitochondrial genome deletion aids in the identification of false- and true-negative prostate needle core biopsy specimens. Am J Clin Pathol. 2008;129:57–66. doi: 10.1309/UJJTH4HFEPWAQ78Q. [DOI] [PubMed] [Google Scholar]
- 191.Mehra N, Penning M, Maas J, van Daal N, Giles RH, Voest EE. Circulating mitochondrial nucleic acids have prognostic value for survival in patients with advanced prostate cancer. Clin Cancer Res. 2007;13:421–426. doi: 10.1158/1078-0432.CCR-06-1087. [DOI] [PubMed] [Google Scholar]
- 192.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Koch W, Califano J. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–2491. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]
- 193.Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;49:719–726. doi: 10.1373/49.5.719. [DOI] [PubMed] [Google Scholar]
- 194.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–152. doi: 10.1093/carcin/bgh282. [DOI] [PubMed] [Google Scholar]
- 195.Liu VW, Yang HJ, Wang Y, Tsang PC, Cheung AN, Chiu PM, Ng TY, Wong LC, Nagley P, Ngan HY. High frequency of mitochondrial genome instability in human endometrial carcinomas. Br J Cancer. 2003;89:697–701. doi: 10.1038/sj.bjc.6601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Laktionov PP, Tamkovich SN, Rykova EY, Bryzgunova OE, Starikov AV, Kuznetsova NP, Vlassov VV. Cell-surface-bound nucleic acids: Free and cell-surface-bound nucleic acids in blood of healthy donors and breast cancer patients. Ann N Y Acad Sci. 2004;1022:221–227. doi: 10.1196/annals.1318.034. [DOI] [PubMed] [Google Scholar]
- 197.Rykova EY, Laktionov PP, Skvortsova TE, Starikov AV, Kuznetsova NP, Vlassov VV. Extracellular DNAin breast cancer: Cell-surface-bound tumor-derived extracellular DNA in blood of patients with breast cancer and nonmalignant tumors. Ann N Y Acad Sci. 2004;1022:217–220. doi: 10.1196/annals.1318.033. [DOI] [PubMed] [Google Scholar]
- 198.Bennett RM, Gabor GT, Merritt MM. DNA binding to human Evidence for a receptor-mediated association leukocytes internalizationand degradation of DNA. J Clin Invest. 1985;76:2182–2190. doi: 10.1172/JCI112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Holdenrieder S, Stieber P, Bodenmuller H, Busch M, Von Pawel J, Schalhorn A, Nagel D, Seidel D. Circulating nucleosomes in serum. Ann N Y Acad Sci. 2001;945:93–102. doi: 10.1111/j.1749-6632.2001.tb03869.x. [DOI] [PubMed] [Google Scholar]
- 200.Lichtenstein AV, Melkonyan HS, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci. 2001;945:239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. [DOI] [PubMed] [Google Scholar]
- 201.Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975;35:2375–2382. [PubMed] [Google Scholar]
- 202.Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, Rossier A, Chen XQ, Anker P. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 203.Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci U S A. 1972;69:1685–1689. doi: 10.1073/pnas.69.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 205.Fournie GJ, Courtin JP, Laval F, Chale JJ, Pourrat JP, Pujazon MC, Lauque D, Carles P. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett. 1995;91:221–227. doi: 10.1016/0304-3835(95)03742-f. [DOI] [PubMed] [Google Scholar]
- 206.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 207.Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, Roden R, Sokoll LJ, Chan DW, Shih Ie M. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- 208.Ellinger J, Muller SC, Wernert N, von Ruecker A, Bastian PJ. Mitochondrial DNA in serum of patients with prostate cancer: a predictor of biochemical recurrence after prostatectomy. BJU Int. 2008;102:628–632. doi: 10.1111/j.1464-410X.2008.07613.x. [DOI] [PubMed] [Google Scholar]
- 209.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA. Roles of the Raf/MEK/ERK pathway in cell growth malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. Induction of an epithelial to mesenchymal transition in human immortal malignant keratinocytes by TGF-beta1 involves MAPK Smad and AP-1 signalling pathways. J Cell Biochem. 2005;95:918–931. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 212.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 213.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 214.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 216.Tsuruzoe K, Araki E, Furukawa N, Shirotani T, Matsumoto K, Kaneko K, Motoshima H, Yoshizato K, Shirakami A, Kishikawa H, Miyazaki J, Shichiri M. Creation characterization of a mitochondrial DNA-depleted pancreatic beta-cell line: impaired insulin secretion induced by glucose leucine and sulfonylureas. Diabetes. 1998;47:621–631. doi: 10.2337/diabetes.47.4.621. [DOI] [PubMed] [Google Scholar]
- 217.Spelbrink JN, Van Galen MJ, Zwart R, Bakker HD, Rovio A, Jacobs HT, Van den Bogert C. Familial mitochondrial DNA depletion in liver: haplotype analysis of candidate genes. Hum Genet. 1998;102:327–331. doi: 10.1007/s004390050700. [DOI] [PubMed] [Google Scholar]
- 218.Rotig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim Biophys Acta. 2009;1792:1103–1108. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 219.Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]