Abstract
AIM
The most common causes of variability in drug response include differences in drug metabolism, especially when the hepatic cytochrome P450 (CYP) enzymes are involved. The current study was conducted to assess the differences in CYP activities in human liver microsomes (HLM) of Chinese or Caucasian origin.
METHODS
The metabolic capabilities of CYP enzymes in 30 Chinese liver microsomal samples were compared with those of 30 Caucasian samples utilizing enzyme kinetics. Phenacetin O-deethylation, coumarin 7-hydroxylation, bupropion hydroxylation, amodiaquine N-desethylation, diclofenac 4′-hydroxylation (S)-mephenytoin 4′-hydroxylation, dextromethorphan O-demethylation, chlorzoxazone 6-hydroxylation and midazolam 1′-hydroxylation/testosterone 6β-hydroxylation were used as probes for activities of CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A, respectively. Mann-Whitney U test was used to assess the differences.
RESULTS
The samples of the two ethnic groups were not significantly different in cytochrome-b5 concentrations but were significantly different in total CYP concentrations and NADPH-P450 reductase activity (P < 0.05). Significant ethnic differences in intrinsic clearance were observed for CYP1A2, CYP2C9, CYP2C19 and CYP2E1; the median values of the Chinese group were 54, 58, 26, and 35% of the corresponding values of the Caucasian group, respectively. These differences were associated with differences in Michaelis constant or maximum velocity. Despite negligible difference in intrinsic clearance, the Michaelis constant of CYP2B6 appeared to have a significant ethnic difference. No ethnic difference was observed for CYP2A6, CYP2C8, CYP2D6 and CYP3A.
CONCLUSIONS
These data extend our knowledge on the ethnic differences in CYP enzymes and will have implications for drug discovery and drug therapy for patients from different ethnic origins.
Keywords: cytochrome P450, ethnic difference, human liver microsomes
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
The mean catalytic activities of CYP1A2, CYP2A6, CYP2D6, CYP2E1 and CYP3A4 in human liver microsomes (HLM) of Japanese origin were found to be lower than the corresponding mean values of Caucasian HLM, but the distribution patterns of data in these studies were not well addressed.
Interindividual variations in metabolic activities of Chinese HLM samples were reported for CYP1A2, CYP2C9, CYP2D6 and CYP3A4, but little is known about the difference in CYP activities between Chinese and Caucasian HLM samples.
The vast majority of the preceding studies were based on the measurement of metabolite formation rates at a certain fixed substrate concentration, rather than enzyme kinetic analyses.
WHAT THIS STUDY ADDS
Significant differences in metabolic capability were observed for CYP1A2, CYP2C9, CYP2C19 and CYP2E1 with the median values lower for the Chinese HLM samples vs. the Caucasian samples, which were associated with differences in Michaelis constant or maximum velocity.
Despite negligible differences in metabolic capability for CYP2A6, CYP2B6, CYP2C8, CYP2D6 and CYP3A, Michaelis constant of CYP2B6 appeared to have a significant ethnic difference.
Introduction
Drug efficacy and toxicity can vary among ethnic groups [1–4]. Recognizing the relevance of the interethnic differences in drug response, the International Conference on Harmonization (ICH) has issued a guideline entitled ‘Ethnic Factors in the Acceptability of Foreign Clinical Data’ (http://www.ich.org/products/guidelines/efficacy/efficacy-single/article/ethnic-factors-in-the-acceptability-of-foreign-clinical-data.html) to facilitate the registration of medicines among ICH regions. Interethnic differences in drug response involve one or more of three mechanisms, i.e. drug metabolism/transport, drug targets and the disease pathways. The variation in drug metabolism is considered to be profoundly important because it affects the systemic exposure and disposition of drugs.
Cytochromes P450 (CYPs), highly concentrated in the liver and also expressed in many other tissues, are the most prominent family of drug metabolizing enzymes in humans. CYPs are located in the smooth endoplasmic reticulum membrane of cells. Thus they can be recovered in microsomal fractions. Knowledge of the human CYPs responsible for drug metabolism, primarily found in CYP families 1–3 (CYP1–3), is fundamental for drug development and drug therapy. The most significant drug metabolizing CYPs in humans include CYP2C9, CYP2C19, CYP2D6 and CYP3A4, whereas CYP1A2, CYP2C8, CYP2A6, CYP2B6 and CYP2E1 are also responsible for metabolism of some drugs and/or metabolic activation of procarcinogens [5]. CYP genes in the families 1–3 are known to be genetically polymorphic and the genetic variants can alter the activity or expression of the drug metabolizing enzymes. Variations associated with CYPs affecting drug concentration and response occurred at varying allele frequencies among populations of different ethnic origin [6]. Poor metabolizer frequencies of several major CYP enzymes are significantly different between Asians and Caucasians [7]. Understanding the interethnic differences in the metabolic capacity of CYPs is a valuable tool for drug development that can potentially lead to improved safety and efficacy of drug therapy for patients of different ethnic backgrounds.
The process of drug discovery and development has become increasingly reliant on the use of human-derived test systems (e.g. human liver microsomes, hepatocytes, etc.) to screen drug candidates for their liability of CYP inhibition or induction and to determine the involvement of drug metabolizing enzymes and transporters. Interethnic differences should be taken into account when performing in vitro drug metabolism studies. Recent results have indicated that testing CYP2D6-metabolized candidates or investigational drugs for allele-specific metabolism/interactions may be important [8]. However, to what extent ethnicity can affect the use of human-derived test systems remains largely unknown and requires more studies. The current study aimed to assess and understand the interethnic differences in the activities of major CYPs in human liver microsomes (HLM) of Chinese or Caucasian origin. Therefore the appropriate HLM can be utilized for future in vitro drug metabolism studies, and data would allow the prediction of interethnic variability in the clinical studies by applying these results to physiologically based pharmacokinetic modelling. Nine CYP enzymes, including CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A, in HLM derived from 30 Chinese samples were assessed in terms of catalytic activity in comparison with those from 30 Caucasians samples.
Methods
Samples of human liver microsomes
As shown in Table 1, 30 Chinese samples of HLM, including 24 male samples and six female samples, were obtained from the Research Institute for Liver Diseases (Shanghai) Co. Ltd (Shanghai, China) following the donation regulations in China with fully informed consent, and under the approval of an independent Institutional Review Board. Fourteen HLM samples of Caucasian origin (11 male samples and three female samples) were obtained from the Medical College of Wisconsin under a protocol approved by the Committee for the Conduct of Human Research. Twelve Caucasian samples (seven male samples and five female samples) were purchased from Celsis In Vitro Inc (Baltimore, MD, USA), and four male Caucasian samples were purchased from BD Gentest (Woburn, MA, USA). The HLM samples were prepared by differential centrifugation. Because detailed histories of some of the donors were not known, the CYP variations with respect to the 60 HLM samples were analyzed without considerations of disease states, drug intake, smoking, alcohol drinking and other dietary habits. Pooled HLM, obtained from Xenotech (Lenexa, KS, USA), were also used for the development of enzyme activity assays.
Table 1.
Information about HLM of test Chinese and Caucasian samples
| Sample ID | Age (years) | Gender | Cause of death/drug history/social history |
|---|---|---|---|
| Chinese group (n = 30) | |||
| am01 | 31 | Male | Hepatolithiasis |
| am02 | 70 | Male | Hepatic cyst |
| am03 | 31 | Male | Trauma/smoker |
| am04 | 21 | Male | Trauma |
| am05 | 24 | Male | Trauma |
| am06 | 26 | Male | Trauma |
| am07 | 28 | Male | Trauma/smoker |
| am08 | 24 | Male | Trauma/smoker |
| am09 | 29 | Male | Trauma/smoker |
| am10 | 38 | Male | Trauma/smoker |
| am11 | 25 | Male | Trauma/smoker |
| am12 | 36 | Male | Trauma |
| am13 | 51 | Male | Trauma/smoker |
| am14 | 48 | Male | Trauma/smoker |
| am15 | 32 | Male | Trauma |
| am16 | 36 | Male | Trauma/smoker |
| am17 | 27 | Male | Trauma/smoker |
| am18 | 39 | Male | Trauma |
| am19 | 44 | Male | Trauma |
| am20 | 46 | Male | Trauma/smoker |
| am21 | 28 | Male | Trauma |
| am22 | 33 | Male | Trauma |
| am23 | 41 | Male | Trauma/smoker |
| am24 | 33 | Male | Trauma/smoker |
| aw25 | 53 | Female | Hepatic cyst |
| aw26 | 51 | Female | Hepatolithiasis |
| aw27 | 68 | Female | Giant hepatic haemangiomas |
| aw28 | 65 | Female | Cholangiocarcinoma |
| aw29 | 53 | Female | Hepatic cyst |
| aw30 | 78 | Female | Primary carcinoma of liver |
| Caucasian group (n = 30) | |||
| cm01 | 55 | Male | Pulmonary fibrosis/ethanol |
| cm02 | 50 | Male | Unknown |
| cm03 | 22 | Male | Unspecified drug abuse |
| cm04 | 31 | Male | Unknown |
| cm05 | 25 | Male | Unknown |
| cm06 | 55 | Male | Unknown |
| cm07 | 45 | Male | Lisinopril |
| cm08 | 67 | Male | Unknown |
| cm09 | 63 | Male | Cerebrovascular trauma |
| cm10 | 18 | Male | 0.24% ethanol at time of death |
| cm11 | 82 | Male | Cerebrovascular trauma |
| cm12 | 23 | Male | 0.06% ethanol at time of death |
| cm13 | 59 | Male | Unknown |
| cm14 | 77 | Male | Head trauma/smoker |
| cm15 | 65 | Male | Cardiac arrest |
| cm16 | 54 | Male | Anoxia |
| cm17 | 53 | Male | Cerebrovascular trauma |
| cm18 | 41 | Male | Motorcycle trauma |
| cm19 | 59 | Male | Cerebrovascular trauma/smoker |
| cm20 | 58 | Male | Subarachnoid haemorrhage |
| cm21 | 21 | Male | Diabetic, 0.08% ethanol at time of death |
| cm22 | 47 | Male | Anoxia and cardiopulmonary arrest |
| cw23 | 58 | Female | Poorly controlled hypertension |
| cw24 | 72 | Female | Cerebrovascular trauma |
| cw25 | 28 | Female | Cerebrovascular trauma |
| cw26 | 77 | Female | Chemotherapy for 1 year for colon cancer |
| cw27 | 56 | Female | Cerebrovascular trauma |
| cw28 | 50 | Female | Cerebrovascular trauma |
| cw29 | 49 | Female | Cerebrovascular trauma |
| cw30 | 48 | Female | Chlorpheniramine |
Incubation conditions for measurement of the metabolic capabilities of CYP enzymes in HLM
Pilot studies were performed with each biotransformation to ensure that the comparison of metabolic capabilities of the test CYP enzymes were determined under linear metabolite formation conditions with respect to time and microsomal protein concentration. Incubations were performed in duplicate in 96-well plates in a total assay volume of 100 µl with each well, which contained HLM [at 0.05 mg protein ml−1 for CYP2C8, CYP2C9 and CYP3A (for testosterone 6β-hydroxylation), 0.1 mg protein ml−1 for CYP1A2, CYP2A6, CYP2D6, CYP2E1 and CYP3A (for midazolam 1′-hydroxylation), and 0.2 mg protein ml−1 for CYP2B6 and CYP2C19], substrate (concentration ranges as indicated), 100 mm potassium phosphate buffer (pH 7.4, for CYP1A2, CYP2B6, CYP2C8, CYP2C19, CYP2D6, CYP2E1 and CYP3A)/100 mm tris(hydroxymethyl)aminomethane buffer (pH 7.4, for CYP2A6 and CYP2C9), an NADPH-generating system comprising 3.3 mm magnesium chloride, 3.3 mm of glucose-6-phosphate, 0.5 U ml−1 glucose-6-phosphate dehydrogenase and 1.3 mm NADP. Before commencement of the reaction by adding the NADPH-generating system, the incubation mixture was pre-incubated for 3 min at 37°C. The optimal incubation time was 4 min for midazolam 1′-hydroxylation, 5 min for testosterone 6β-hydroxylation, 10 min for amodiaquine N-desethylation and diclofenac 4′-hydroxylation, 20 min for phenacetin O-deethylation, coumarin 7-hydroxylation, bupropion 1-hydroxylation, chlorzoxazone 6-hydroxylation and dextromethorphan O-demethylation and 30 min for (S)-mephenytoin 4′-hydroxylation. For the biotransformations, seven to ten substrate concentrations were examined over the following ranges: 3.91 to 2000 µm for phenacetin, 0.10 to 50 µm for coumarin, 0.20 to 100 µm for amodiaquine, 0.20 to 100 µm for diclofenac, 1.95 to 1000 µm for (S)-mephenytoin, 0.49 to 250 µm for dextromethorphan, 0.98 to 500 µm for chlorzoxazone, 3.91 to 500 µm for bupropion, 0.05 to 50 µm for midazolam and 3.91 to 250 µm for testosterone. The reactions were terminated by adding 100 µl of ice-cold methanol. The resulting samples were centrifuged at 3000 g for 10 min to remove protein and 10 µl of the supernatant were analyzed by LC-MS/MS.
LC-MS/MS analyses of metabolites
Validated LC-MS/MS-based methods were used to determine metabolite formation rates. The LC-MS/MS system was assembled from a Waters Acquity UPLC separation module (Waters, Milford, MA, USA) and an Applied Biosystems-Sciex API 4000 Q Trap mass spectrometer (Applied Biosystems, Toronto, Canada) with Turbo V ESI-source. Methanol-prepared biosamples were separated using a generic pulse gradient elution [9, 10] on a 5 µm Ultimate XB-C18 column (50 mm × 2.1 mm i.d.; Welch Materials, MD, USA) with mobile phases of CH3CN : H2O (10:490, v/v, containing 0.5 mm HCOOH) and CH3CN : H2O (495:5, v/v, 0.5 mm HCOOH). The retention times of the measured metabolites ranged from 2.0 to 2.5 min. The MS/MS parameters in the positive or negative electrospray ionization mode were optimized to maximize generation of protonated or deprotonated molecules. The precursor-to-product ion transitions were monitored at m/z 152→110, 161→133, 257→238, 328→283, 310→266, 233→133, 258→157, 184→120, 342→324 and 305→91 for acetaminophen, 7-hydroxycoumarin, hydroxybupropion, desethylamodiaquine, 4′-hydroxydiclofenac, 4′-hydroxymephenytoin, dextrorphan, 6-hydroxychlorzoxazone, 1′-hydroxymidazolam and 6β-hydroxytestosterone, respectively. Matrix-matched standard curves of the peak area of a given analyte vs. the nominal concentration in nM were linear and showed correlation coefficients >0.99. The assays were validated according to the US Food and Drug Administration guidance on bioanalytical method validation (http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm070107.pdf). The intra-day and inter-day assay accuracies ranged from 80% to 108% and from 88% to 109%, respectively, whereas the ranges of precision values for the assays were 0.1%–19% and 0.6%–20%, respectively.
Spectral measurement of cytochrome b5 and total CYP concentrations in HLM
Methods by Omura & Sato [11] for measuring concentrations of cytochrome b5 and total CYP in microsomes were slightly modified using 96-well plates. In brief, 200 µl of diluted HLM (1 mg protein ml−1, pH 7.4) were measured for absorbance at 410 and 425 nm using a SpectraMax M2 Multi-detection Reader (Molecular Devices, Sunnyvale, CA, USA) before and after reduction with 5 µl of 10 mm NADH. The cytochrome b5 concentration was calculated from the absorbance difference between 425 and 410 nm, which was based on an extinction coefficient of 185 mm−1 cm−1. Following the measurement of cytochrome b5, the samples were further spiked with 5 µl of 540 mm sodium dithionite and saturated with carbon monoxide gas for 2 min. The total CYP concentration in HLM was determined from the absorbance difference between 450 and 490 nm using an extinction coefficient of 91 mm−1 cm−1.
Measurement of NADPH-P450 reductase activity
NADPH-P450 reductase assay is based on the use of the electron acceptor surrogate cytochrome c [12], which was conducted in a 96-well plate. The reaction solution contained 0.3 m potassium phosphate buffer (pH 7.7), 0.2 mm cytochrome c, 1 mm potassium cyanide, HLM at 0.025 mg ml−1 microsomal protein and 0.5 mm NADPH. Absorbance of the samples at 550 nm was recorded using the SpectraMax M2 Multi-detection Reader in the kinetic mode before and after the addition of NADPH (0–2 min). The rate of reducing cytochrome c was determined as the slope of the initial linear part of the curve describing the time-dependent increase in absorbance, using an extinction coefficient of 0.021 mm−1 cm−1.
Data analysis
Michaelis constant (Km) and maximum velocity (Vmax) values were determined by nonlinear regression analysis of initial rates of metabolite formation as a function of substrate concentration using GraFit software (Version 5.0; Erithacus Software Ltd, Surrey, UK). In vitro intrinsic clearance (CLint) was calculated from the ratio of Vmax to Km.
Shapiro-Wilk test of normality was used to check the distribution shape of CLint, Km and Vmax for each individual CYP enzyme, as well as the distribution shape of cytochrome b5 concentration, total CYP content and NADPH-P450 reductase activity in HLM samples. In addition, the outliers and extreme values were identified according to whether the values were 1.5–3 and >3 box lengths, respectively, from the upper or lower edge of the box of inter-quartile range (IQR). All the outlying values were confirmed as correct values and there were no data entry or recording errors. Median and IQR were used as measures of central tendency and dispersion for non-normally distributed data. All the outliers and extreme values were included in the calculations. Mann-Whitney U test, a rank based non-parametric test, was used to compare data between the two ethnic groups and also used to estimate the effects of gender and age (18–50 and 51–82 years old) on the cytochrome b5 concentration, total CYP concentration and CLint of test individual CYP enzymes. A P value <0.05 was considered to be the minimum level of statistical significance (two-tailed). Before the study was initiated, power calculations were performed using the power analysis program G*power 3 [13] with 80% power and a 0.05 significance level. The results indicated that 30 HLM samples per ethnic group were needed to detect an effect size of 0.6 or greater. As to the gender difference, 24 male and six female HLM samples in the Chinese group would detect an effect size of 1.1 or greater, whereas the detectable effect size would be 1.0 or greater for the Caucasian group containing 22 male and eight female samples. Twenty-two samples of 18–50 years old and eight samples of 51–82 years old comprized the Chinese group, whereas the sample sizes were 14 and 16, respectively, for the Caucasian group. The detectable effect sizes would be 1.0 or greater for the Chinese group and 0.9 or greater for the Caucasian group.
In addition, the CLint values were transformed by dividing the group median for calculation of Kendall's tau-b coefficients to estimate whether a certain CYP enzyme in HLM was positively or negatively related to another coexisting CYP enzyme, as well as the possible influence of the ethnic factor on the association. Correlation was statistically significant at P < 0.05 level (two-tailed). PASW statistics 18 software (SPSS Inc., Chicago, IL, USA) was used for data management and statistical analyses.
Chemicals and reagents
Most CYP substrates and metabolites were obtained from Sigma-Aldrich (St Louis, MO, USA), including phenacetin, acetaminophen, coumarin, 7′-hydroxycoumarin, bupropion, hydroxybupropion, amodiaquine, desethylamodiaquine, diclofenac, 4′-hydroxydiclofenac (S)-mephenytoin (S)-4′-hydroxymephenytoin, dextromethorphan, dextrorphan, chlorzoxazone, 6-hydroxychlorzoxazone and 1′-hydroxymidazolam, except for midazolam and testosterone from the National Institutes for Food and Drug Control (Beijing, China). 6β-hydroxytestosterone was synthesized by the Department of Medicinal Chemistry of Shanghai Institute of Materia Medica (Shanghai, China). Reduced β-nicotinamide adenine dinucleotide (NADH), reduced β-nicotinamide adenine dinucleotide phosphate (NADPH), β-nicotinamide adenine dinucleotide phosphate hydrate, glucose-6-phosphate monosodium salt, glucose-6-phosphate dehydrogenase and cytochrome c were also purchased from Sigma-Aldrich. Organic solvents and other chemicals used were obtained from Sinopharm Chemical Reagent Co. (Shanghai, China). High purity water was prepared with a Millipore Direct-Q 3 u.v. water purifying system (Bedford, MA, USA).
Results
The concentration of cytochrome b5 in HLM was measured to judge the quality of the test HLM samples before examining their metabolic capabilities of CYP enzymes. The cytochrome b5 concentrations in HLM of 30 Chinese samples ranged from 175 to 674 pmol mg−1 protein with a median of 269 pmol mg−1 protein, whereas those for 30 Caucasian samples ranged from 170 to 489 pmol mg−1 protein with a median of 264 pmol mg−1 protein (Figure 1). No significant difference was found between the ethnic groups for cytochrome b5 (P = 0.7). Meanwhile, the total CYP content in HLM ranged from 100 to 715 pmol mg−1 protein for the Chinese samples and from 140 to 723 pmol mg−1 protein for the Caucasian samples. The total CYP concentration for the Chinese samples was significantly lower than that of the Caucasian samples (P = 0.05), but the median values of the two groups were comparable, i.e. the Chinese value being 82% of the Caucasian value. In addition, there was no significant age-related difference in either cytochrome b5 concentration (P = 0.3) or total CYP concentration (P = 0.8) in HLM for the Chinese samples. The age-related difference was neither significant in total CYP concentration (P = 1.0) nor in cytochrome b5 (P = 0.9) for the Caucasian samples. When the influence of gender was examined, a statistically significant difference in cytochrome b5 concentration was found for the Chinese samples (P = 0.01), while no statistically significant gender difference was found for the Caucasian samples (P = 0.6). There was no significant gender-related difference in total CYP concentrations between these two populations (P = 0.3 for the Chinese samples and P = 0.8 for the Caucasian samples).
Figure 1.
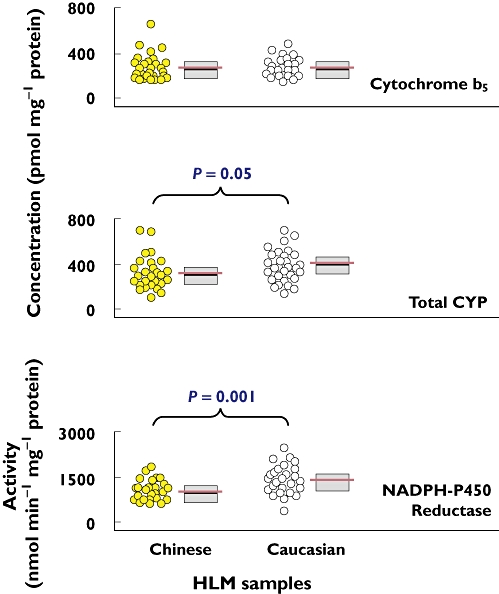
Comparison of the cytochrome b5, total CYP concentrations and NADPH-P450 reductase activity in HLM between Chinese and Caucasian samples. On the right side of the vertical scatter plot, the associated box plot shows the median as the black horizontal line inside the box and the IQR as length of the box. The associated red horizontal line (longer than the median line) represents the mean value
Like cytochrome b5, NADPH-P450 reductase plays important role in CYP-mediated drug metabolism via electron transfer [14]. The activity of NADPH-P450 reductase measured by reduction of cytochrome c showed significant difference between Chinese and Caucasian samples with a P value of 0.001 (Figure 1). The NADPH-P450 reductase activity in the Chinese and Caucasian groups ranged from 598 to 1861 and from 411 to 2548 nmol cytochrome c reduced min−1 mg−1 protein, respectively, and the Chinese median value was 73% of the Caucasian median value. Furthermore, there was neither significant age- nor gender-related differences for NADPH-P450 reductase activity (P = 0.4–1) in these populations.
Phenacetin O-deethylation, coumarin 7-hydroxylation, bupropion hydroxylation, amodiaquine N-desethylation, diclofenac 4′-hydroxylation, (S)-mephenytoin 4′-hydroxylation, dextromethorphan O-demethylation and chlorzoxazone 6-hydroxylation were used as indicators of the activities of CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6 and CYP2E1 in HLM (Figure 2), respectively, which are known to be selective for the CYP enzymes [15, 16]. Both midazolam 1′-hydroxylation and testosterone 6β-hydroxylation were used as measures of CYP3A activity in the current study. An earlier study indicated that recombinant CYP3A4 and CYP3A5 have similar CLint towards midazolam metabolism (≤60 µm), whereas CYP3A4 is about 40 times better at forming the 6β-hydroxyl metabolite of testosterone than is CYP3A5 [17].
Figure 2.
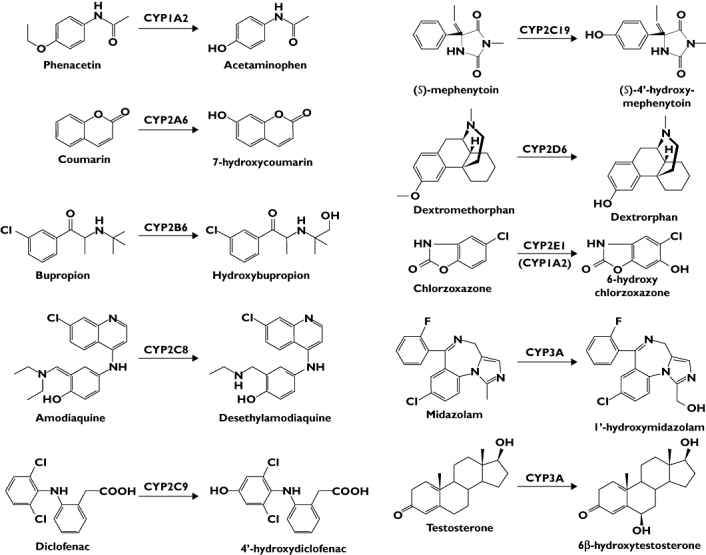
Probe reactions catalyzed by the test human CYP enzymes
Figure 3 shows interethnic differences between the Chinese and Caucasian samples in the CLint values in respect of each individual CYP enzyme. The Shapiro-Wilk test showed that most distributions in CLint for both ethnic groups were significantly different from a normal distribution (P < 0.05), except for CYP2A6 for the Chinese samples (P = 0.6) and the Caucasian samples (P = 0.2). Accordingly, non-parametric statistical tests were applied to the data of the current study. Figure 4 depicts comparative profiles of 60 human samples of two ethnic groups in terms of the relative CLint of individual CYP enzymes, which were normalized to the respective median values of the entire sample set.
Figure 3.
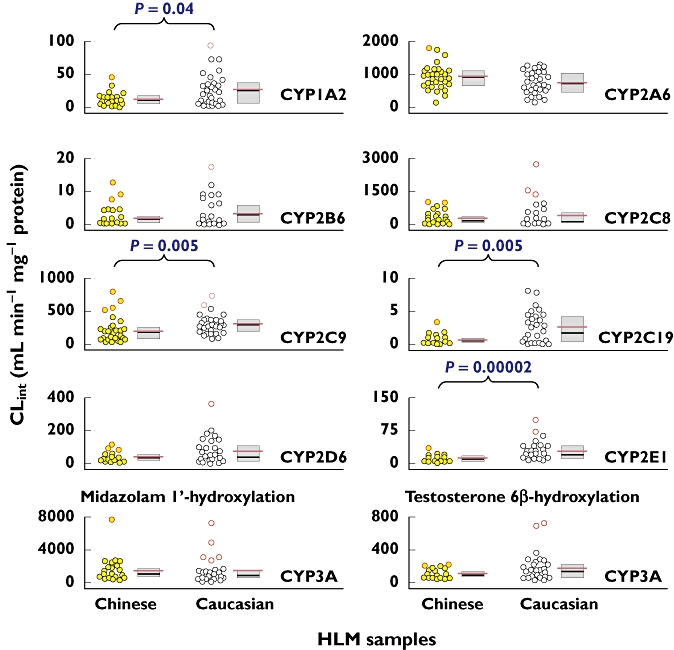
Comparison of the metabolic capabilities (CLint) of various CYP enzymes in HLM between Chinese and Caucasian samples. On the right side of the vertical scatter plot, the associated box plot shows the median as the black horizontal line inside the box and the IQR as length of the box. The associated red horizontal line (longer than the median line) represents the mean value. The outliers and extreme values in each ethnic data set are shown as red circles and are mentioned in the Results section
Figure 4.
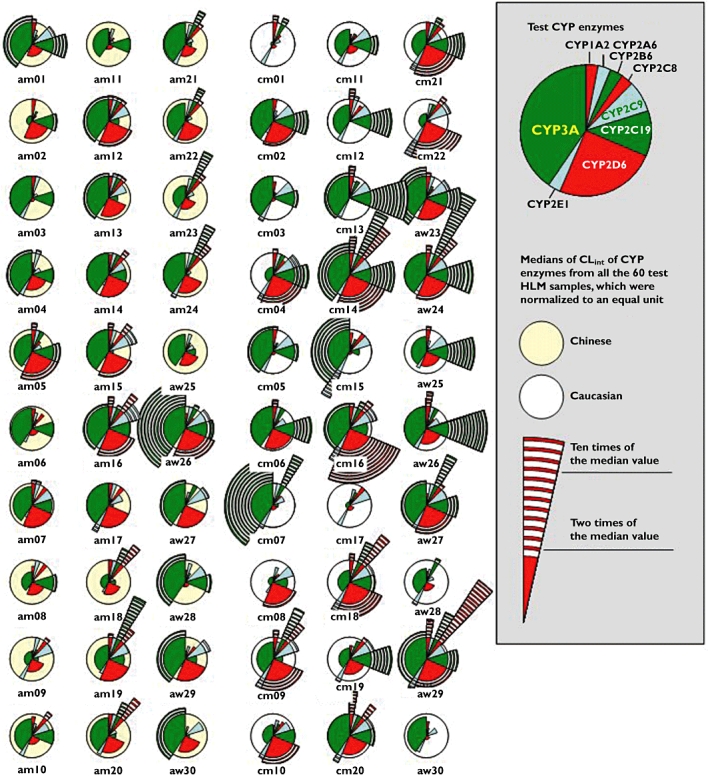
Profiles of multiple CYP enzymes in HLM of the test Chinese or Caucasian samples with respect to metabolic capacities. The data of CYP3A were derived from the CLint towards midazolam 1′-hydroxylation
Within the full data set of both the races, the greatest interindividual difference in CLint was found for the activity of CYP2C19, followed by that of CYP2C8, CYP2B6, CYP1A2, CYP2D6, CYP2E1, CYP2C9 and CYP2A6, demonstrating the percent IQR values (calculated as the IQR as a percentage of the associated median) 321%, 290%, 270%, 204%, 190%, 125%, 105% and 65%, respectively. The interindividual differences for CYP3A towards midazolam 1′-hydroxylation and towards testosterone 6β-hydroxylation were quite comparable, i.e. 91% and 105%, respectively. In addition, there was no significant age-related difference in CLint for the two ethnic groups, except for CYP2C8 (P = 0.004, median of 18–50 years old vs. median of 51–82 years old 261 vs. 44 µl min−1 mg−1 protein), CYP2E1 (P = 0.000004, median of 18–50 years old vs. median of 51–82 years old 8.82 vs. 2.58 µl min−1 mg−1 protein), CYP3A-m (P = 0.01, median of 18–50 years old vs. median of 51–82 years old 726 vs. 2121 µl min−1 mg−1 protein) for the Chinese samples. Meanwhile, gender-related differences in CLint were not significant either, except for CYP1A2 (P = 0.05, male median vs. female median, 12.0 vs. 4.2 µl min−1 mg−1 protein), CYP2C8 (P = 0.003, male median vs. female median, 261 vs. 42 µl min−1 mg−1 protein), CYP2C9 (P = 0.02, male median vs. female median, 101 vs. 321 µl min−1 mg−1 protein), CYP2E1 (P = 0.001, male median vs. female median, 7.54 vs. 2.58 µl min−1 mg−1 protein), and CYP3A towards midazolam metabolism (P = 0.0008, male vs. female median 726 vs. 2463 µl min−1 mg−1 protein) for the Chinese data set.
Table 2 shows the value ranges of CLint, Km and Vmax for each individual CYP enzyme of the Chinese and the Caucasian groups, as well as their associated medians and IQRs. The individual kinetic data for the test CYP enzymes are shown in Tables S1–S10. Similar to CLint, most values of Km and Vmax were also found to be not normally distributed in the Chinese and the Caucasian groups (P < 0.05).
Table 2.
CLint, Km and Vmax for the individual CYP enzymes in HLM
| Chinese group (n = 30) | Caucasian group (n = 30) | ||||||
|---|---|---|---|---|---|---|---|
| CYP | CLint (µl min−1 mg−1 protein) | Km (µm) | Vmax (pmol min−1 mg−1 protein) | CLint (µl min−1 mg−1 protein) | Km (µm) | Vmax (pmol min−1 mg−1 protein) | |
| 1A2 | Range | 0.6–51.4* | 31–379 | 214–2245* | 2.9–92.7 | 30–142 | 182–2772 |
| Median | 10.9 | 85 | 819 | 20.1 | 62 | 1314 | |
| IQR | 12.7 | 80 | 656 | 31.1 | 64 | 1135 | |
| 2A6 | Range | 150–1776 | 0.29–2.64 | 125–3143 | 140–1315 | 0.49–3.75 | 134–4372 |
| Median | 933 | 1.12 | 939 | 663 | 1.58 | 882 | |
| IQR | 468 | 0.61 | 1072 | 610 | 1.01 | 1353 | |
| 2B6 | Range | 0.1–12.6 | 13–175* | 3–953 | 0.1–17.4 | 11–297 | 2–765 |
| Median | 0.5 | 76 | 38 | 0.9 | 32 | 51 | |
| IQR | 2.2 | 80 | 175 | 3.1 | 57 | 291 | |
| 2C8 | Range | 14–1003 | 0.8–81.0 | 372–2042 | 14–2745 | 0.8–53.2 | 100–2235 |
| Median | 194 | 3.9 | 791 | 104 | 7.3 | 861 | |
| IQR | 340 | 11.6 | 582 | 333 | 12.6 | 683 | |
| 2C9 | Range | 15–790* | 4.4–28.5* | 385–4704 | 77–715 | 3.2–14.8 | 823–4067 |
| Median | 166 | 11.6 | 1565 | 287 | 7.0 | 1916 | |
| IQR | 180 | 12.8 | 1271 | 166 | 4.5 | 943 | |
| 2C19 | Range | 0.03–3.36* | 25–288 | 9–118* | 0.02–8.10 | 9–630 | 7–302 |
| Median | 0.43 | 93 | 29 | 1.64 | 60 | 72 | |
| IQR | 0.70 | 65 | 42 | 3.84 | 90 | 110 | |
| 2D6 | Range | 3–119 | 2.0–18.2 | 41–637 | 0.2–358 | 1.9–47.0 | 8–1272 |
| Median | 25 | 8.9 | 215 | 28 | 6.1 | 196 | |
| IQR | 20 | 5.5 | 175 | 99 | 7.7 | 351 | |
| 2E1 | Range | 0.5–34.1* | 12–117* | 32–945 | 4.1–96.3 | 6.4–28.4 | 62–1383 |
| Median | 5.8 | 34 | 215 | 16.8 | 14.4 | 230 | |
| IQR | 9.0 | 40 | 300 | 19.5 | 6.5 | 196 | |
| 3A-m | Range | 191–7505 | 0.62–6.83 | 495–5892 | 90–7089 | 0.8–12.7 | 231–12926 |
| Median | 906 | 2.65 | 2211 | 748 | 2.6 | 2003 | |
| IQR | 902 | 1.42 | 1294 | 847 | 2.4 | 3639 | |
| 3A-t | Range | 35–210 | 17.8–62.2 | 883–7735* | 32–728 | 15–181 | 699–31566 |
| Median | 90 | 24.2 | 2498 | 123 | 30 | 3236 | |
| IQR | 63 | 16.2 | 2311 | 158 | 31 | 6728 | |
‘3A-m’ and ‘3A-t’ represent CYP3A towards midazolam 1′-hydroxylation and towards testosterone 6β-hydroxylation, respectively.
Ethnic differences were statistically significant with a P value of <0.05 (two-tailed).
CYP1A2
The CLint values for phenacetin O-deethylation by CYP1A2 in HLM from the Chinese samples ranged from 0.6 to 51.4 µl min−1 mg−1 protein, whereas those from the Caucasian samples were from 2.9 to 92.7 µl min−1 mg−1 protein. For CLint of the Chinese samples, am16 (51.4 µl min−1 mg−1 protein) was identified as an outlier. For the Caucasian samples, cm20 was also identified as an outlier (92.7 µl min−1 mg−1 protein). The Mann-Whitney U test suggested the difference between the two groups was significant (P = 0.04) with the median CLint for the Caucasian samples 1.8 times greater than that for the Chinese samples. The observed difference in the CLint between Chinese and Caucasian samples tended to be mainly associated with differences in Vmax, rather than in Km. There was a significant difference between the two ethnic groups in Vmax (P = 0.02), whereas the difference in Km was not significant (P = 0.3).
CYP2A6
The CLint values for CYP2A6 towards coumarin 7-hydroxylation ranged from 150 to 1776 µl min−1 mg−1 protein for Chinese samples and 140 to 1315 µl min−1 mg−1 protein for Caucasian samples. For CLint of the Chinese samples, am12 (1776 µl min−1 mg−1 protein) was an outlier. There were no outlying values for the Caucasian samples. Although the two ethnic groups were not considered to be significantly different in CLint (P = 0.07), the median value of the Chinese group (933 µl min−1 mg−1 protein) tended to be slightly greater than that of the Caucasian group (663 µl min−1 mg−1 protein). The Vmax was not significantly different between the two ethnic groups (P = 0.8). Meanwhile, the median Km of the Chinese samples (1.12 µm) tended to be lower than that of the Caucasian samples (1.58 µm) with a P value of 0.1.
CYP2B6
The CLint values for bupropion hydroxylation by CYP2B6 for the Chinese samples ranged from 0.1 to 12.6 µl min−1 mg−1 protein, whereas those for the Caucasian samples were from 0.1 to 17.4 µl min−1 mg−1 protein. There were three extreme values (am19, am24 and am23 with CLint values of 12.6, 9.21 and 7.74 µl min−1 mg−1 protein, respectively) in the Chinese data set. The CLint value of cw24 (17.4 µl min−1 mg−1 protein) was an outlier for the Caucasian samples. Although the median CLint for the Chinese samples (0.5 µl min−1 mg−1 protein) was lower than that for the Caucasian samples (0.9 µl min−1 mg−1 protein), the difference between the two ethnic groups was not statistically significant (P = 0.4). Interethnic difference in Vmax was also negligible (P = 0.5), whereas the median Km of the Chinese samples (76 µm) was significantly higher than that of the Caucasian samples (32 µm) with a P value of 0.02.
CYP2C8
The CLint values for CYP2C8 for amodiaquine N-desethylation varied from 14 to 1003 µl min−1 mg−1 protein for the Chinese samples and from 14 to 2745 µl min−1 mg−1 protein for the Caucasian samples. There were two outliers (am18 and am20, with CLint values of 1003 and 997 µl min−1 mg−1 protein, respectively) in the Chinese data set, whereas an extreme value (cw29, 2745 µl min−1 mg−1 protein) and two outliers (cm14 and cm18, 1532 and 1364 µl min−1 mg−1 protein, respectively) were observed in the Caucasian data set. Although the median CLint for the Chinese data set (194 µl min−1 mg−1 protein) was higher than that for the Caucasian data set (104 µl min−1 mg−1 protein), no statistically significant difference was found between the two ethnic groups (P = 0.6). There was no significant interethnic difference found in Km (P = 0.6) or in Vmax (P = 0.5), which was consistent with the observation of negligible difference in CLint.
CYP2C9
The CLint values for diclofenac 4′-hydroxylation by CYP2C9 ranged from 15 to 790 µl min−1 mg−1 protein for the Chinese samples and 77 to 715 µl min−1 mg−1 protein for the Caucasian samples. There was an extreme value (am16 with CLint values of 790 µl min−1 mg−1 protein) and three outliers (am15, aw26 and aw29, with CLint values of 655, 535 and 516 µl min−1 mg−1 protein, respectively) in the Chinese data set and two outliers (cm04 and cm16 with CLint values of 715 and 587 µl min−1 mg−1 protein, respectively) in the Caucasian data set. The difference between the two ethnic groups was highly significant with a P value of 0.005 and the median CLint for the Caucasian group was 1.7 times greater than that for the Chinese group. In contrast to CYP1A2, the interethnic difference in CLint for CYP2C9 appeared to be mainly attributed to the difference in Km, rather than that of Vmax. There was highly significant difference between the two ethnic groups in Km (P = 0.002), whereas the difference in Vmax was not significant (P = 0.3).
CYP2C19
The CLint values for CYP2C19 for (S)-mephenytoin 4′-hydroxylation varied from 0.03 to 3.36 µl min−1 mg−1 protein for the Chinese data set and 0.02 to 8.10 µl min−1 mg−1 protein for the Caucasian data set. There was an outlier (am01, 3.36 µl min−1 mg−1 protein) in the Chinese data set. No outlying values were found in the Caucasian data set. The interethnic difference in CLint was highly significant between Chinese and Caucasian groups with a P value of 0.005. The median CLint for the Caucasian group was 3.8 times greater than that for the Chinese group. Similar to CYP1A2, the CLint difference between the Chinese and Caucasian groups was mainly associated with the difference in Vmax, vs. a difference in Km. The difference in Vmax was significant between the two ethnic groups (P = 0.006), whereas the difference in Km was not considered to be significant (P = 0.1).
CYP2D6
The CLint values for dextromethorphan O-demethylation by CYP2D6 were from 3 to 119 µl min−1 mg−1 protein for the Chinese group and 0.2 to 358 µl min−1 mg−1 protein for the Caucasian group. There were two extreme values (aw27 and am16 with CLint values of 119 and 97 µl min−1 mg−1 protein, respectively) and an outlier (am05, 82 µl min−1 mg−1 protein) observed in the Chinese data set of CLint and an extreme value (cm16, 358 µl min−1 mg−1 protein) in the Caucasian data set. The median CLint for the Chinese data set was comparable with that of the Caucasian group and there was no statistically significant difference between the two ethnic groups (P = 0.6). Neither Km nor Vmax were found to have a significant difference between the two ethnic groups with P values of 0.3 and 0.9, respectively.
CYP2E1
The CLint for CYP2E1 towards chlorzoxazone 6-hydroxylation varied from 0.5 to 34.1 µl min−1 mg−1 protein for the Chinese group and 4.1 to 96.3 µl min−1 mg−1 protein for the Caucasian group. There was an outlier (am17, 34.1 µl min−1 mg−1 protein) in Chinese data set and one extreme value (cm01, 96.3 µl min−1 mg−1 protein) and an outlier (cm22, 58.8 µl min−1 mg−1 protein) in the Caucasian data set. The difference in CLint between the two ethnic groups was highly significant (P = 0.00002) with the median CLint for the Caucasian group 2.9 times greater than that for the Chinese group. It is worth noting that there is no ideal probe reaction for CYP2E1 activity. Although chlorzoxazone 6-hydroxylation was used in the current studies as an indicator of the activity of CYP2E1 in HLM, the concomitant expression of CYP1A2 in HLM would also considerably contribute to the reaction for some samples. The difference in CLint for chlorzoxazone 6-hydroxylation between Chinese and Caucasian groups tended to be mainly attributed to the difference in Km, rather than that of Vmax. The difference between the two ethnic groups in Km was highly significant (P = 0.000000003), whereas the difference in Vmax was not significant (P = 1).
CYP3A
The ranges of CLint for midazolam 1′-hydroxylation for the Chinese and Caucasian groups were 191 to 7505 and 90 to 7089 µl min−1 mg−1 protein, respectively, whereas the ranges for testosterone 6β-hydroxylation were 35 to 210 and 32 to 728 µl min−1 mg−1 protein, respectively. In the Chinese data set, the sample aw27 (7505 µl min−1·mg protein−1) was characterized as an extreme value for midazolam 1′-hydroxylation, while am19 (210 µl min−1 mg−1 protein) and am05 (206 µl min−1 mg−1 protein) were characterized as outliers for testosterone metabolism. Among the Caucasian samples, cm07 exhibited an extreme value of 7089 µl min−1 mg−1 protein, cm01, cm14, cw30 and cw29 gave outliers of 4790, 2945, 2904, and 2611 µl min−1 mg−1 protein, respectively, for midazolam metabolism, whereas cw29 gave an extreme value of 728 µL min−1 mg−1 protein and cm14 gave an outlier of 700 µl min−1 mg−1·protein for testosterone metabolism. No statistically significant difference was found between the two ethnic groups in CYP3A metabolic capability for midazolam 1′-hydroxylation (P = 0.5). Although the median CLint for testosterone 6β-hydroxylation for the Caucasian group was 1.4 times greater than that for the Chinese group, the difference between the two ethnic groups was not considered to be significant (P = 0.2). Neither Km nor Vmax for testosterone 6β-hydroxylation were found to have significant difference between the two ethnic groups with P values of 0.4 and 0.06, respectively.
Correlations among CLint of individual CYP enzymes and those between cytochrome b5 concentration or NADPH-P450 reductase activity and CYP enzyme activity
A comparison of the Kendall's tau-b correlations in Table 3 showed that there were statistically significant associations of the activity of CYP3A with the activities associated with CYP1A2, CYP2A6, CYP2B6 or CYP2C9. These associations were both CYP3A activity indicator dependent and ethnically dependent. The correlations of CYP3A for testosterone 6β-hydroxylation with CYP2B6 activity were statistically significant for both the Chinese (P = 0.02) and the Caucasian (P = 0.006) groups, while the correlations with CYP2A6 activity, CYP2C8 activity, CYP2C19 activity and CYP3A for midazolam 1′-hydroxylation were significant for only the Caucasian groups, with P values of 0.002, 0.02, 0.05 and 0.01, respectively. Meanwhile, the correlations of CYP3A for midazolam 1′-hydroxylation and CYP1A2 activity was statistically significant in only the Caucasian data set (P = 0.04). The correlation of CYP3A for midazolam 1′-hydroxylation and CYP2C8, CYP2C9 activities was significant for only the Chinese group, with P values of 0.04 and 0.001, respectively.
Table 3.
Associations between individual CYP enzymes in transformed CLint
| Kendall's tau-b coefficient of CYP enzyme | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C19 | 2D6 | 2E1 | 3A-m | 3A-t |
| 1A2 | 0.22 | 0.07 | 0.24 | 0.08 | 0.25 | 0.21 | 0.03 | −0.09 | 0.21 |
| (0.21) | (0.20) | (0.20) | (0.09) | (0.38*) | (0.02) | (0.02) | (0.27*) | (0.23) | |
| 2A6 | 0.19 | 0.30* | 0.08 | −0.06 | 0.27* | 0.27* | 0.03 | 0.03 | |
| (0.34*) | (0.30*) | (0.09) | (0.27*) | (0.26*) | (0.10) | (0.25) | (0.39*) | ||
| 2B6 | 0.44* | 0.02 | −0.26* | 0.17 | 0.28* | −0.13 | 0.30* | ||
| (0.52*) | (0.02) | (−0.01) | (0.32*) | (0.33*) | (0.17) | (0.36*) | |||
| 2C8 | 0.01 | −0.26* | 0.12 | 0.30* | −0.26* | 0.14 | |||
| (0.30*) | (0.05) | (0.46*) | (0.45*) | (0.06) | (0.31*) | ||||
| 2C9 | −0.04 | 0.38* | −0.11 | 0.42* | 0.17 | ||||
| (0.10) | (0.27*) | (0.16) | (0.02) | (−0.10) | |||||
| 2C19 | −0.12 | −0.22 | −0.04 | 0.15 | |||||
| (0.02) | (−0.20) | (0.20) | (0.25*) | ||||||
| 2D6 | −0.01 | 0.15 | 0.04 | ||||||
| (0.27*) | (−0.07) | (0.14) | |||||||
| 2E1 | −0.16 | 0.13 | |||||||
| (0.02) | (0.07) | ||||||||
| 3A-m | −0.01 | ||||||||
| (0.32*) | |||||||||
CLint values were transformed by dividing by the group median for calculation of Kendall's tau-b coefficients that were used to measure the associations. In addition to the data for the Chinese samples, the data for the Caucasian samples were also exhibited in parentheses. ‘3A-m’ and ‘3A-t’ represent the CYP3A towards midazolam metabolism and testosterone metabolism, respectively.
Correlations were statistically significant with a P value of <0.05 (two-tailed).
Notable interethnic differences were found for the activity of CYP2Cs associated with other CYP enzyme activities. CYP2C8 activity was significantly correlated with those of CYP2A6 (P = 0.02), CYP2B6 (P = 0.0007) and CYP2E1 (P = 0.02) in the Chinese data set. Similar associations in activity were also found for the Caucasian group with P values of 0.03, 0.00006, and 0.0005, respectively. In addition, CYP2C8 activity was significantly correlated with that of CYP2C19 for the Chinese samples (P = 0.05) and with that of CYP2C9 and CYP2D6 for the Caucasian samples with P values of 0.02 and 0.0003, respectively. The enzyme activity of CYP2C9 was also associated with that of CYP2D6 for both the Chinese samples (P = 0.03) and the Caucasian samples (P = 0.04). With regard to CYP2C19, the association in activity with CYP2B6 was statistically significant in only the Chinese data set (P = 0.05) and the association with CYP1A2 and CYP2A6 was significant in only the Caucasian data set, with P values of 0.003 and 0.04, respectively. In addition, the enzyme activity of CYP2E1 was associated with CYP2A6 (P = 0.04) or CYP2B6 (P = 0.03) for the Chinese samples, and the enzyme activity of CYP2E1 was associated with CYP2D6 activity (P = 0.04) and that of CYP2B6 was associated with the activities of CYP2A6 (P = 0.01), CYP2D6 (P = 0.01) or CYP2E1 (P = 0.01) for the Caucasian samples. The correlations of enzyme activity of CYP2D6 with that of CYP2A6 were statistically significant for both the Chinese (P = 0.04) and the Caucasian (P = 0.04) groups.
There was a significant correlation between cytochrome b5 concentration and CYP enzyme activity for CYP2C9 and CYP3A in the Chinese data set and CYP2E1 in the Caucasian data set, but no significant correlation was observed for other CYPs. Similarly, only CYP2C9 and CYP3A correlated significantly with NADPH-P450 reductase activity in Chinese data set, while there was no significant correlation found for the other CYP enzymes.
Discussion
Statistical tests can be either parametric or non-parametric. In the current study, utilizing a relatively large population of 60 HLM samples, most of the data for CLint, Km, Vmax, cytochrome b5 concentrations, total CYP concentrations and NADPH-P450 reductase activity were found to be non-normally distributed. Thus, the non-parametric Mann-Whitney U test and Kendall's tau-b correlation analysis were used in the analyses of the data. It is worth pointing out that the use of non-parametric tests would avoid the outliers biasing the summary statistics and P values [18]. Several earlier studies compared the CYP activities and concentrations in HLM between Japanese and Caucasian samples [19–23]. However, the distribution shapes of the results in these studies were not well addressed.
CYP1A2 is highly inducible by dietary and environmental factors, which may mask genetic variation in enzyme activity [24]. The current work found that CYP1A2 activity exhibited large interindividual variation in CLint. The results suggested lower CYP1A2 activity in HLM from the Chinese samples than that from Caucasians. This is consistent with the results of an earlier clinical study in Swedes and Koreans using the plasma paraxanthine : caffeine ratio to indicate CYP1A2 activity [25]. In addition, in vitro CYP1A2 activity towards phenacetin O-deethylation was observed here to be lower within the Chinese female samples than the Chinese male samples, but such a gender difference was not significant with the Caucasian samples. In earlier human studies, CYP1A2 activity towards caffeine metabolism appeared to be lower for women than men in Chinese [26] and Caucasians [27].
Interethnic differences in CYP2A6-mediated metabolism tend to be mainly attributed to genetic polymorphism [28]. The defective CYP2A6 variants in Asians include CYP2A6*4, CYP2A6*7 and CYP2A6*9, whereas in Caucasians there is only CYP2A6*9. In the current study, no significant interethnic difference between Chinese and Caucasian samples was observed with CLint for CYP2A6. However, Shimada et al. [20] reported that both the CYP2A6 protein concentration in HLM and the coumarin 7-hydroxylase activity were lower in Japanese HLM samples than Caucasian samples. In addition, the difference between Japanese subjects and other Asians populations was also reported [29]. Japanese subjects had a significantly lower metabolic ratio of cotinine to nicotine than Korean subjects, but there was no difference between Caucasian and Korean subjects. Japanese had a higher frequency of CYP2A6*4 (∼20%) than Koreans (∼10%). Consistent with the Korean population, Chinese have been reported to have a lower frequency of CYP2A6*4 (∼5%) than Japanese (∼20%), but a higher frequency of CYP2A6*1 (∼80%) than Japanese (∼45%) [29, 30]. These results are consistent with the insignificant difference in the activity of CYP2A6 between Chinese and Caucasian HLM samples. Further, the results reported here and those of the previous studies suggest differences between the Chinese, Japanese and Korean populations that requires additional studies to be confirmed.
Genetic polymorphisms with CYP2B6 are common and the frequencies of the various alleles of CYP2B6 for Asians appear to be comparable with those in Caucasians, albeit with some differences [31, 32]. In the current study, no significant difference in CLint for CYP2B6 towards bupropion hydroxylation was found between the Chinese and the Caucasian samples. However, the associated Km values were observed to be of statistically significant difference between the ethnic groups with the median Km of Chinese samples 2.4 times greater than that of Caucasian samples. Jinno et al. [33] compared the metabolic capabilities of several cDNA recombinant CYP2B6 allelic variants towards 7-ethoxy-4-trifluoromethylcoumarin O-deethylation, including CYP2B6.1, CYP2B6.2, CYP2B6.3, CYP2B6.4, CYP2B6.5, CYP2B6.6 and CYP2B6.7, and found that the tested CYP2B6 allelic variants possessed comparable Km values ranging from 5.5 to 7.7 µm. The reason for our observed ethnic difference in Km is unknown.
CYP2C8 is a slightly inducible CYP enzyme [34]. Little is known about the effects of CYP2C8 variant alleles on drug metabolism. CYP2C8*3 and *4 are primarily found in Caucasians at frequencies of 10–17% and 5–8%, whereas they are extremely rare in Asians [35]. However, neither clinical nor in vitro data show substantial effect of CYP2C8*3 or CYP2C8*4 alleles on paclitaxel 6α-hydroxylation [36, 37]. In the current report, no significant interethnic differences were observed in Km, Vmax or CLint values for CYP2C8 toward amodiaquine metabolism. It is worth noting that the outlying values of CLint in the Chinese and Caucasian data sets appeared to be mainly the result of low Km values.
CYP2C9 is a quantitatively significant CYP enzyme in human liver and metabolizes about 15% of clinically important drugs. Most studies on CYP2C9 genetic variations have been associated with the alleles CYP2C9*2 and CYP2C9*3, which are found in Caucasians with frequencies 8–19% and 3–10%, respectively, but virtually absent in Asians [35]. The enzyme encoded by CYP2C9*2 for diclofenac 4′-hydroxylation has a 59% CLint, a 76% Km and a 44% Vmax of the corresponding values of CYP2C9*1, while the enzyme of CYP2C9*3 possesses a 31% CLint, a three times larger Km and an equal Vmax as compared with the wild type enzyme [38]. Such genotype specific enzyme kinetic data appear to be substrate-dependent, i.e. the CLint ranging from 7% to 111% (the median CLint 60%, excluding the outlier values), Km from 17% to 157% (the median Km 94%) and Vmax from 8% to 100% (the median Vmax 53%) for CYP2C9.2 and the CLint from 2% to 57% (the median CLint 10%), Km from 18% to 825% (the median Km 325%) and Vmax from 4% to 130% (the median Vmax 30%) for CYP2C9.3 as compared with the respective data of CYP2C9.1 [39]. Contrary to the expectation from these in vitro enzyme kinetic data, however, the results of the current study indicate a significantly lower diclofenac 4′-hydroxylase activity in Chinese HLM than Caucasian HLM, which was mainly associated with a difference in Km. Consistent with the results reported here, Chinese and Japanese patients take lower warfarin doses than Caucasians to maintain the same therapeutic efficacy [40, 41]. The lower warfarin dose in Asians suggests that future studies on additional defective CYP2C9 alleles in Asians or on other factors affecting the activity of CYP2C9 are needed. Recently Gu et al. [42] reported that Chinese patients with the CYP2C9 IVS3-65 GC or CC genotype received lower doses of warfarin (0.9 ± 0.2 mg day−1) than those with the CYP2C9*1/*1 genotype (2.6 ± 1.4 mg day−1). Still, the plasma total and unbound warfarin concentrations were significantly lower in the patients with the CYP2C9*1/*1 genotype. The frequencies and intrinsic enzyme activities of CYP2C9 IVS3-65 GC and CC in Chinese and Caucasian populations remain to be better understood.
Genetic differences are a major reason for the large interindividual and interethnic variations in CYP2C19 activity. CYP2C19*2 and CYP2C19*3, two null variants, are responsible for 99% of the poor metabolizer phenotype [43]. The CYP2C19*2 and CYP2C19*3 allele frequencies are higher in Asians (30 and 10%, respectively) than in Caucasians (14 and 0%, respectively) [44]. In addition, CYP2C19*17 is a recently identified allele causing ultrarapid or extensive metabolism of CYP2C19 substrates and the frequency in the Caucasian population is four times higher than that of the Asians (<5%) [45, 46]. Herein and consistent with the known genetic differences, significantly decreased metabolic capability of CYP2C19 in HLM was observed with the Chinese samples compared with the Caucasian samples, which was mainly attributed to the difference in Vmax.
Despite its relatively low abundance in human liver, CYP2D6 is responsible for the metabolism of 25–30% of clinically important drugs. CYP2D6 is not known to be significantly inducible and the genetic variation contributes largely to the interindividual and interethnic variations in metabolic activity [47]. The appearance of poor metabolizers is lower in Asians (∼1%) than Caucasians (5–10%), whereas enzyme activity is lower in Asian extensive metabolizers than in Caucasian counterparts. CYP2D6*3, CYP2D6*4, CYP2D6*5 and CYP2D6*6 account for over 95% of Caucasian poor metabolizers. CYP2D6*10 is specific variant in Asians at a frequency of 40–60%. Compared with the wild-type, the enzyme encoded by CYP2D6*10 shows decreased metabolic capacity in a substrate dependent fashion [8] and Japanese HLM homozygous for CYP2D6*10 tend to have a lower CYP2D6 expression level [21]. In the current study, dextromethorphan O-demethylase activity in HLM showed a large interindividual difference, which appeared to mask the interethnic difference.
Unlike CYP2B6, CYP2C9, CYP2C19 and CYP2D6 that have significant polymorphisms, CYP2E1 is relatively conserved. This CYP enzyme is also inducible [47]. In the current work, a significantly lower chlorzoxazone 6-hydroxylase activity (CLint) was observed in Chinese HLM than Caucasian HLM. The interethnic difference in CLint appeared to be mainly associated with the different Km. Consistent with the current results, the data by Kim et al. [22] suggested that chlorzoxazone 6-hydroxylase activity (measured at one substrate concentration) in Japanese HLM samples was lower than that of Caucasian samples. In addition, the Japanese samples had lower protein concentration of CYP2E1 than the Caucasian samples.
The CYP3A subfamily, mainly CYP3A4 and CYP3A5, represents the most abundant CYP enzymes in human liver and intestine and is associated with the metabolism of ∼50% of drugs. The clinical relevance of CYP3A4 genotypes appears to be relatively limited due to the strong environmental impact via induction or inhibition. In addition, the effect of CYP3A5 polymorphisms on the total CYP3A activity appears to depend on the concomitant expression of CYP3A4. Testosterone and midazolam show some selectivity for CYP3A4 and CYP3A5 [17]. In the current study, no difference for CLint, as well as that of Km or Vmax, of CYP3A was observed between the Chinese and the Caucasian samples.
Recent data have shown co-induction of CYP2C and CYP2B with CYP3A [48, 49]. The nuclear receptors pregnane X receptor and constitutive androstane receptor mediate the induction of CYP3A4, CYP2B and CYP2C enzymes. Wortham et al. found significant correlations among mRNA levels of CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19 and NADPH-P450 reductase, suggesting that these xenobiotic metabolism genes are coregulated at the transcriptional level [50]. In addition, they also observed correlations among CYP enzyme activities (the metabolite formation rate at a single fixed substrate concentration) with 20 HLM samples (11 male and six female Caucasian samples, two male Hispanic samples and one male African American sample), i.e. four correlations for CYP3A (with CYP2A6, CYP2B6, CYP2C8 and CYP2C9), three additional for CYP2C8 (with CYP2A6, CYP2B6 and CYP2C9), four additional for CYP2C9 (with CYP2A6, CYP2B6, CYP2D6 and CYP2E1), two additional for CYP2A6 (with CYP2B6 and CYP2E1), and one additional for CYP2B6 (with CYP2D6). In the current study, significant correlations were found among CLint of CYPs, including six correlations for CYP3A (with CYP2B6 and CYP2C8 in both the ethnic groups, CYP1A2, CYP2A6 and CYP2C19 in the Caucasian group and CYP2C9 in the Chinese group), six additional for CYP2C8 (with CYP2A6, CYP2B6 and CYP2E1 in both the ethnic groups, CYP2C19 in the Chinese group, CYP2C9 and CYP2D6 in the Caucasian groups), one additional for CYP2C9 (with CYP2D6 in both the ethnic groups), three additional for CYP2C19 (with CYP1A2 and CYP2A6 in the Caucasian group and CYP2B6 in the Chinese group), three additional for CYP2A6 (with CYP2D6 in both the ethnic groups, CYP2B6 in the Caucasian group and CYP2E1 in the Chinese group), and two additional for CYP2B6 (with CYP2E1 in both the ethnic groups and CYP2D6 in the Caucasian group) and one additional for CYP2D6 (with CYP2E1 in the Caucasian group). According to these data, the correlations appeared to be ethnically dependent. The data presented here with the Caucasian group are largely consistent with the data by Wortham et al. with respect to the various correlations. The inconsistent results might be due to the difference in HLM samples assessed. In addition, differences in CYP enzyme activity substrates might also have some influence on the results.
Gaining an understanding of interethnic differences in CYP activities is important in order to select the right doses for clinical studies and to avoid unnecessary toxicity due to over-exposure or absence of efficacy due to under-exposure to the drug candidates. In today's environment, multinational pharmaceutical companies are developing drugs that can be used in equally safe and efficient ways in the different regions where they are marketed. Detailed knowledge about interindividual and interethnic variations of the CYP enzymes will facilitate the use of tailored therapy for drugs predominantly eliminated by CYP enzymes and having narrow therapeutic indices. For example, the detailed enzyme kinetics and ranges of values generated in the current study should be useful in improving physiologically based pharmacokinetic models predicting the pharmacokinetics of compounds in development by incorporating ethnic differences and variability. Accordingly, the information about hepatic CYP activities (Figure 3) will be particularly useful for rational choices of drug candidates and predicted doses.
In summary, the metabolic capabilities of nine drug metabolizing CYP enzymes were compared between Chinese HLM and Caucasian HLM. As shown in Table 4, the notable findings include that the Chinese samples had significantly lower CLint for substrates of CYP1A2, CYP2C9, CYP2C19 and CYP2E1 than the Caucasian samples (the median values of the Chinese group being 54, 58, 26, and 35% of the corresponding values of the Caucasian group, respectively), which were mainly attributed to the different Km or Vmax values. However, no significant ethnic difference in CLint was observed for CYP2A6, CYP2B6, CYP2C8, CYP2D6, CYP3A for midazolam 1′-hydroxylation, or CYP3A for testosterone 6β-hydroxylation. The median values of the Chinese group were 141, 56, 187, 89, 121 or 73% of the corresponding values of the Caucasian group, respectively. Despite insignificant interethnic differences in CLint, CYP2B6 had significantly different Km values. The data of the current study significantly extend the knowledge on the ethnic differences in CYP enzymes and have implications for drug discovery and drug therapy (especially for drugs with a narrow therapeutic window) for patients of different ethnic origins. However, since the Chinese and Caucasian samples were not matched for genotype, medication/disease history and diet, the causes or sources of the observed differences are not well understood. Additional studies are planned to look at the genetic differences between Chinese and Caucasian liver samples and match them with microsomal CYP activities.
Table 4.
Summary of interethnic differences in CLint, Km and Vmax of CYP enzymes in HLM samples of Chinese origin (n = 30) compared with those of Caucasian samples (n = 30)
| CYP | CLint | Km | Vmax |
|---|---|---|---|
| 1A2 | ↓ | ↔ | ↓ |
| 2A6 | ↔ | ↔ | ↔ |
| 2B6 | ↔ | ↑ | ↔ |
| 2C8 | ↔ | ↔ | ↔ |
| 2C9 | ↓ | ↑ | ↔ |
| 2C19 | ↓ | ↔ | ↓ |
| 2D6 | ↔ | ↔ | ↔ |
| 2E1 | ↓ | ↑ | ↔ |
| 3A-m | ↔ | ↔ | ↔ |
| 3A-t | ↔ | ↔ | ↔ |
The marks ‘↑’ or ‘↓’ denotes a significantly higher or lower value for the Chinese group (P < 0.05), whereas ‘↔’ implies no statistically significant difference.
Acknowledgments
This work was supported in part by Grant 2009ZX09304-002 from the National Science and Technology Major Project of China ‘Key New Drug Creation and Manufacturing Program’, Grant 30925044 from the National Science Fund of China for Distinguished Young Scholars, Grant 08DZ1980200 from the Shanghai Science and Technology Major Project and Grant KSCX2-YW-R-191 from the Knowledge Innovation Program of the Chinese Academy of Sciences. The work was also supported in part by Eli Lilly and Company.
Competing Interests
There are no competing interests to declare.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1
Individual kinetic data for CYP1A2
Table S2
Individual kinetic data for CYP2A6
Table S3
Individual kinetic data for CYP2B6
Table S4
Individual kinetic data for CYP2C8
Table S5
Individual kinetic data for CYP2C9
Table S6
Individual kinetic data for CYP2C19
Table S7
Individual kinetic data for CYP2D6
Table S8
Individual kinetic data for CYP2E1
Table S9
Individual kinetic data for CYP3A-m
Table S10
Individual kinetic data for CYP3A-t
REFERENCES
- 1.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–71. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 2.Wilson JF, Weale ME, Smith AC, Gratrix F, Fletcher B, Thomas MG, Bradman N, Goldstein DB. Population genetic structure of variable drug response. Nat Genet. 2001;29:265–9. doi: 10.1038/ng761. [DOI] [PubMed] [Google Scholar]
- 3.Burroughs VJ, Maxey RW, Levy RA. Racial and ethnic differences in response to medicines: towards individualized pharmaceutical treatment. J Natl Med Assoc. 2002;94:1–26. [PMC free article] [PubMed] [Google Scholar]
- 4.Weinshilboum R. Inheritance and drug response. N Eng J Med. 2003;348:529–37. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 5.Wrighton SA, Stevens JC. The human hepatic cytochromes P450 involved in drug metabolism. Crit Rev Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 6.Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functionaldiversity. Pharmaconomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 7.Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metab Rev. 2003;35:99–106. doi: 10.1081/dmr-120023681. [DOI] [PubMed] [Google Scholar]
- 8.Shen H-W, He MM, Liu H-F, Wrighton SA, Wang L, Guo B, Li C. Comparative metabolic capabilities and inhibitory profiles of CYP2D6.1, CYP2D6.10, and CYP2D6.17. Drug Metab Dispos. 2007;35:1292–300. doi: 10.1124/dmd.107.015354. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Sun Y, Du F-F, Niu W, Lu T, Kan J-M, Xu F, Yuan K, Qin T, Liu C, Li C. ‘LC-electrolyte effects’ improve the bioanalytical performance of liquid chromatography/tandem mass spectrometric assays in supporting pharmacokinetic study for drug discovery. Rapid Commun Mass Spectrom. 2007;21:2573–84. doi: 10.1002/rcm.3129. [DOI] [PubMed] [Google Scholar]
- 10.Li Y-F, Sun Y, Du F-F, Yuan K-H, Li C. Pulse gradient, large-volume injection, high-throughput ultra-performance liquid chromatographic/tandem mass spectrometry bioanalysis for measurement of plasma amrubicin and its metabolite amrubicinol. J Chromatogr A. 2008;1193:109–16. doi: 10.1016/j.chroma.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. J Biol Chem. 1964;230:2370–8. [PubMed] [Google Scholar]
- 12.Guengerich FP, Martin MV, Sohl CD, Cheng Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc. 2009;4:1245–51. doi: 10.1038/nprot.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 14.Hart SH, Zhong X-B. P450 oxidoreductase: genetic polymorphisms and implications for drug metabolism and toxicity. Expert Opin Drug Metab Toxicol. 2008;4:439–52. doi: 10.1517/17425255.4.4.439. [DOI] [PubMed] [Google Scholar]
- 15.Yuan R, Madani S, Wei X-X, Reynolds K, Huang S-M. Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos. 2002;30:1311–9. doi: 10.1124/dmd.30.12.1311. [DOI] [PubMed] [Google Scholar]
- 16.Walsky RL, Obach RS. Validated assays for human cytochrome P450 activities. Drug Metab Dispos. 2004;32:647–60. doi: 10.1124/dmd.32.6.647. [DOI] [PubMed] [Google Scholar]
- 17.Williams JA, Ring BJ, Cantrell VE, Jones DR, Ecksten J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30:883–91. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 18.Peat J, Barton B. Medical Statistics: a Guide to Data Analysis and Critical Appraisal. Oxford: Blackwell Publishing; 2005. [Google Scholar]
- 19.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–23. [PubMed] [Google Scholar]
- 20.Shimada T, Yamazaki H, Guengerich FP. Ethnic-related differences in coumarin 7-hydroxylation activities catalyzed by cytochrome P450 2A6 in liver microsomes of Japanese and Caucasian populations. Xenobiotica. 1996;26:395–403. doi: 10.3109/00498259609046718. [DOI] [PubMed] [Google Scholar]
- 21.Shimada T, Tsumura F, Yamazaki H, Guengerich FP, Inoue K. Characterization of (±)-bufuralol hydroxylation activities in liver microsomes of Japanese and Caucasian subjects genotyped for CYP2D6. Pharmacogenetics. 2001;11:143–56. doi: 10.1097/00008571-200103000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Kim RB, Yamazaki H, Chiba K, O'shea D, Minura M, Guengerich FP, Ishizaki T, Shimada T, Wilkinson GR. In vivo and in vitro characterization of CYP2E1 activity in Japanese and Caucasians. J Pharmacol Exp Ther. 1996;279:4–11. [PubMed] [Google Scholar]
- 23.Inoue K, Yamazaki H, Shimada T. Characterization of liver microsomal 7-ethoxycoumarin O-deethylation and chlorzoxazone 6-hydroxylation activities in Japanese and Caucasian subjects genotyped for CYP2E1 gene. Arch Toxicol. 2000;74:372–78. doi: 10.1007/s002040000151. [DOI] [PubMed] [Google Scholar]
- 24.Gunes A, Dahl ML. Variation in CYP1A2 activity and its clinical implications: influence of environmental factors and genetic polymorphisms. Pharmacogenomics. 2008;9:625–37. doi: 10.2217/14622416.9.5.625. [DOI] [PubMed] [Google Scholar]
- 25.Ghotbi R, Christensen M, Roh H-K, Ingelman-Sundberg M, Aklillu E, Bertilsson L. Comparisons of CYP1A2 genetic polymorphisms, enzyme activity and the genotype-phenotype relationship in Swedes and Koreans. Eur J Clin Pharmacol. 2007;63:537–46. doi: 10.1007/s00228-007-0288-2. [DOI] [PubMed] [Google Scholar]
- 26.Ou-Yang D-S, Huang S-L, Wang W, Xie H-G, Xu Z-H, Shu Y, Zhou H-H. Phenotypic polymorphism and gender-related differences of CYP1A2 activity in a Chinese population. Br J Clin Pharmacol. 2000;49:145–51. doi: 10.1046/j.1365-2125.2000.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrillo JA, Benitez J. CYP1A2 activity, gender and smoking, as variables influencing the toxicity of caffeine. Br J Clin Pharmacol. 1996;41:605–8. doi: 10.1046/j.1365-2125.1996.35418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther. 2005;77:145–58. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, Kwon JT, McLeod HL, Yokoi T. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80:282–97. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Nurfadhlina M, Foong K. The LK, Tan SC, Zaki SM, Ismail R. CYP2A6 polymorphisms in Malays, Chinese and Indians. Xenobiotica. 2006;36:684–92. doi: 10.1080/00498250600715932. [DOI] [PubMed] [Google Scholar]
- 31.Xie H-J, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, Rane A. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J. 2003;3:53–61. doi: 10.1038/sj.tpj.6500157. [DOI] [PubMed] [Google Scholar]
- 32.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck WH, Eichelbaum M, Chwab M, Zanger UM. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics. 2005;15:861–73. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Jinno H, Tanaka-Kagawa T, Ohno A, Makino Y, Matsushima E, Hanioka N, Ando M. Functional characterization of cytochrome P450 2B6 allelic variants. Drug Metab Dispos. 2003;31:398–403. doi: 10.1124/dmd.31.4.398. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson SS, Chen Y-P, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4α. Mol Pharmacol. 2005;68:747–57. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Martin E, Martinez C, Ladero JM, Agúndez JAG. Interethinic and intraethnic variability of CYP2C8 and CYP2C9 polymorphisms in healthy individuals. Mol Diag Ther. 2006;10:29–40. doi: 10.1007/BF03256440. [DOI] [PubMed] [Google Scholar]
- 36.Daily EB, Aquilante CL. Cytochrome P450 2C8 pharmacogenetics: a review of clinical studies. Pharmacogenomics. 2009;20:1489–510. doi: 10.2217/pgs.09.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, Houdt JV, Hendrickx J, Mannens G, Bohets H, Williams FM, Armstrong M, Crespi CL, Daly AK. CYP2C8 polymorphisms in Caucasian and their relationship with paclitaxel 6α-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–89. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 38.Yasar U, Tybring G, Hidestrand M, Oscarson M, Ingelman-Sundberg M, Dahl M-L, Eliasson E. Role of CYP2C9 polymorphism in losartan oxidation. Drug Metab Dispos. 2001;29:1051–6. [PubMed] [Google Scholar]
- 39.Kirchheiner J, Tsahuridu M, Jabrane W, Roots I, Brockmőller J. The CYP2C9 polymorphism: from enzyme kinetics to clinical dose recommendations. Personalized Med. 2004;1:63–84. doi: 10.1517/17410541.1.1.63. [DOI] [PubMed] [Google Scholar]
- 40.Yu HC, Chan TY, Critchley JA, Woo KS. Factors determining the maintenance doses of warfarin in Chinese patients. QJM. 1996;89:127–35. doi: 10.1093/qjmed/89.2.127. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi H, Kashima T, Nomizo Y, Muramoto N, Shimizu T, Nasu K, Kubota T, Kimura S, Echizen H. Metabolism of warfarin enantiomers in Japanese patients with heart disease having different CYP2C9 and CYP2C19 genotypes. Clin Pharmacol Ther. 1998;63:519–28. doi: 10.1016/S0009-9236(98)90103-5. [DOI] [PubMed] [Google Scholar]
- 42.Gu Q, Kong K, Schneede J, Xiao Y-B, Chen L, Zhong Q-J, Wang X-F, Hao J, Chen B-C, Chen J-J. VKORC1-1639G > A, CYP2C9, EPHX1691A > G genotype, body weight, and age are important predictors for warfarin maintenance doses in patients with mechanical heart valve prostheses in southwest China. Eur J Clin Pharmacol. 2010;66:1217–27. doi: 10.1007/s00228-010-0863-9. [DOI] [PubMed] [Google Scholar]
- 43.Kim K, Johnson JA, Derendorf H. Differences in drug pharmacokinetics between East Asians and Caucasians and the role of genetic polymorphisms. J Clin Pharmacol. 2004;44:1083–105. doi: 10.1177/0091270004268128. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu T, Ochiai H, Åsell F, Shimizu H, Saitoh R, Hama Y, Katada J, Hashimoto M, Matsui H, Taki K, Kaminuma T, Yamamoto M, Aida Y, Ohashi A, Ozawa N. Bioinformatics research on inter-ethnic difference in drug metabolism I. Analysis on frequencies of mutant alleles and poor metabolizer on CYP2D6 and CYP2C19. Drug Metab Pharmacokin. 2003;18:48–70. doi: 10.2133/dmpk.18.48. [DOI] [PubMed] [Google Scholar]
- 45.Ragia G, Arvanitidis KI, Tavridou A, Manolopoulos VG. Need for reassessment of reported CYP2C19 allele frequencies in various populations in view of CYP2C19*17 discovery: the case of Greece. Pharmacogenomics. 2009;10:43–9. doi: 10.2217/14622416.10.1.43. [DOI] [PubMed] [Google Scholar]
- 46.Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol. 2010;69:222–30. doi: 10.1111/j.1365-2125.2009.03578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trafalis DT, Panteli ES, Grivas A, Tsigris C, Karamanakos PN. CYP2E1 and risk of chemically mediated cancers. Expert Opin Drug Metab Toxicol. 2010;6:307–19. doi: 10.1517/17425250903540238. [DOI] [PubMed] [Google Scholar]
- 48.Lin JHCY. P induction-mediated drug interactions: in vitro assessment and clinical implications. Pharm Res. 2006;23:1089–116. doi: 10.1007/s11095-006-0277-7. [DOI] [PubMed] [Google Scholar]
- 49.Hewitt NJ, Lecluyse EL, Ferguson SS. Induction of hepatic cytochromes P450 enzymes: methods, mechanisms, recommendations, and in vitroin vivo correlations. Xenobiotica. 2007;37:1196–224. doi: 10.1080/00498250701534893. [DOI] [PubMed] [Google Scholar]
- 50.Wortham M, Czerwinski M, He L, Parkinson A, Wan Y-J. Expression of constitutive androstane receptor, hepatic nuclear factor 4α, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos. 2007;35:1700–10. doi: 10.1124/dmd.107.016436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


