Abstract
Interfacial peptides and proteins are critical in many biological processes and thus are of interest to various research fields. To study these processes, surface sensitive techniques are required to completely describe different interfacial interactions intrinsic to many complicated processes. Sum frequency generation (SFG) spectroscopy has been developed into a powerful tool to investigate these interactions and mechanisms of a variety of interfacial peptides and proteins. It has been shown that SFG has intrinsic surface sensitivity and the ability to acquire conformation, orientation and ordering information about these systems. This paper reviews recent studies on peptide/protein-substrate interactions, peptide/protein-membrane interactions and protein complexes at interfaces and demonstrates the ability of SFG on unveiling the molecular pictures of complicated interfacial biological processes.
Introduction
Studies on peptides and proteins at interfaces have been found to be extremely important in the applications of chemical, biological, medical science and engineering.1–7 Interfacial peptides and proteins contribute to various biological functions such as cell signaling, immune responses, cell adhesion, and catalytic reactions. Understanding the structures and properties of these peptides and proteins at interfaces can aid in resolving the mechanisms they follow during different biological processes. Although it is still a challenging topic, the study of these interfacial molecules has becoming one of the hot-spots of today’s scientific research.8–10
A variety of surface sensitive analytical techniques and methodologies have been applied to explore conformation, ordering and orientation perspectives about the interfacial peptides and proteins. Ellipsometry, which measures the change of polarization of a circular-polarized incident light, is very sensitive to surface changes, allowing for the ability to follow proteins’ adsorption kinetics and coverage.11–13 Surface Plasmon Resonance (SPR) is sensitive to local refractive index changes and therefore provides real-time information of interactions including adsorption/desorption between biomolecules and the substrate surfaces (typically self-assembled monolayers).14–16 Secondary Ion Mass Spectrometry (SIMS) uses a focused ion beam to fragment the sample surface and mass-to-charge ratios of these ejected species are used to map out the structures of surface components. It has been employed to monitor structural changes of peptides and proteins on surfaces.17–19 X-ray Photoelectron Spectroscopy (XPS) irradiates the sample surfaces, ejecting electrons from the surface atoms. It analyzes the binding energy between the atoms to determine their chemical environment, allowing the study of biomolecule interactions.10,20,21 Atomic Force Microscopy (AFM) collects the feedback signal from an AFM tip interacting with a sample surface. It is important in investigating the protein packing and ordering state.22–24
In addition, many techniques focus on using intrinsic vibrational modes of various chemical species to determine composition and orientation of biomolecules. Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) spectroscopy is a vibrational spectroscopy frequently used due to the shallow penetration depth from the evanescent wave produced by total internal reflection of IR light. Using different-polarized incident light, ATR-FTIR can be used to study orientation of interfacial peptide/proteins.25–27 Surface-enhanced Raman Spectroscopy (SERS) is another useful spectroscopic technique which enhances Raman scattering signal of molecules absorbed on metal substrates such as gold or silver. It is exceptionally sensitive and has been used to follow structural changes in various biological processes.28,29 These experimental tools have the ability to acquire a qualitative and quantitative understanding at the molecular-level of interfacial peptides/proteins and their functions. However, it is still very difficult to probe molecular level structural information of peptides and proteins in situ (e.g., at the solid/liquid interface) with a monolayer surface sensitivity. As a result, further questions about the molecular interactions still require more accurate in situ measurements.
Sum Frequency Generation vibrational spectroscopy (SFG) is a nonlinear optical technique which is intrinsically surface specific. It requires small amount of samples and probing can be done in situ and in real time. It has been developed into a powerful tool since the 1980s, demonstrating its ability to resolve crucial questions in the areas of surface catalysis, electrochemistry, and polymer science.30–43 Recently, SFG has also been employed to study biomolecules such as lipids, peptides and proteins which further progressed this research field.44–61 This article presents an overview of studies that have been done in our group that sheds light on elucidating molecular level information about interfacial peptides/proteins involved in different biological environments as well as processes.
SFG Technique
1. Main Characteristics
The theoretical background and the experimental parameters detailing SFG have been extensively described elsewhere30–43,62–64 and will not be reiterated here. In this review, we want to focus on a few unique characteristics of SFG in order to illustrate why it is suitable for the study of interfacial peptides/proteins.
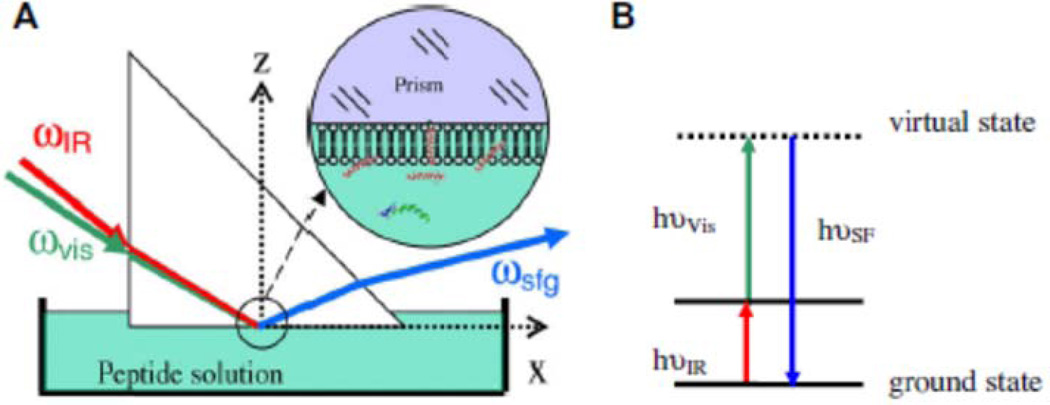
SFG is a second order nonlinear optical process in which two laser beams of frequencies ωIR and ωVis are overlapped both spatially and temporally in a medium generating the sum frequency (SF) beam, ωSF = ωIR + ωVis (Figure 1). Conventionally, the frequency (or wavelength) of the visible beam is fixed while that of the IR beam is tunable. The sum frequency process is resonantly enhanced when the frequency of the tunable IR beam matches a vibrational transition of a functional group in the probed material (Figure 1), generating a vibrational spectrum characteristic of the sample. Recently, broadband SFG systems using a femtosecond laser to generate a visible input beam with a narrow frequency width and an IR input beam with a broad frequency spectrum have been implemented. All molecular vibrational signatures are enhanced simultaneously and the resulting SFG signal is spectrally spread and can be collected by a CCD camera. Furthher details have been reported in previous publications and will not be repeated here.65,66
Figure 1.
A: The near-total-reflection geometry used in SFG experiment; B: SFG energy level diagram (Reproduced with permission from J. Struct. Biol. 20009, 168, 61–77. Copyright 2009, Elsevier)
The most important property that makes SFG an appropriate tool to study interfacial peptides and proteins is from its selection rules. SFG is forbidden in materials with inversion symmetry under the electric dipole approximation. Most bulk materials have inversion symmetry and thus do not generate an SFG signal. However, at a surface or interface, inversion symmetry is broken allowing for SFG to occur. As a result, bulk signal is usually negligible compared to signal from the surface/interface. For peptides or proteins, we have demonstrated that a thin film model can be applied which allows the study of their structures at interfaces.67 Since SFG can only detect signals from ordered peptides and proteins at interfaces, it is difficult to measure adsorption amounts of peptides and proteins at interfaces. If such interfacial coverage can be measured with complementary techniques, e.g., surface plasmon resonance or reflection infrared spectroscopy, then SFG signals can be used to determine more detailed structural information of peptides and proteins at interfaces (e.g., orientation distribution).
SFG provides characteristic spectral features of different vibrational stretching modes. These representative peaks or features within each spectrum are assigned to different functional groups being probed and therefore can paint a molecular picture of the system. Additionally, SFG is an optical spectroscopy so any materials accessible by laser light can be studied. In this way, in situ experiment can be performed in real time. In addition to surface sensitivity and in situ ability, SFG also has very low detection limits which are comparable to biologically relevant concentrations of peptides and proteins.
2. SFG Analysis
2.1 C-H Stretching and Amide I Signals
Strong SFG signals can be found in the C-H stretching frequency region (i.e. 2800~3100 cm−1), which consists of contributions from the side chains of peptides or proteins containing alkyl, aromatic or cyclic groups.58,63,68 They can be used to monitor the structures of hydrophobic side chains which are associated with backbone structures.58,63,68 Conversely, SFG signals in the amide I region (i.e. 1600~1700 cm−1), which are mainly from the polypeptide backbone C=O stretching modes, tell us about the structures of protein backbones.69,70 Different secondary structures exhibit different SFG spectral signatures, as those detected in the previous FTIR and Raman studies. For example, our group has successfully determined the SFG peak centers of α-helix, 310-helix and β-sheet secondary structures by fitting the amide I band.59,70,71 SFG signal is affected by the surface coverage, orientation and ordering of functional groups and composition of functional groups (secondary structure in case of amide I band). As a result, SFG can provide information about the conformation, orientation and orientation distribution (ordering) of peptides/proteins at interfaces based on spectral features.
SFG signals come from an interface with no inversion symmetry which is usually infinitely sharp. But an adsorbed protein layer is usually several or tens of nanometers thick and that is much thicker than an infinitely sharp interface. For this situation, we have successfully shown that a thin film model can be employed to treat the probed peptides/proteins.67 Under this model, SFG signals are considered to be generated from the entire adsorbed layer (the entire peptide/protein) in order to deduce interfacial structure.
2.2 Orientation Analysis
From the previous section, we know that SFG is able to detect signals from the side chains (C-H region) and the main backbones (amide I region) of peptides/proteins. Our group has done extensive studies on orientation analysis of functional groups in the C-H stretching frequency region.63,68,72,73 Also, compared to the side chains, backbone structure is a better indicator for the orientation of the entire peptide/protein. As a result, this review will only discuss about the orientation analysis of amide I band.
In our previous publications, we have developed methodologies to determine the orientation of different secondary structures of peptide/protein backbones such as α-helix, 310-helix and β-sheet secondary structures.71,74–78 Since α-helix is the most common secondary structure in peptides and proteins, this review will mainly focus on how to obtain the orientation information of an α-helix.
The orientation information can be obtained by using group theory and projection operators when analyzing the SFG spectra collected with different polarization combinations.75 It has been shown that both amide I A mode and amide I E1 mode are SFG active.74–77 Using the near-total-reflection geometry (shown in Figure 1A), ssp (s-polarized sum frequency signal beam, s-polarized input visible beam, p-polarized input IR beam) and ppp spectra of the amide I band can be collected corresponding to the χyyz and χzzz second order nonlinear optical susceptibility components respectively.74 The dependence of χyyz and χzzz susceptibility components on the molecular hyperpolarizability is described by the following equations:74
For the A mode,
where r = βaac/βccc
For the E1 mode,
where βccc and βaca are the molecular hyperpolarizability elements, Ns is the number density of an ideal α-helix, and “< >” means average. The A mode and E1 mode cannot be completely resolved in the frequency domain because of the limitation of the SFG spectral resolution. The total susceptibility is often assumed to be the sum of the susceptibilities from these two modes.74 The hyperpolarizability elements of an α-helix are the product of the components of the Raman polarizability and IR transition dipole moment. Theoretically, we deduced the ratios r=βaac / βccc = 0.59 and βaca / βccc = 0.31.74 Also, if we assume all α-helical structures at the surface/interface adopt the same orientation, <cos θ> and <cos3 θ> can be substituted by cos θ and cos3 θ. Therefore, we can deduce a function relating χyyz / χzzz with the orientation angle for an ideal α-helix. Experimentally, χyyz/χzzz can be measured. Combining these values and the equations above, the orientation angle θ of an α-helix on a surface or at an interface should be able to be deduced.
There are some deviations from this ideal situation.74 First of all, the calculation model discussed above is perfect α-helical structure which either has a unit cell with 18 peptide units or is infinitely long. In nature, it is not always the case. To address this problem, we have done a thorough discussion in previous publications and have shown that “extra” or “missing” amino acid residues from the 18 (or multiple of 18)-unit cell play minimal role in data analysis. Also the deviations from an ideal helix can be corrected by considering the helix length in the orientation analysis.74 On the other hand, one peptide or protein may have more α-helical segments pointing at different directions. We have successfully calculated the overall hyperpolarizability of such bent helical components, which can be used to determine the orientation of entire proteins.79 Finally, peptides or proteins do not necessarily adopt a single orientation in biological systems; they can stay in multiple orientations. For this situation, the maximum entropy function has been introduced to calculate the orientation distribution of the target peptide or protein in combination of both SFG and ATR-FTIR orientation analysis results.76,77
Example Applications
1. C-H Stretching Frequency Region
Early SFG studies of interfacial peptides/proteins looked at the C-H stretching frequency region which is believed to be generated from the amino acid side chains of peptides or proteins. Because SFG signals are related to the number density, ordering and orientation of the functional groups, our group has demonstrated interfacial peptide/protein responses such as conformational changes in interfacial environments of different hydrophobicity and of different pH values in situ.58,63,68
Alternatively, C-H stretching signals from the species that peptides/proteins interact with can also provide crucial information about the properties of the acting peptides/proteins. The interaction between peptides/proteins and lipid bilayers is a great example. By monitoring the lipid bilayer signals in real time, we were able to show the integrity, deformation, and flip-flop of the lipid bilayers.80–82
For both of the aspects mentioned above, isotope-labeling from C-H to C-D is important in order to differentiate spectral confusion from different components of the system under study. We used isotope-labeled proteins and polymers to study protein adsorption behavior onto different polymeric substrates.83,84 Using uniform labeling (isotope throughout the protein), we can see a protein in a site-independent manner. Using selective labeling, we can specifically monitor one single residue of a protein.83 Isotope-labeling is frequently used to study the interaction between peptides/proteins and lipid bilayers. Through deuteration, we have been able to probe either the bilayer behavior or the response from each leaflet.82,85
2. Amide I Band
Further developments of SFG have extended the detectable range past C-H stretching frequency region. In 2003, our group, for the first time, demonstrated that SFG can be used to detect amide I band which contains various information about the secondary structures of interfacial peptides and proteins at solid/liquid interfaces.69
A near-total-reflection geometry, as shown in Figure 1A, was used in the study. SFG is a noninvasive and surface sensitive vibrational technique having several unique characteristics: (1) Background subtraction is not required to generate an amide I spectrum because water bending mode does not contribute noticeable signal in this range. (2) Near-total-reflection geometry generates very strong SFG amide I signals allowing for easy and reliable SFG data analysis.
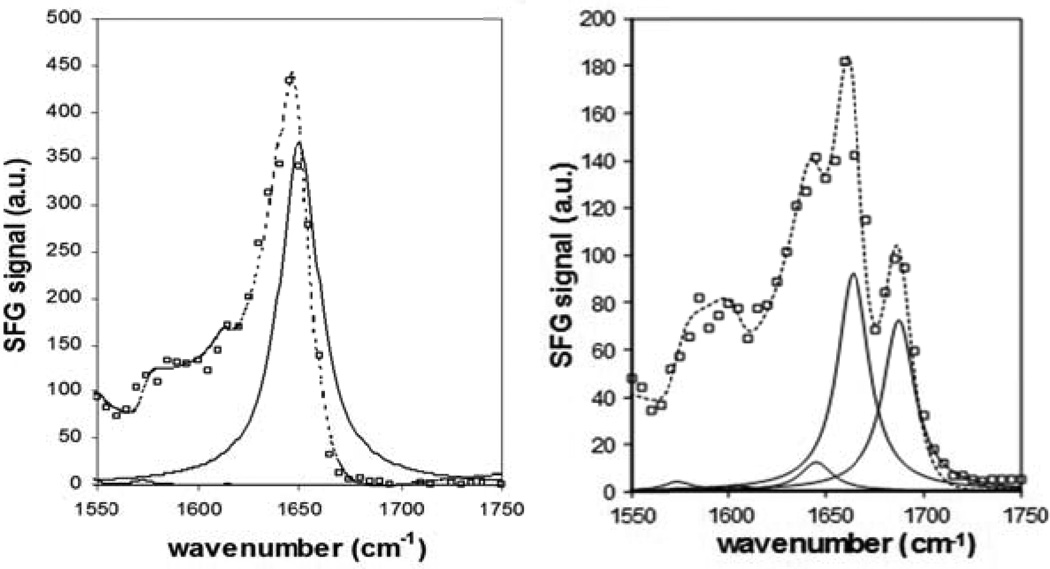
Based on IR and Raman studies, we were able to assign the SFG amide I signals to different secondary structures of peptides/proteins.70 A peak centered around 1650 cm−1 is attributed to α-helical structures. β-sheet structure has characteristic peaks at 1635 cm−1 and 1685 cm−1 (Figure 2).70 A recent study on alamethicin suggested that peaks centered 1635 cm−1 and 1675 cm−1 are from α-helical/310 helical structures.71 SFG is known for its ability to provide orientation information from polarization sensitive measurements. Our group has shown the orientation analyses of different secondary structures of the interfacial peptides and proteins.71,74–78 But due to space constraint, we will focus on α-helical structures in this review.
Figure 2.
SFG spectra and fitting results for 0. 1mg/ml MSI594 (left) and 0.1mg/ml Tachyplesin I (right) adsorbed at the peptide solution/PS interface and with squares representing the actual spectra, dotted line the fitted spectra and solid lines the component peaks used to fit the spectra (Reproduced with permission from Langmuir 2005, 21, 2662–2664. Copyright 2005, Am. Chem. Soc.).
3. Peptide/Protein Surface Interactions
3.1 Interactions between Peptides/Proteins and Polymer Substrates
3.1.1 Physical Adsorption
Peptide/protein adsorption onto the substrate surface is the first reaction between a biomaterial and a biological system. The subsequent interactions such as inflammation or blood coagulation are greatly affected or controlled by this first adsorption step. It has been shown that the changes during adsorption process and the final structure of the peptide/protein on biomaterials affect whether a material can perform its intended function or not. A lot of effort has been put into probing the adsorption process but due to instrumental constraints, few details about the molecular changes occurring at the interfaces have been obtained.
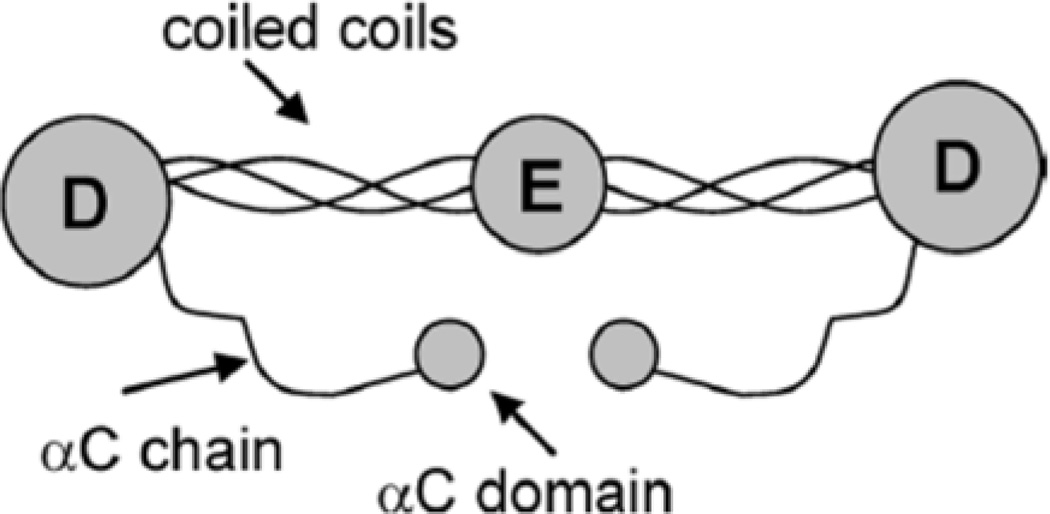
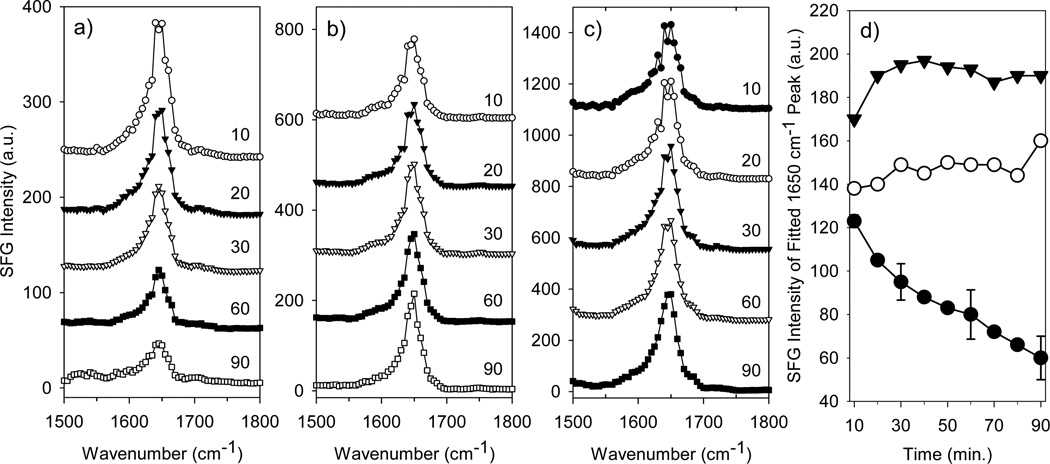
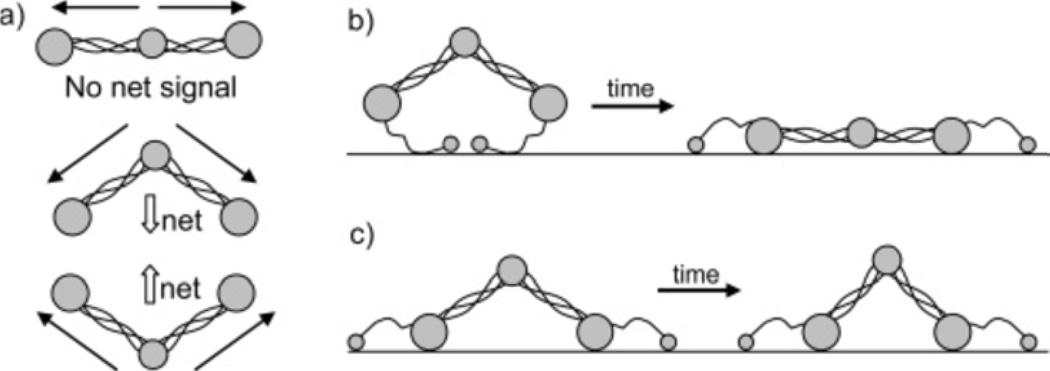
Our group has been able to provide valuable information on the adsorbed peptides/proteins with the use of SFG. Fibrinogen is a common blood protein and its conformation after adsorption onto biomaterials is thought to be greatly related to the further reactions that lead to thrombosis.86 Our group studied adsorption of fibrinogen onto a series of polymer surfaces using SFG.86 This study focused on Amide I signals generated from the secondary structures of fibrinogen. The major peak observed around 1650 cm−1 in the spectra was attributed to the α-helices of the coiled coils within the fibrinogen molecule. Due to the intrinsic inversion symmetry possessed by the molecule (as shown in Figure 3), it was deduced that fibrinogen must adopt a bent structure when adsorbed to the polymer surfaces. By varying the hydrophobicity of polymer, time-dependent changes in α-helical signal were observed (Figure 4). Based on the results from complementary techniques, we concluded that this difference was attributed to different orientational changes of fibrinogen on the polymers. It is believed that fibrinogen interacts with hydrophobic surfaces via the D domains and interacts with hydrophilic surfaces via the αC domains as shown in Figures 5b and 5c. SFG results demonstrate the differences in the initial binding and post-adsorption changes of fibrinogen onto substrates of different hydrophobicity (Figure 5).
Figure 3.
Structural representation of fibrinogen (Reproduced with permission from J. Phys. Chem. B 2005, 109, 22027–22035. Copyright 2005, Am. Chem. Soc.)
Figure 4.
SFG spectra of fibrinogen adsorbed to (a) a polyurethane, (b) a polyurethane-silicone copolymer and (c) a fluorinated polymer in PBS buffer in the amide I range collected at different times (in minutes). Alpha-helix SFG signal as a function of time (d) from fitting SFG spectra for fibrinogen adsorbed to the polyurethane (open circles), the polyurethane-silicone copolymer (closed circles) and the fluorinated polymer (closed triangles). Representative error is shown for the fibrinogen/polyurethane sample (Reproduced with permission from J. Phys. Chem. B 2005, 109, 22027–22035. Copyright 2005, Am. Chem. Soc.).
Figure 5.
a: Net transition dipole moments of fibrinogen in different configurations. Schematic of fibrinogen structural changes with time after adsorption onto b: a polyurethane and c: a polyurethane-silicone copolymer and a fluorinated polymer (Reproduced with permission from J. Phys. Chem. B 2005, 109, 22027–22035. Copyright 2005, Am. Chem. Soc.).
Further study on fibrinogen adsorption was carried out in both amide I and C-H stretching frequency regions using a deuterated polystyrene surface.87 Adding to the information from the previous research, this study demonstrated that the time dependent signal first increased due to an increase in fibrinogen adsorption to the surface and then decreassed because fibrinogen adopted a more linear structure. Comparing spectral changes of C-H and amide I band, we found the changes in hydrophobic side chains were more localized. It does not affect the secondary structure and it reorients to a stable state much faster than the secondary structure.
3.1.2 Chemical Immobilization
Peptide/protein immobilization is the process of binding peptides or proteins onto solid substrates in order to add biological functionalities to these substrates. The structure and orientation of the immobilized peptides/proteins control the biological functions. As a result, molecular-level control and characterization of such interfacial peptides and proteins are important to the development of many biological and biomedical applications such as implant devices and biosensors.
SFG has demonstrated to be a suitable technique to investigate this interfacial phenomenon and it has recently been employed in our group to study the immobilization between peptides/proteins and solid substrates. A study on the immobilization of an α-helical antimicrobial peptide (AMP) cecropin P1 (CP1) was performed.88 CP1 has been used to capture and sense bacterial pathogens and as such, the fabrication of it onto substrates is of great importance to biosensor development. In this study, a polystyrene (PS) surface and a maleimide-functionalized polystyrene (PS-MA) surface were used as solid substrates. The c-terminus of CP1 was modified by a cysteine residue so that it can be chemically bound to PS-MA and only physically adsorbed to PS. The SFG spectra in the amide I region were dominated by the α-helical peak centered at 1650 cm−1. After performing a washing procedure which removed the loosely bound peptides, the signal retained for the chemically immobilized CP1 and almost disappeared for the physically adsorbed CP1. These results directly indicate that physical adsorption of CP1 creates a weak interaction with its substrate and therefore is not ideal for immobilization due to poor stability. On the other hand, the chemically tethered peptides develop a strong interaction with their substrate, allowing for a well-defined orientation. Therefore chemical immobilization leads to a more stable and effective biofunctionalized surface. Also, we showed that different chemical environments (e.g. air and water) resulted in different peptide orientations.
To better understand how the chemical environment affects peptide immobilization process, we further researched the effects of peptide concentration, solvent composition and assembly state (monomer or dimer).89 Chemical immobilization was stressed in this study using the same procedures as mentioned above. Taking the ratios between amide I spectra collected under ppp and ssp polarization combinations, we were able to deduce that as peptide concentration increased, CP1 tended to stand up more which was attributed to the peptide-peptide interaction. We also demonstrated that immobilization reached equilibrium faster with higher peptide concentration through the time-dependent spectra. By using solvents with different ratios between 2,2,2-trifluoroethanol (TFE) and phosphate buffer (PB), we found that TFE induced α-helical content in solution which made the immobilization simpler than in PB solution alone. This resulted in a preferential orientation rather than multiple orientations.89 From the time-dependent spectra in TFE environments, it was clear that CP1 molecules underwent orientational changes (tilted more to the surface) after immobilized to the substrates.
3.2 Peptide-Cell Membrane Interactions
Interactions between cellular membranes and peptides have been well studied due to their importance in antimicrobial activity, material transport and cell signaling. Every peptide is designed to interact with membrane bilayers in a specific manner. Certain peptides such as AMPs are designed to interact strongly with certain types of membrane bilayers and effectively disrupt bilayer function.82 It is of fundamental importance to gain a molecular understanding of how membrane lipid bilayers behave during such interactions. With the ability for submonolayer sensitivity, SFG has shown to be very successful in monitoring these interactions.
One intrinsic difficulty involved in studying such interactions is the extreme sensitivity required for a biophysical/analytical technique to follow the interactions at a molecular level in real time. The minimum inhibitory concentration (MIC) of a peptide, or the concentration where it is an effective AMP, is near the µM range, which may be below the detection limit for some widely used spectroscopies such as Fourier transform infrared (FTIR) spectroscopy and nuclear magnetic resonance (NMR) spectroscopy. It has been demonstrated with various types of membrane-associating peptides that SFG has the capability to not only provide a complete picture of the interactions and mechanisms of these various systems, but is capable of doing this in real time in situ for concentrations representative of typical biological interactions.
Various experimental techniques have been used with SFG to determine the action mode of various peptides. Since SFG is a vibrational spectroscopy and therefore signal is a result of intrinsic molecular vibrations in the system studied, it is important that shared species between lipid and peptide can be independently measured and analyzed. The most common technique to resolve this issue is by using isotope-labeling schemes to selectively decouple the spectra generated by the lipid bilayer from that of the peptide or between two leaflets of one lipid bilayer. For example, this labeling scheme was used in the investigation of melittin.80
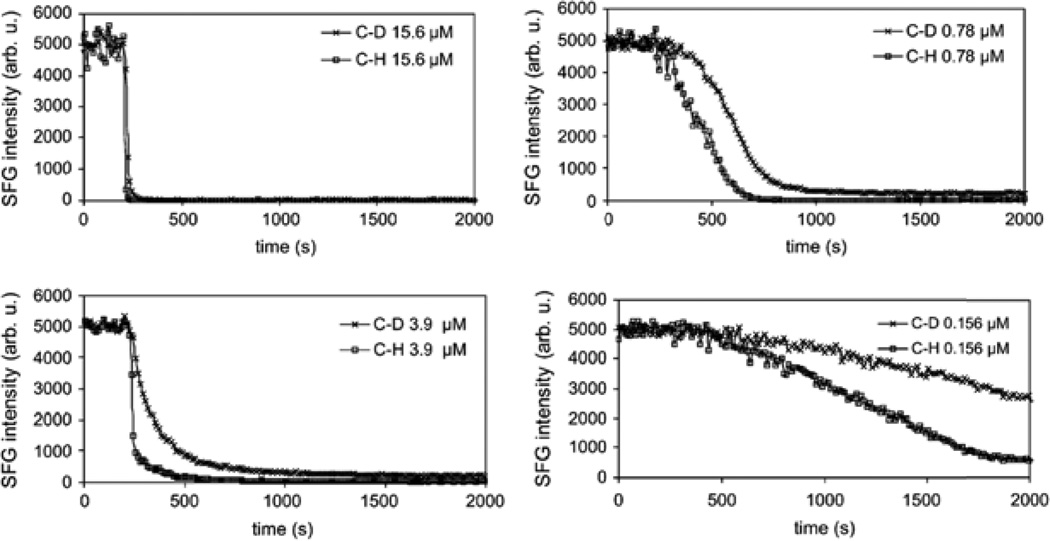
In this study, hydrogenated and deuterated 1,2-dipalmitoyl(D62)-sn-glycero-3-phosphoglycerol (DPPG and d-DPPG) were used to make asymmetric bilayers as model cell membranes. Melittin is composed of 26 amino acid residues with the 20 residues starting from the N-terminus having hydrophobic properties and the remaining 7 amino acids having highly charged groups. This makes melittin a potent AMP. When the DPPG (or d-DPPG) leaflet was disrupted by melittin, the SFG C-H (or C-D) stretching signal would decrease. Time dependent SFG intensity measurements shown in Figure 6 demonstrate the concentration dependence of melittin. By individually isolating each leaflet, it is clear that melittin has a weak interaction at low concentrations (0.156 µM and 0.78 µM, Signals decreased slowly). Intermediate concentration (3.9 µM) show that melittin interacts with each leaflet individually (or signal decrease from one leaflet was slower than the other), indicative of toroidal pore mechanisms. At high concentration (15.6 µM), melittin has detergent-like interactions with DPPG/d-DPPG, disrupting both leaflets immediately after the interactions.
Figure 6.
Signal intensity change at 2070 cm−1 and 2875 cm−1 monitoring melittin interacting with d-DPPG/DPPG bilayers on CaF2. Melittin stock solution was injected into the subphase at 200 s. Four solution concentrations were used, and dramatically different patterns were observed (Reproduced with permission from Biophys. J. 2007, 93, 866–875. Copyright 2007, Elsevier).
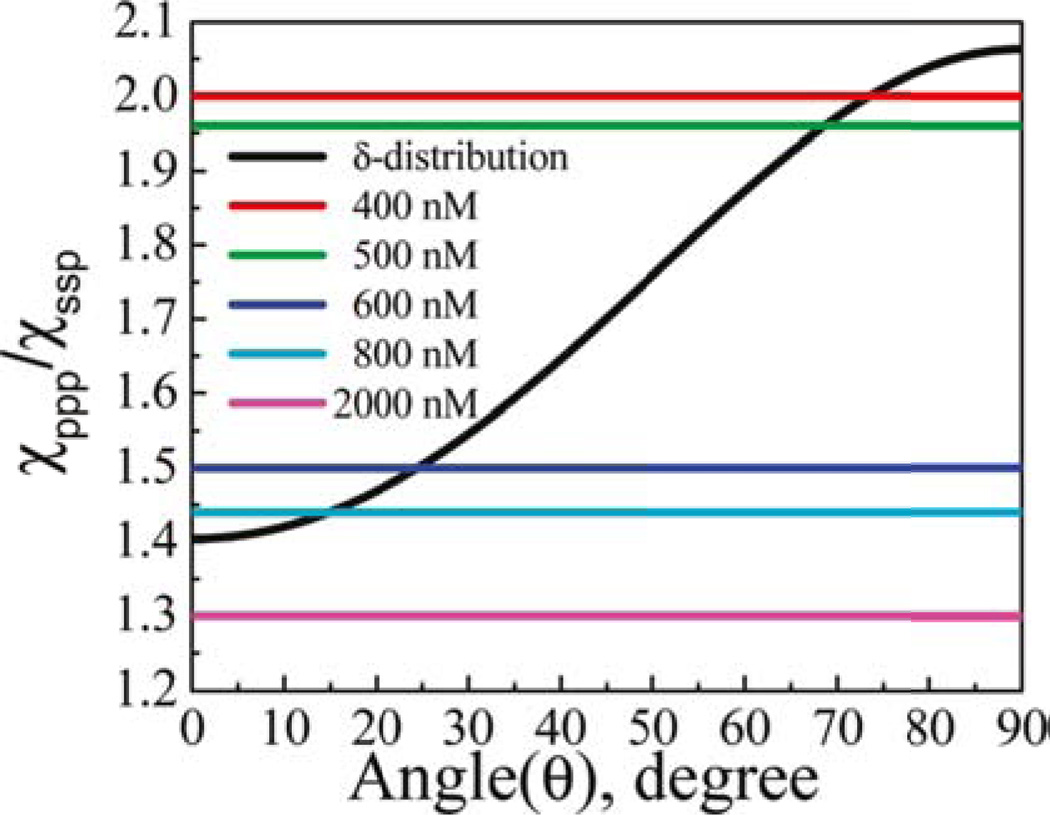
In addition to isotope-labeling schemes, it is also possible to determine the orientation of peptides as they interact with various bilayers by collecting polarized SFG spectra. A more thorough discussion about the calculations necessary to analyze such spectra has been added into section 2.2. To review, SFG measures <cosθ> and <cos3θ> where “< >” represents average value.90 By taking the ratio of two measured second order nonlinear optical susceptibility component values at different polarizations, it is possible to generate a curve as shown in Figure 7. The results shown in this figure are representative of MSI-78,91 an analog of magainin 2,81 both of which are AMPs that target charged bilayers characteristic of bacterial cell membranes. From this plot, it shows that at low concentrations of MSI-78 (400 nM and 500 nM), the peptide adopts a near parallel orientation and lays on the surface of the bilayer. With the addition of more peptide (600 nM and 800 nM), the AMP starts to insert into the bilayer. Finally after reaching the critical concentration, MSI-78 begins to form toroidal pores within DPPG bilayers (e.g., 2000 nM).91 In contrast, bilayers prepared with zwitterionic 1,2-dipalmitoyl-sn-glycero-3-phosphocholin (DPPC) showed almost no interaction with MSI-78.76
Figure 7.
Relation between the χppp/χssp ratio and the orientation angle of MSI-78 molecules in a DPPG/d-DPPG bilayer (Reproduced with permission from Langmuir 2011, 27, 7760–7767. Copyright 2011, Am. Chem. Soc.).
Situations may arise where one measurement is insufficient to adequately explain the data. As shown in Figure 7, at much higher concentrations of MSI-78, around 2000 nM, a δ-distribution does not accurately describe the possible orientation values. A more complicated distribution curve (e.g., a Gaussian, or a dual δ-distribution) need to be used. However, the determination of such a more complicated distribution curve requires more measurements. The use of ATR-FTIR spectroscopy not only provides complementary spectra, but when taken with polarized light, can also be used as a tool for orientational analysis. It measures <cos2θ>, which provides an independent measurement from SFG results.90 For the studies on melittin, a dual δ-distribution was used and the combined SFG and ATR-FTIR results indicated that around one quarter of melittin peptides would insert into the bilayer while the rest remained at the surface.76 This result was confirmed when using a maximum entropy trial function to determine the melittin orientation distribution.76 A proposed model is shown in Figure 8.
Figure 8.
Proposed mechanism of melittin interaction with a DPPG bilayer (Reproduced with permission from J. Am. Chem. Soc. 2007, 129, 1420–1427. Copyright 2007, Am. Chem. Soc.).
From this discussion, SFG has shown to be a powerful tool to characterize interactions of peptides with lipid bilayers. A few of the techniques used in experimental design and analysis were discussed above. These include techniques such as isotopic labeling, polarization measurements and using complementary spectroscopies. Various AMPs were looked at to determine their modes of action and their orientations near their MIC. Analysis methods used for peptides can be building blocks for larger proteins discussed in the next section.
3.3 Protein Complexes at Interfaces
It is possible to use these same tools and techniques discussed above to look at larger systems such as protein complexes. Proteins themselves can be quite complex with many amino acids and structures spanning from globlular to very ordrered. However, substantial fundamental questions regarding function and orientation are still obtainable by SFG.
G proteins are membrane associated proteins that are involved in a wide variety of biological processes. Little is known about how they interact and order at the bilayer surface.79 SFG has been used to characterize the orientation of the Gβγ subunit of the G protein.92
By monitoring the amide I SFG signal, orientational analysis on all the α-helical components in the protein complex discussed above can be used to determine how Gβγ orients at the surface. Figure 9 below shows both the collected SFG spectra in different polarization combinations and a proposed orientation deduced on the SFG intensity ratios from such polarized SFG spectra, by assuming the twist angle of the Gβγ is fixed.92 Recently, to deduce the orientation of complicated proteins with many α-helices, a computer program was developed. Along with collected SFG spectra, this software was used to study the orientation of Gβγ-GRK2 protein complex associated with the lipid bilayer.79 This research demonstrated the feasibility to determine the possible orientation range of large protein complex associated with cell membrane using SFG.79 Similarly, we developed a computer program to determine orientation range of interfacial proteins according to polarized ATR-FTIR measurements. By combining the possible ranges calculated from the software based on SFG and ATR-FTIR results, we can quantify the spatial orientation of large protein complexes in the future.
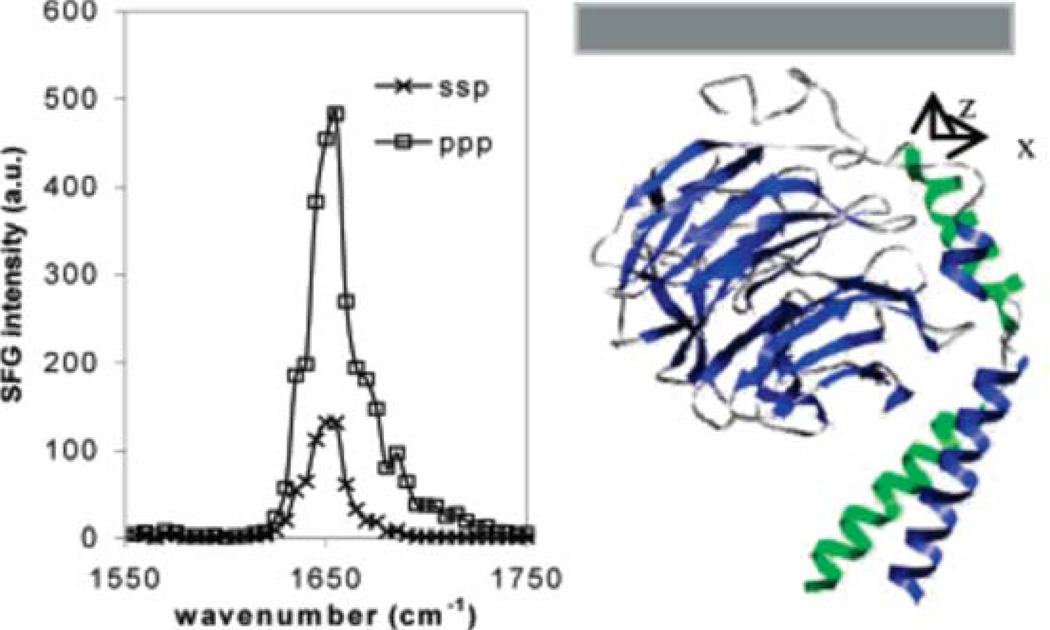
Figure 9.
(Left) SFG amide I spectra of interfacial Gβγ (25 µg/mL) adsorbed into a POPC/POPC bilayer; (right) Gβγ orientation deduced based on the SFG intensity ratio. Summary (Reproduced with permission from J. Amm. Chem. Soc. 2007, 129, 12658–12659. Copyright 2007, Am. Chem. Soc.).
In this review, we summarized recent applications of SFG techniques on the studies of interfacial peptides and proteins. SFG is proven to be a versatile tool to elucidate the molecular interactions involving peptides and proteins because molecular level understanding such as conformation and orientation of the interfacial peptides and proteins can be provided without harsh treatment or tedious sample preparation. Most importantly, these signals can be monitored in situ and in real time, allowing for experiments to be performed in near biological conditions. Both SFG signals in the C-H stretching and amide I frequency regions were introduced and analyzed. This review has focused on analysis of α-helical structures, however SFG has been successful in studying other secondary structures as well such as β-sheets and 310-helices.59,71,78 These SFG results provide important information in determining the working mechanisms and conditions of the peptides/proteins.
Acknowledgments
This research is supported by National Institutes of Health (NIH) Grant GM081655, Office of Naval Research (ONR) Grant N00014-08-1-1211, U.S. Army Natick Soldier Center Grant W911QY-10-C-0001, Defense Threat Reduction Agency (DTRA) Grant HDTRA1-11-1-0019, and Army Research Office (ARO) Grant W911NF-11-1-0251.
References
- 1.Horbett TA, Brash JL, editors. Proteins at Interfaces II: Fundamentals and Applications (ACS Symposium) Washington, DC: American Chemical Society; 1995. [Google Scholar]
- 2.Giangaspero A, Sandri L, Tossi A. Eur. J. Biochem. 2001;268:5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 3.Morris MC, Deshayes S, Heitz F, Divita G. Biol. Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 4.Tweedle MF. Acc. Chem. Res. 2009;42:958–968. doi: 10.1021/ar800215p. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Xu Z, Peng L, Fang X, Yin X, Xu N, Cen P. Peptides. 2006;27:931–940. doi: 10.1016/j.peptides.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Jin W. Anal. Chim. Acta. 2002;461:1–36. [Google Scholar]
- 7.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 8.Schuck P. Annu. Rev. Biophys. Biomol. Struct. 1997;26:541–566. doi: 10.1146/annurev.biophys.26.1.541. [DOI] [PubMed] [Google Scholar]
- 9.Marx KA. Biomacromolecules. 2003;4:1099–1120. doi: 10.1021/bm020116i. [DOI] [PubMed] [Google Scholar]
- 10.Roach P, Parker T, Gadegaard N, Alexander MR. Surf. Sci. Rep. 2010;65:145–173. [Google Scholar]
- 11.Elwing H. Biomaterials. 1998;19:397–406. doi: 10.1016/s0142-9612(97)00112-9. [DOI] [PubMed] [Google Scholar]
- 12.Tengvall P, Lundström I, Liedberg B. Biomaterials. 1998;19:407–422. doi: 10.1016/s0142-9612(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 13.Santos O, Kosoric J, Hector MP, Anderson P, Lindh L. J Colloid Interface Sci. 2008;318:175–182. doi: 10.1016/j.jcis.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Mozsolits H, Aguilar M-I. Biopolymers. 2002;66:3–18. doi: 10.1002/bip.10200. [DOI] [PubMed] [Google Scholar]
- 15.Campbell CT, Kim G. Biomaterials. 2007;28:2380–2392. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 16.Maynard JA, Lindquist NC, Sutherland JN, Lesuffleur A, Warrington AE, Rodriguez M, Oh S-H. Biotechnol. J. 2009;4:1542–1558. doi: 10.1002/biot.200900195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingshott P, McArthur S, Thissen H, Castner DG, Griesser HJ. Biomaterials. 2002;23:4775–4785. doi: 10.1016/s0142-9612(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 18.Henry M, Dupont-Gillain C, Bertrand P. Langmuir. 2003;19:6271–6276. doi: 10.1021/la701850p. [DOI] [PubMed] [Google Scholar]
- 19.Baugh L, Weidner T, Baio JE, Nguyen PT, Gamble LJ, Stayton PS, Castner DG. Langmuir. 2010;21:16434–16441. doi: 10.1021/la1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanea E, Simon V. Appl. Surf. Sci. 2011;257:2346–2352. [Google Scholar]
- 21.Wagner MS, McArthur SL, Shen M, Horbett TA, Castner DG. J. Biomater. Sci. Polymer Edn. 2002;13:407–428. doi: 10.1163/156856202320253938. [DOI] [PubMed] [Google Scholar]
- 22.Muller DJ, Sass HJ, Muller SA, Buldt G, Engel A. J. Mol. Biol. 1999;285:1903–1909. doi: 10.1006/jmbi.1998.2441. [DOI] [PubMed] [Google Scholar]
- 23.Berquand A, Mingeot-Leclercq M-P, Dufrêne YF. Biochim. Biophys. Acta. 2004;1664:198–205. doi: 10.1016/j.bbamem.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Milhiet PE, Gubellini F, Berquand A, Dosset P, Rigaud JL, Le Grimellec C, Levy D. Biophys. J. 2006;91:3268–3275. doi: 10.1529/biophysj.106.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axelsen PH, Citra MJ. Prog. Biophys. Mol. Biol. 1996;66:227–253. doi: 10.1016/s0079-6107(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 26.Le Coutre J, Kaback HR, Patel CKN, Heginbotham L, Miller C. Proc. Natl. Acad. Sci. USA. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinchurkar MS, Chen KHC, Yu SSF, Kuo SJ, Chiu HC, Chien SH, Chan SI. Biochemistry. 2004;43:13283–13292. doi: 10.1021/bi049016i. [DOI] [PubMed] [Google Scholar]
- 28.Ahern AM, Garrell RL. Langmuir. 1991;7:254–261. [Google Scholar]
- 29.Wen Z-Q, Li G, Ren D. Appl. Spectrosc. 2011;65:514–521. doi: 10.1366/10-06089. [DOI] [PubMed] [Google Scholar]
- 30.Shen Y. Nature. 1989;337:519–525. [Google Scholar]
- 31.Zhuang X, Miranda PB, Kim D, Shen YR. Phys. Rev. B. 1999;59:12632–12640. [Google Scholar]
- 32.Chen Z, Shen YR, Somorjai GA. Ann. Rev. Phys. Chem. 2002;53:437–465. doi: 10.1146/annurev.physchem.53.091801.115126. [DOI] [PubMed] [Google Scholar]
- 33.Eisenthal KB. Chem. Rev. 1996;96:1343–1360. doi: 10.1021/cr9502211. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z. Prog. Polym. Sci. 2010;35:1376–1402. doi: 10.1016/j.progpolymsci.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z. Polym. Inter. 2007;56:577–587. [Google Scholar]
- 36.Kim J, Somorjai GA. J. Am. Chem. Soc. 2003;125:3150–3158. doi: 10.1021/ja028987n. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Cremer PS. ChemPhysChem. 2001;2:543–546. doi: 10.1002/1439-7641(20010917)2:8/9<543::AID-CPHC543>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Li G, Ye S, Morita S, Nishida T, Osawa M. J. Am. Chem. Soc. 2004;126:12198–12199. doi: 10.1021/ja046183x. [DOI] [PubMed] [Google Scholar]
- 39.Voges AB, Al-Abadleh HA, Musorrariti MJ, Bertin PA, Nguyen ST, Geiger FM. J. Phys. Chem. B. 2004;108:18675–18682. [Google Scholar]
- 40.Li QF, Hua R, Chea IJ, Chou KC. J. Phys. Chem. B. 2008;112:694–697. doi: 10.1021/jp072147j. [DOI] [PubMed] [Google Scholar]
- 41.Ye HK, Gu ZY, Gracias DH. Langmuir. 2006;22:1863–1868. doi: 10.1021/la052030r. [DOI] [PubMed] [Google Scholar]
- 42.Yatawara AK, Tiruchinapally G, Bordenyuk AN, Andreana PR, Benderskii AV. Langmuir. 2009;25:1901–1904. doi: 10.1021/la803113h. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Conboy JC. Biophys. J. 2005;89:2522–2532. doi: 10.1529/biophysj.105.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Ward R, Tian Y, Malizia F, Gracias DH, Shen YR, Somorjai GA. J. Biomed. Mater. Res. 2002;62:254–264. doi: 10.1002/jbm.10075. [DOI] [PubMed] [Google Scholar]
- 45.Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. J. Am. Chem. Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- 46.Phillips DC, York RL, Mermut O, McCrea KR, Ward RS, Somorjai GA. J. Phys. Chem. C. 2007;111:255–261. [Google Scholar]
- 47.York RL, Browne WK, Geissler PL, Somorjai GA. Isr. J. Chem. 2007;47:51–58. [Google Scholar]
- 48.Weidner T, Apte JS, Gamble LJ, Castner DG. Langmuir. 2010;26:3433–3440. doi: 10.1021/la903267x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidner T, Breen NF, Li K, Drohny GP, Castner DG. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13288–13293. doi: 10.1073/pnas.1003832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu L, Ma G, Yan EC. J. Am. Chem. Soc. 2010;132:5405–5412. doi: 10.1021/ja909546b. [DOI] [PubMed] [Google Scholar]
- 51.Fu L, Liu J, Yan EC. J. Am. Chem. Soc. 2011;133:8094–8097. doi: 10.1021/ja201575e. [DOI] [PubMed] [Google Scholar]
- 52.Weidner T, Breen NF, Drobny GP, Castner DG. J. Phys. Chem. B. 2009;113:15423–15426. doi: 10.1021/jp908773c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anglin TC, Liu J, Conboy JC. Biophys. J. 2007;92:L01–L03. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anglin TC, Brown KL, Conboy JC. J. Struct. Biol. 2009;168:37–52. doi: 10.1016/j.jsb.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung SY, Lim SM, Albertorio F, Kim G, Gurau MC, Yang RD, Holden MA, Cremer PS. J. Am. Chem. Soc. 2003;125:12782–12786. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- 56.Chen X, Sagle LB, Cremer PS. J. Am. Chem. Soc. 2007;129:15104–15105. doi: 10.1021/ja075034m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall SA, Jena KC, Trudeau TG, Hore DK. J. Phys. Chem. C. 2011;115:11216–11225. [Google Scholar]
- 58.Wang J, Buck SM, Chen Z. Analyst. 2003;128:773–778. doi: 10.1039/b212551j. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Chen X, Clarke ML, Chen Z. Proc. Natl. Acad. Sci. USA. 2005;102:4978–4983. doi: 10.1073/pnas.0501206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreesen L, Humbert C, Sartenaer Y, Caudano Y, Volcke C, Mani AA, Peremans A, Thiry PA, Hanique S, Frere JM. Langmuir. 2004;20:7201–7207. doi: 10.1021/la0488001. [DOI] [PubMed] [Google Scholar]
- 61.Dressen L, Sartenaer Y, Humbert C, Mani AA, Methivier C, Pradier CM, Thiry PA, Peremans A. ChemPhysChem. 2004;5:1719–1725. doi: 10.1002/cphc.200400213. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Chen C, Buck SM, Chen Z. J. Phys. Chem. B. 2001;105:12118–12125. [Google Scholar]
- 63.Wang J, Buck SM, Even MA, Chen Z. J. Am. Chem. Soc. 2002;124:13302–13305. doi: 10.1021/ja026881m. [DOI] [PubMed] [Google Scholar]
- 64.Miranda PB, Shen YR. J. Phys. Chem. B. 1999:3292–3307. [Google Scholar]
- 65.Smith JP, Hinson-Smith V. Anal. Chem. 2004;76:287A–290A. doi: 10.1021/ac041610a. [DOI] [PubMed] [Google Scholar]
- 66.Ye S, Osawa M. Chem. Lett. 2009;38:386–391. [Google Scholar]
- 67.Wang J, Paszti Z, Even MA, Chen Z. J. Phys. Chem. B. 2004;108:3625–3632. [Google Scholar]
- 68.Wang J, Buck SM, Chen Z. J. Phys. Chem. B. 2002;106:11666–11672. [Google Scholar]
- 69.Wang J, Even MA, Chen X, Schmaier AH, Waite JH, Chen Z. J. Am. Chem. Soc. 2003;125:9914–9915. doi: 10.1021/ja036373s. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Wang J, Sniadecki JJ, Even MA, Chen Z. Langmuir. 2005;21:2662–2664. doi: 10.1021/la050048w. [DOI] [PubMed] [Google Scholar]
- 71.Ye S, Nguyen KT, Chen Z. J. Phys. Chem. B. 2010;114:3334–3340. doi: 10.1021/jp911174d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Paszti Z, Even MA, Chen Z. J. Am. Chem. Soc. 2002;124:7016–7023. doi: 10.1021/ja012387r. [DOI] [PubMed] [Google Scholar]
- 73.Chen C, Clarke ML, Wang J, Chen Z. Phys. Chem. Chem Phys. 2005;7:2357–2363. doi: 10.1039/b501910a. [DOI] [PubMed] [Google Scholar]
- 74.Nguyen KT, Le Clair SV, Ye S, Chen Z. J. Phys. Chem. B. 2009;113:12169–12180. doi: 10.1021/jp904153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee S, Wang J, Krimm S, Chen Z. J. Phys. Chem. A. 2006;110:7035–7044. doi: 10.1021/jp057200n. [DOI] [PubMed] [Google Scholar]
- 76.Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. J. Am. Chem. Soc. 2007;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, Lee SH, Chen Z. J. Phys. Chem. B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen KT, King JT, Chen Z. J. Phys. Chem. B. 2010;114:8291–8300. doi: 10.1021/jp102343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boughton AP, Yang P, Tesmer VM, Ding B, Tesmer JJG, Chen Z. Proc. Natl. Acad. Sci. 2011;108:E667–E673. doi: 10.1073/pnas.1108236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X, Wang J, Kristalyn CB, Chen Z. Biophys. J. 2007;93:866–875. doi: 10.1529/biophysj.106.099739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen KT, Le Clair SV, Ye S, Chen Z. J. Phys. Chem. B. 2009;113:12358–12363. doi: 10.1021/jp904154w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen X, Chen Z. Biochim. Biophys. Acta. 2006;1758:1257–1273. doi: 10.1016/j.bbamem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Clarke M, Zhang Y, Chen X. Langmuir. 2003;19:7862–7866. [Google Scholar]
- 84.Clarke ML, Chen Z. Langmuir. 2006;22:8627–8630. doi: 10.1021/la0607656. [DOI] [PubMed] [Google Scholar]
- 85.Ye S, Nguyen KT, Le Clair SV, Chen Z. J. Struct. Biol. 2009;168:61–77. doi: 10.1016/j.jsb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clarke ML, Wang J, Chen Z. J. Phys. Chem. B. 2005;109:22027–22035. doi: 10.1021/jp054456k. [DOI] [PubMed] [Google Scholar]
- 87.Wang J, Chen X, Clarke ML, Chen Z. J. Phys. Chem. B. 2006;110:5017–5024. doi: 10.1021/jp0534683. [DOI] [PubMed] [Google Scholar]
- 88.Ye S, Nguyen KT, Boughton AP, Mello CM, Chen Z. Langmuir. 2010;26:6471–6477. doi: 10.1021/la903932w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han X, Soblosky L, Slutsky M, Mello CM, Chen Z. Langmuir. 2011;27:7042–7051. doi: 10.1021/la200388y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J, Paszti Z, Clarke ML, Chen X, Chen Z. J. Phys. Chem. B. 2007;111:6088–6095. doi: 10.1021/jp070383o. [DOI] [PubMed] [Google Scholar]
- 91.Yang P, Ramamoorthy A, Chen Z. Langmuir. 2011;27:7760–7767. doi: 10.1021/la201048t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen X, Boughton AP, Tesmer JJG, Chen Z. J. Am. Chem. Soc. 2007;129:12658–12659. doi: 10.1021/ja075542w. [DOI] [PubMed] [Google Scholar]