Abstract
We report an algorithm to detect structural variation and indels from 1 base pair to 1 megabase pair within exome sequence datasets. Splitread uses one-end anchored placements to cluster the mappings of subsequences of unanchored ends to identify the size, content and location of variants with good specificity and high sensitivity. The algorithm discovers indels, structural variants, de novo events and copy-number polymorphic processed pseudogenes missed by other methods.
Although the proportion of structural variants (SVs) and small insertions and deletions (indels; shorter than 50 bp) detected in sequence databases have increased exponentially1,2, recent comparisons of both experimental and computational methods suggest that the false negative rate remains high3,4. In addition to whole-genome sequencing, the widespread use of exome-capture technologies that target genomic protein-coding regions provides a rich resource to discover potentially impactful SVs and indels associated with disease. The nature of the capture methods, limited size of coding regions, and non-uniform distribution of the reads pose significant computational challenges. As a result, variants greater than 15 bp have rarely been reported in exome studies5,6. Discovery has been based largely on sequence alignment gaps limited to uniquely mapped regions of the genome (GATK7 or SAMtools8). Here, we detail a general combinatorial algorithm (Splitread) and validate its utility to discover indels and SVs in exome datasets.
We developed Splitread to detect SVs and indels based on the computational prediction of breakpoints (see online Methods and Supplementary Note for details). Similar to Pindel9, which is another split read based approach for detecting breakpoints of indels via a regional search around the anchored reads within the maximum event size, our algorithm searches for clusters of mate pairs where one end maps to the reference genome but the other end does not because it traverses a breakpoint creating a mapping inconsistency with respect to the reference sequence (Fig. 1a). We initially map reads using mrsFAST10, which guarantees all possible placements within a given Hamming distance (reflecting the number of allowed mismatches). Next, we decompose the unmapped end into subsequences of either equal length (balanced splits) or unequal length (unbalanced splits). Unlike Pindel which uses pattern growth for optimal matching in the target region, we reiteratively search for clusters of split reads using the balanced splits as seeds (Fig. 1a), which refine the location and size of the indel or SV event. We apply weighted set-cover approximation (Supplementary Note) to minimize the number of possible breakpoints, which essentially provides a maximum parsimony framework for all the mappings at the breakpoints.
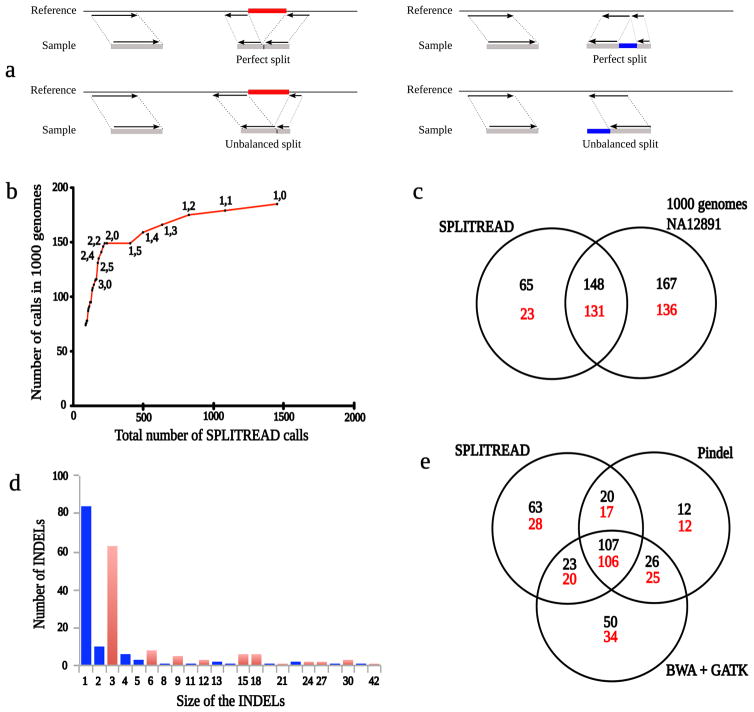
Figure 1. Splitread definition and analyses.
(A) Schematic diagrams for the mapping of paired-end sequences in cases where an individual has either a deletion (red) or an insertion (blue) with respect to the reference sequence. In each case, one-end anchored sequence is used to map one read in a pair. The second (unmapped) read is then decomposed into either two equal subsequences (balanced split) or two unequal subsequences (unbalanced split). (B) Number of Splitread predictions called by 1000 Genomes plotted against the total number of Splitread predictions using the indicated threshold numbers of balanced and unbalanced reads, respectively. A threshold of two balanced and two unbalanced splits maximizes intersection with 1000 Genomes Project calls without losing any positive predictive value. (C) A Venn diagram comparing variants detected by Splitread exome analysis versus whole-genome sequence analysis of NA12891 (black) or all variants within dbSNP130 (red). In order to intersect, variants must be at the same position and within 10 base pairs of the predicted size. (D) Length distribution of insertions and deletions mapping within the coding region of NA12891 as predicted by Splitread. Events with multiples of three base pairs (red) are compared to those that would disrupt the frame (blue). (E) A Venn diagram comparing Pindel, GATK and Splitread call sets on NA12891. The total number of events (black) is compared to those previously detected (red) as part of dbSNP130 and/or the 1000 Genomes Project.
We tested different thresholds for the number of balanced and unbalanced splits required to support a call. For each configuration, we plotted the proportion of events called by the 1000 Genomes Project (http://www.1000genomes.org) that were predicted by Splitread for sample NA12891 (Fig 1b and Supplementary Table 1). The slope provides the positive predictive value (PPV) and we could maximize sensitivity (number of corroborated predictions) without any loss of specificity by selecting the local maximum of this line. At a threshold of at least two balanced and two unbalanced splits, we predicted a total of 213 indel events less than 50 bp in the NA12891 exome, of which 69% (148) intersect with whole-genome sequence analysis (Fig. 1c) and 72% (154) intersect with dbSNP1302. As expected for protein-coding sequence11, indel sizes were predominantly in multiples of three resulting in no disruption of the protein-coding frame (47% or 100/213; Fig. 1d). If we exclude 1 bp indels, this fraction increases to 78% (100/129). We applied this threshold for the remainder of our analysis for calling the final events.
We identified an additional 63 SV events ( > 50 bp) after excluding annotated processed pseudogenes (Supplementary Table 2). Although only four of these were predicted by the 1000 Genomes Project, nine of the remaining events intersect with SVs from dbSNP130 with sizes varying from 51 bp to 3,584 bp. We predict that 48 of these variants are common (observed in multiple HapMap samples we analyzed) with only 21 variants being specific to NA12891. Several correspond to genes known to carry complex insertion and deletion polymorphisms or variable number of tandem repeats (VNTRs) such as MUC6, DSPP and MUC1612.
We compared Splitread with alternative indel detection methods Pindel9 and GATK7 (see Supplementary Note for comparison to CREST). 70% of Splitread calls are predicted by one of the other methods but a substantial fraction of calls are unique to each method. As expected, events called by two or more methods show the best corroboration with dbSNP and 1000 Genomes calls (Fig. 1e). We selected 19 events uniquely called by Splitread and previously not reported by dbSNP or 1000 Genomes for PCR-based validation. Thirteen of 19 events were validated (Supplementary Table 1), giving an estimated PPV of 68%. Most map within low complexity regions and correspond to repeat expansions and deletions (Supplementary Table 1). If we include previously reported events, Splitread accuracy rises to 87% (41/47).
We extended our analyses by generating exome sequence data from 11 HapMap samples whose genomes were sequenced at 3- to 4-fold coverage by the 1000 Genomes Project (Supplementary Table 3). Using Splitread, we observed an average of 325 events for each sample, including 286 indels and 39 SVs (5:1 ratio). Approximately 68% and 70% of the calls intersected 1000 Genomes and dbSNP130 predictions, respectively. From the 11 samples, we identified 192 novel SVs, 93 of which were observed two or more times; an average of nine events that disrupt genes are unique to each individual (Supplementary Tables 2,3).
As a final test, we applied Splitread to published exome data from 20 parent-child trios affected with sporadic autism spectrum disorder6. We identified an average of 191 indels and 57 SVs in this dataset (Supplementary Table 4). To test the accuracy of our calling method, we randomly selected indels and SVs not found in either dbSNP or the control individuals as part of the Exome Sequencing Project (http://esp.gs.washington.edu). We confirmed 10/12 events by PCR and sequencing, giving an estimated PPV of 83% (Supplementary Table 5). This included bona fide variation within repetitive and low-complexity regions such as a triplet and 12-mer insertion within a low-complexity coding portion of SHROOM4 (Supplementary Fig. 1) missed by Pindel9 and GATK7.
An important goal of parent-child trio sequencing is to discover potentially disruptive de novo events. This is challenging since the selection of potential de novo events will either enrich in false-positives or represent inherited variants that were not detected (false negatives) in one of the parents. In this study, we were only able to detect and confirm one previously reported de novo variant, in FOXP16. The remaining events were either present in a parent or were false positives (Supplementary Table 1). We sought to increase our confidence in predicting de novo events by filtering via read-depth. Because our method uses Hamming distance to align reads, SV and indel breakpoints should cause fewer reads to map in the affected child if the event is truly de novo (Supplementary Note). We added this functionality as a filter which normalizes the read-depth of coding regions based on coverage and then compares proband and parents to flag regions of reduced depth. The filter is applied specifically at predicted breakpoints to minimize false positives (Supplementary Fig. 2).
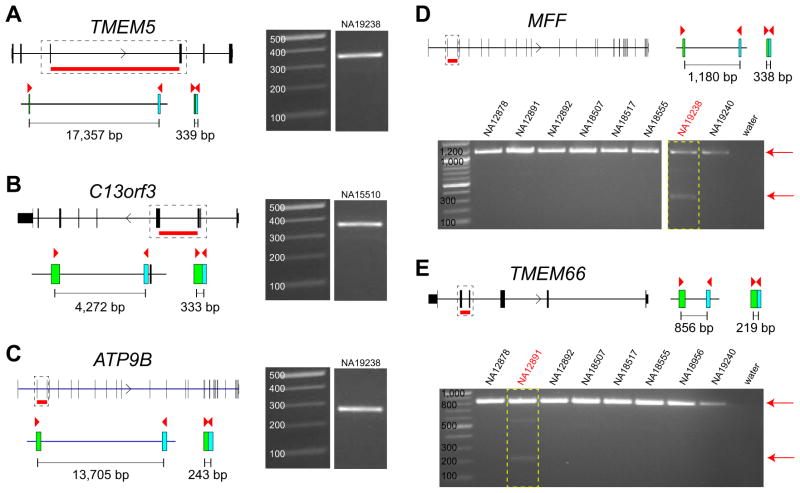
During our analysis of exome datasets, we routinely detected putative deletion events where an intron was precisely removed such that flanking exons were perfectly abutting. The structure of these events suggested uncharacterized processed pseudogenes as opposed to allelic deletions. These arise as a result of retrotransposition of spliced mRNA back into the genome. We discovered 25 such events in the 11 HapMap exomes (Supplementary Table 6), 14 of which could not be identified by BLAST searches against the reference genome (GRCh37). DNA amplification of flanking exons yielded 16 products consistent with a processed pseudogene in the affected individual while the other nine appear to be polymorphic in the population (Fig. 2). Since pseudogenes can create potential Splitread artifacts we created a modified exome reference for mapping that includes known processed pseudogenes, segmental duplications, and copy-number polymorphic pseudogenes. Compared to a whole-genome reference, this modified exome reference increases speed by 10-fold with only a 2% difference in the number of calls. Thus, Splitread can be applied to a large number of exomes in a computationally efficient manner to generate a database of bona fide exonic indels and SVs.
Figure 2. Validation of processed pseudogenes.
Gene models and predicted intron deletions of the processed pseudogenes are shown. Primers (red triangles) are designed in the coding region of the genes and the expected product size for the processed pseudogenes are shown for (A) TMEM5, (B) C13orf3, (C) ATP9B, (D) MFF, and (E) TMEM66. Gel images show the size of the amplified product. We were able to detect the processed version of these genes in our PCR experiments. In D-E we genotyped the processed pseudogenes MFF and TMEM66 within eight HapMap samples and show that each is amplified only in the predicted sample [boxed in red: NA19238 (MFF) and NA12891 (TMEM66)]. All PCRs amplify the normal gene (signal on the top) with only one sample each amplifying the processed gene.
To test the applicability of Splitread to whole genome datasets we analyzed the genome of a patient (ND06769) with a hexanucleotide repeat expansion (GGCCCC) in the C9orf72 gene. Renton et al.13 demonstrated that this is the causal variant of 9p21-associated Amyotrophic Lateral Sclerosis with frontotemporal dementia (ALS-FTD). This repeat expansion was missed by GATK and was discovered only through manual inspection of the read alignments13. Although the insertion is too long to be fully characterized by a split-read method (estimated 1.5 kilobase pairs), our algorithm was able to discover the approximate breakpoint of the expansion and supported the call with read-depth analysis. Splitread can detect insertions and deletions without any size limitation. The size spectrum of the insertions that can be accurately characterized by Splitread is bound by the read length; however it is possible to detect the approximate breakpoints of larger insertions using one-end anchored reads.
Many validated events detected exclusively by Splitread involve microsatellite, low complexity, or polynucleotide tracts (Supplementary Table 1 and Supplementary Fig. 1). Such regions are subject to higher mutation rates, due in part to their greater potential for replication slippage14. Variation of this type, especially within coding regions, has frequently been associated with diseases including triplet repeat instability14. Our increased PPV for this class of variant stems from the fact that we consider multiple mappings frequently discarded by other methods. There is, however, genetic variation that we clearly missed (Fig. 1) emphasizing that no single approach is comprehensive in capturing all genetic variation3. One limitation of the Splitread is the dependence on the balanced splits to seed an event, which is directly dependent on the coverage. Given 76 bp reads, the chance of detecting a heterozygous event is 55% at 20X coverage, but rises to > 90% at 60X coverage. The sensitivity estimate increases from 79% at 20X coverage to 98% at 60X coverage. Such median sequence coverage is not uncommon in many exome sequencing projects.
An unexpected consequence of our exome analysis has been the discovery of a substantial number of processed pseudogenes that are polymorphic but not represented in the human reference genome (Supplementary Table 6). Most of these variants were seen more than once, ranging in frequency from 3% to 72% based on an assessment of 51 exomes (Supplementary Table 6). Using read-pair information, we were able to map the location of all of these polymorphisms using a one-end anchored mapping strategy15. A comprehensive catalog of the most common of these will be important for correctly interpreting disease-causing variants discovered in exome studies.
Since different computational methods vary in their sensitivity and specificity depending on the size, class, and context of variants, multiple approaches must be considered to maximize variant discovery. While most efforts are focused on the detection of point mutations within coding sequence, there is an opportunity to explore the landscape of intermediate and larger genetic variation, which is more likely to be gene disruptive. It is critical to include this type of variation in future analyses to correctly interpret the causes of disease. Re-examining exome datasets for larger and more complex variation may be particularly relevant when the causal variants for seemingly Mendelian diseases remain undiscovered.
Supplementary Material
Supplementary Figure 1 Validation of a complex indel.
Supplementary Figure 2 Read depth filtering for de novo events.
Supplementary Table 1 Splitread Validation for the NA12891 exome.
Supplementary Table 2 The list of all structural variants and the frequency of these variants among 11 samples.
Supplementary Table 3 Summary for Splitread analysis of 11 HapMap Exomes.
Supplementary Table 4 Analysis of the 63 individuals from autism trio data.
Supplementary Table 5 Splitread Validation from Autism Trios.
Supplementary Table 6 Copy-number polymorphic processed pseudogenes. Supplementary Note
Acknowledgments
We thank Tonia Brown and S. Girirajan for helpful comments during manuscript preparation. This work was supported by a Simons Foundation Autism Research Initiative Award SFARI191889 (E.E.E) and National Institutes of Health grants HD065285 (E.E.E.), HHSN273200800010C (D.A.N.), HL 102926 (D.A.N.). E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
E.K. designed and implemented the Splitread algorithm, E.K. and C.A. analyzed data, B.J.O., L.V., M.J.R, and D.A.N. generated sequencing data, M.D. and K.M. performed validation experiments and analyzed processed pseudogenes, E.K., C.A., and E.E.E. wrote the manuscript.
Competing Financial Interests
E.E.E. is a member of the Scientific Advisory Board of Pacific Biosciences.
Accession codes. Short read archive (SRA): SRA039053.
References
- 1.Church DM, et al. Public data archives for genomic structural variation. Nature genetics. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills RE, et al. Mapping copy number variation at fine scale by population scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics (Oxford, England) 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nature methods. 2010;7:576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills RE, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome research. 2011;21:830–839. doi: 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen TV, et al. Short mucin 6 alleles are associated with H pylori infection. World J Gastroenterol. 2006;12:6021–6025. doi: 10.3748/wjg.v12.i37.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renton AE, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011 doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nature reviews. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 15.Kidd JM, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nature methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics (Oxford, England) 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nature methods. 2010;7:576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamming RW. Error-detecting and error-correcting codes. Bell System Technical Journal. 1950;29:147–160. [Google Scholar]
- 19.Kidd JM, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nature methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajirasouliha I, et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics (Oxford, England) 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karp RM. In: Complexity of Computer Computations. Miller JWTRE, editor. Plenum; New York: 1972. pp. 85–103. [Google Scholar]
- 22.Chvatal V. A Greedy Heuristic for the Set-Covering Problem. Mathematics of Operations Research. 1979;4:233–235. [Google Scholar]
- 23.International HapMap Cconsortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Validation of a complex indel.
Supplementary Figure 2 Read depth filtering for de novo events.
Supplementary Table 1 Splitread Validation for the NA12891 exome.
Supplementary Table 2 The list of all structural variants and the frequency of these variants among 11 samples.
Supplementary Table 3 Summary for Splitread analysis of 11 HapMap Exomes.
Supplementary Table 4 Analysis of the 63 individuals from autism trio data.
Supplementary Table 5 Splitread Validation from Autism Trios.
Supplementary Table 6 Copy-number polymorphic processed pseudogenes. Supplementary Note