Abstract
AIM
To determine whether the SNP rs4149056 in SLCO1B1 alters the pharmacodynamics of pravastatin.
METHODS
rs4149056 was genotyped in 626 pravastatin-treated participants in the WOSCOPS trial and the response after 1 year of treatment was compared between the different genotypes.
RESULTS
Pravastatin reduced serum LDL cholesterol by 22.2% in TT homozygotes, by 22.2% in TC heterozygotes and by 17.7% in CC homozygotes (TT + TC vs. CC P value 0.33). There were no significant differences in the response of total cholesterol, LDL, HDL, triglycerides or CRP to pravastatin between the genotypes.
CONCLUSION
The rs4149056 SNP did not significantly affect the pharmacodynamics of pravastatin.
Keywords: OATP1B1, pravastatin, rs4149056, SLCO1B1, statins, WOSCOPS
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
The SNP rs4149056 is associated with altered statin pharmacokinetics and increased risk of statin intolerance. LDL-cholesterol lowering by simvastatin is also affected by rs4149056 but differences between the genotypes are small. Studies of the effect of rs4149056 on pravastatin pharmacodynamics have been inconclusive.
WHAT THIS PAPER ADDS
rs4149056 was not associated with an altered lipid or CRP response to 40 mg day−1 pravastatin in more than 600 high risk men from Scotland, suggesting that the rs4149056 genotype does not significantly affect pravastatin efficacy.
Introduction
The hepatic anion transporter OATP1B1 encoded by the gene SLCO1B1 facilitates the entry of statins along with a variety of other drugs and endogenous anions into hepatocytes [1]. Statins target HMG-CoA reductase in the liver and are largely excreted in the bile so entry into hepatocytes is required for both their action and excretion. A recent genome-wide screen for single nucleotide polymorphisms (SNPs) that confer increased risk of simvastatin-induced myopathy found that the SNP rs4149056 in the coding region of SLCO1B1 is significantly associated with increased risk [2]. Two copies of the high risk C-allele conferred an odds ratio of 16.9 for myopathy, and the association was replicated in participants in the Heart Protection Study [2]. The C-allele of rs4149056 encodes a reduced activity variant of OATP1B1 with a valine to alanine substitution at position 174. Carriers of the rs4149056 SNP develop a significantly higher plasma area under the concentration−time curve (AUC) than non-carriers after dosing with pravastatin, atorvastatin, rosuvastatin and simvastatin [1].
As the pharmacokinetics of statins are profoundly altered by rs4149056 it seems likely that the pharmacodynamics are also affected, but the evidence to date has been inconsistent. The largest study was by the SEARCH group who showed a small although statistically significant decrease in the LDL-cholesterol lowering response in carriers of the rs4149056 rare C allele (LDL reductions 1.28% ± 0.25% smaller per copy of the SNP, P < 0.0001) after dosing with 40 mg simvastatin [2]. This contrasts with three small studies of pravastatin showing larger effects of rs4149056 on the lipid lowering response [3–5]. Simvastatin is given as a lactone prodrug but only the acid form of the drug is transported by OATP1B1 [6]. Pravastatin is given in the acid form so rs4149056 may have a larger effect with pravastatin than with simvastatin. The purpose of this study was to further investigate the effects of rs4149056 on pravastatin pharmacodynamics.
Methods
Sample
The sample was 626 pravastatin-treated (40 mg day−1) participants in the West of Scotland Coronary Prevention Study (WOSCOPS) [7]. The sample set was as previously described [8, 9] although here only the pravastatin-treated participants were examined. The trial participants were men aged 45–64 years with mean baseline total cholesterol of 7 mmol l−1. Serum measurements of lipid fractions and CRP were as previously described [7, 8]. The ethics committee of the Glasgow Royal Infirmary approved this study.
Genotyping rs4149056
DNA was extracted as previously described [9]. rs4149056 was genotyped by PCR restriction fragment length polymorphism analysis using the following primers:
Forward primer: TGGGTCATACATGTGGATATACG
Reverse primer: CCAAGAATGCATGGTTCTTATTC
There were no natural restriction sites that overlapped the SNP so a site was generated with a single base mismatch in the forward primer. The common T-allele of rs4149056 produced a PCR fragment with a single ACGT cutting site for the HypCH4IV restriction endonuclease, whereas the PCR product amplified from the rare C-allele did not cut with HypCH4IV. DNA (15 ng) from each study participant was amplified with 0.4 U BIOTAQ DNA polymerase (Bioline Ltd, London, UK) in a 20 µl reaction mix containing each primer at 400 nm (MWG Operon, Ebersberg, Germany). The PCR products were digested with HypCH4IV (New England Biolabs, Beverley, MA, USA) and fragments were separated on 7.5% polyacrylamide MADGE electrophoresis gels [10] and visualized by ethidium bromide staining. The genotypes were read by two independent observers and any equivocal results were reanalyzed. All plates included a DNA sample heterozygous for the SNP as a positive control. Six samples (two for each genotype) were confirmed by sequencing.
Statistical analysis
The concentrations of lipid fractions and CRP at baseline and after 1 year of treatment with pravastatin were compared in participants with the different rs4149056 genotypes. P values were from analysis of variance. Values were adjusted for BMI, systolic blood pressure, diastolic blood pressure, HDL, LDL, log triglycerides, nitrate use, history of angina, history of hypertension and history of diabetes. The results for CRP and triglycerides were skewed so the P values are from log-transformed data.
Results
Pravastatin reduced the total cholesterol concentration by 16.6% in participants homozygous for the T allele (n = 449), by 16.3% in TC heterozygotes (n = 164) and by 13.1% in homozygous carriers of the high risk C allele (n = 13) (TT + TC vs. CC P value 0.35) (Table 1, Figure 1). The LDL-cholesterol concentration was reduced by 22.2% in T allele homozygotes, by 22.2% in TC heterozygotes and by 17.7% in homozygous carriers of the C allele (TT + TC vs. CC P value 0.33). Although the CC homozygotes had a reduced response to pravastatin this did not reach statistical significance. There were no significant differences in the response of HDL, triglycerides or CRP to pravastatin between the different genotypes.
Table 1.
Difference between baseline and 1 year measurements of lipid fractions and CRP by rs4149056 genotype
| Genotypes | ||||||
|---|---|---|---|---|---|---|
| TT/TC/CC n | TT Mean (SE) | TC Mean (SE) | CC Mean (SE) | Overall P value | CC vs. TC + TT P value | |
| Total cholesterol (mmol l−1) | 449/164/13 | |||||
| Baseline | 7.10 (0.042) | 7.08 (0.045) | 7.21 (0.070) | 0.088 | 0.040‡ | |
| 1 year | 5.88 (0.189) | 5.90 (0.201) | 6.13 (0.322) | 0.65 | 0.36 | |
| Difference† | −1.18 (0.189) | −1.16 (0.201) | −0.96 (0.323) | 0.71 | 0.42 | |
| % change† | −16.6%(2.68) | −16.3% (2.86) | −13.1% (4.58) | 0.62 | 0.35 | |
| HDL cholesterol (mmol l−1) | 449/164/13 | |||||
| Baseline | 1.15 (0.045) | 1.15 (0.048) | 1.13 (0.075) | 0.93 | 0.82 | |
| 1 year | 1.23 (0.056) | 1.24 (0.060) | 1.21 (0.095) | 0.92 | 0.74 | |
| Difference† | 0.09 (0.034) | 0.09 (0.037) | 0.07 (0.059) | 0.93 | 0.77 | |
| % change† | 7.16% (2.98) | 7.70% (3.17) | 5.05% (5.07) | 0.80 | 0.59 | |
| LDL cholesterol (mmol l−1) | 449/164/13 | |||||
| Baseline | 5.06 (0.090) | 5.06 (0.096) | 4.91 (0.150) | 0.45 | 0.21 | |
| 1 year | 3.93 (0.175) | 3.93 (0.187) | 4.04 (0.299) | 0.89 | 0.64 | |
| Difference† | −1.11 (0.167) | −1.11 (0.178) | −0.92 (0.285) | 0.73 | 0.43 | |
| % change† | −22.2% (3.38) | −22.2% (3.61) | −17.7% (5.77) | 0.62 | 0.33 | |
| Triglycerides (mmol l−1)* | 449/164/13 | |||||
| Baseline | 1.88 (1.074) | 1.99 (1.078) | 1.62 (1.126) | 0.048‡ | 0.085 | |
| 1 year | 1.73 (1.094) | 1.75 (1.100) | 1.60 (1.164) | 0.77 | 0.51 | |
| Ratio† | 0.94 (1.074) | 0.91 (1.079) | 0.95 (1.130) | 0.68 | 0.87 | |
| CRP (mg l−1)* | 398/139/11 | |||||
| Baseline | 2.08 (1.224) | 2.06 (1.241) | 1.93 (1.413) | 0.97 | 0.80 | |
| 1 year | 1.92 (1.249) | 2.13 (1.264) | 2.28 (1.439) | 0.49 | 0.62 | |
| Ratio† | 0.91 (1.186) | 1.01 (1.198) | 1.25 (1.332) | 0.15 | 0.21 | |
Results are presented as adjusted mean (SE) except
which are adjusted geometric mean (SE).
In addition adjusted for baseline level. n indicates number of participants for whom baseline and 1 year data were available.
Similar differences in baseline total cholesterol and triglycerides were not seen in placebo treated participants and are probably due to chance.
Figure 1.
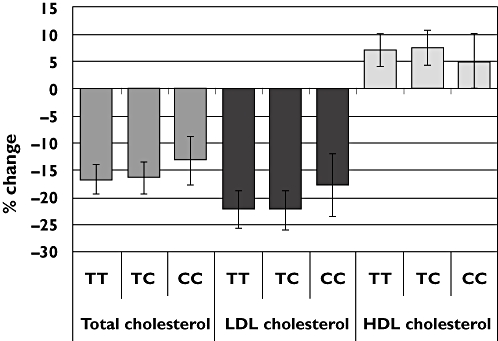
The percentage change of cholesterol fractions after 1 year of pravastatin treatment by rs4149056 genotype. Error bars are SE and results are adjusted for confounding covariates and baseline level
Discussion
Our results on the effect of rs4149056 on the pharmacodynamics of pravastatin were similar to those shown by the SEARCH group with simvastatin [2], although our study was not powered to detect the small differences they demonstrated (LDL reductions 1.28% ± 0.25% smaller per copy of the SNP, P < 0.0001). The attenuation of response in the CC homozygotes was larger than that seen in the SEARCH study but the number of CC participants was small and the differences were not statistically significant. Our results contrast with three studies of pravastatin that showed larger attenuations of the pravastatin response in the TC heterozygotes (total cholesterol reductions of 13.1% in TCs vs. 19.1% in TTs [3], 9.8% in TCs vs. 20.9% in TTs [4] and 14.5% in TCs vs. 22.4% in TTs [5]), which were statistically significant in two out of the three studies [3–5]. Differences in the timing of lipid measurements and the ethnicity of the study populations may have contributed to the contrast in results. None of the three studies of pravastatin contained any CC homozygotes so comparisons with the CC results cannot be made. A recently published physiology-based pharmacokinetic model of pravastatin predicted that variants of OATP1B1 with reduced transporter activity will have a large effect on the plasma AUC but only a minimal effect on the liver AUC [11]. This is consistent with our findings, as it is the liver AUC that governs the lipid-lowering response, and this is the likely explanation for the lack of effect of rs4149056 on pravastatin pharmacodynamics, even though an effect on pravastatin plasma concentrations is well established [1].
Other SNPs in SLCO1B1 including the coding SNP rs2306283 and the promoter SNP rs4149015 have been linked with altered statin transport, but genotyping these SNPs in this sample set is unlikely to be useful. Analyses of pravastatin pharmacokinetics consistently show rs4149056 to be responsible for most of the difference between SLCO1B1 haplotypes [1, 12]. Moreover, rs2306283 had only half the effect of rs4149056 on the LDL response to simvastatin in the SEARCH study [2]. Although there may be larger differences between rare haplotypes containing multiple SNPs, the inevitable reduction in the number of participants in each group means the differences are unlikely to be statistically significant unless a larger study is performed.
One potentially confounding factor in this study is the use of co-medication by trial participants as some drugs including antidiabetics, valsartan and immunosuppressives inhibit OATP1B1 [12]. It is unlikely that this affected the results as serious illness was an exclusion criterion in WOSCOPS and the use of co-medication was probably low. Moreover, results were corrected for history of diabetes and hypertension, and valsartan was not in use until after the trial period.
This study was well controlled and provides good evidence that there is little difference in the response of TT and TC participants to pravastatin 1 year after dosing. The low number of CC participants meant only large differences in the response of CC compared with TT participants could have been detected. A retrospective power calculation shows that the sample available could have detected a 13.1% difference in LDL response between the CC and TC + TT subjects with an 80% power and 5% significance, but only a 4.5% difference was seen, and a larger study is required to investigate this group further. The inclusion of additional time points after treating with pravastatin would also be useful as one study suggested that rs4149056 affects the timing of cholesterol reduction after treatment [4].
In summary, rs4149056 was not significantly associated with a difference in the lipid-lowering response to pravastatin. This suggests that all individuals no matter what their genotype will benefit equally from statin treatment although C-allele carriers are at higher risk of adverse drug reactions.
Acknowledgments
This work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. SEH, WP and KWL are supported by the British Heart Foundation (RG2008/008). WOSCOPS was supported by a research grant from the Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, N.J.
Competing Interests
There are no competing interests to declare.
REFERENCES
- 1.Romaine SP, Bailey KM, Hall AS, Balmforth AJ. The influence of SLCO1B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenomics J. 2010;10:1–11. doi: 10.1038/tpj.2009.54. [DOI] [PubMed] [Google Scholar]
- 2.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N Engl J Med. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 3.Igel M, Arnold KA, Niemi M, Hofmann U, Schwab M, Lutjohann D, von Bergmann K, Eichelbaum M, Kivisto KT. Impact of the SLCO1B1 polymorphism on the pharmacokinetics and lipid-lowering efficacy of multiple-dose pravastatin. Clin Pharmacol Ther. 2006;79:419–26. doi: 10.1016/j.clpt.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Takane H, Miyata M, Burioka N, Shigemasa C, Shimizu E, Otsubo K, Ieiri I. Pharmacogenetic determinants of variability in lipid-lowering response to pravastatin therapy. J Hum Genet. 2006;51:822–6. doi: 10.1007/s10038-006-0025-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Chen BL, Ozdemir V, He YJ, Zhou G, Peng DD, Deng S, Xie QY, Xie W, Xu LY, Wang LC, Fan L, Wang A, Zhou HH. SLCO1B1 521T->C functional genetic polymorphism and lipid-lowering efficacy of multiple-dose pravastatin in Chinese coronary heart disease patients. Br J Clin Pharmacol. 2007;64:346–52. doi: 10.1111/j.1365-2125.2007.02892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–9. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd J, Cobbe SM, Ford I, Isles CG, Lorimer AR, MacFarlane PW, McKillop JH, Packard CJ. Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. West of Scotland Coronary Prevention Study Group. N Engl J Med. 1995;333:1301–7. doi: 10.1056/NEJM199511163332001. [DOI] [PubMed] [Google Scholar]
- 8.Basso F, Lowe GD, Rumley A, McMahon AD, Humphries SE. Interleukin-6 -174G>C polymorphism and risk of coronary heart disease in West of Scotland coronary prevention study (WOSCOPS) Arterioscler Thromb Vasc Biol. 2002;22:599–604. doi: 10.1161/01.atv.0000013283.84306.1a. [DOI] [PubMed] [Google Scholar]
- 9.Zito F, Lowe GD, Rumley A, McMahon AD, Humphries SE. Association of the factor XII 46C>T polymorphism with risk of coronary heart disease (CHD) in the WOSCOPS study. Atherosclerosis. 2002;165:153–8. doi: 10.1016/s0021-9150(02)00196-x. [DOI] [PubMed] [Google Scholar]
- 10.Day IN, Humphries SE. Electrophoresis for genotyping: microtiter array diagonal gel electrophoresis on horizontal polyacrylamide gels, hydrolink, or agarose. Anal Biochem. 1994;222:389–95. doi: 10.1006/abio.1994.1507. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Kusuhara H, Maeda K, Shitara Y, Sugiyama Y. Physiologically based pharmacokinetic modeling to predict transporter-mediated clearance and distribution of pravastatin in humans. J Pharmacol Exp Ther. 2009;328:652–62. doi: 10.1124/jpet.108.146647. [DOI] [PubMed] [Google Scholar]
- 12.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


