Abstract
Marine bioresources produce a great variety of specific and potent bioactive molecules including natural organic compounds such as fatty acids, polysaccharides, polyether, peptides, proteins, and enzymes. Lectins are also one of the promising candidates for useful therapeutic agents because they can recognize the specific carbohydrate structures such as proteoglycans, glycoproteins, and glycolipids, resulting in the regulation of various cells via glycoconjugates and their physiological and pathological phenomenon through the host-pathogen interactions and cell-cell communications. Here, we review the multiple lectins from marine resources including fishes and sea invertebrate in terms of their structure-activity relationships and molecular evolution. Especially, we focus on the unique structural properties and molecular evolution of C-type lectins, galectin, F-type lectin, and rhamnose-binding lectin families.
1. Introduction
Marine bioresources such as marine cyanobacteria, algae, invertebrate animals, and fishes produce a great variety of specific and potent bioactive molecules including natural organic compounds such as fatty acids, polysaccharides, polyether, peptides, proteins, and enzymes. To date, many researchers focused on the marine natural products and their various pharmacological functions to develop new potent drugs including antimicrobials, anti-human immunodeficiency virus (HIV), anticancer, and Alzheimer's therapeutics. There are several excellent reviews that described the potent therapeutic agents derived from marine resources and their medicinal applications [1–3]. In the drug discovery from natural resources, lectins are one of the promising candidates for useful therapeutic agents because carbohydrate structures such as proteoglycans, glycoproteins, and glycolipids have been implicated in certain cell types and their physiological and pathological functions including host-pathogen interactions and cell-cell communications. For example, griffithsin (GRFT), a lectin isolated from red algae Griffithsia sp., showed a strong anti-HIV activity with half maximal effective concentration (EC50) of 0.043–0.63 μM via specific binding to gp120, which is envelop glycoprotein anchored to the HIV membrane and involved in viral entry into cells by recognition of CD4 [4]. Cyanovirin (CN-V) isolated from Nostoc ellipsosporum cyanobacteria has been reported as potent lectin with anti-HIV activity [5].
Lectins are group of sugar-binding proteins except for antibodies and enzymes that recognize specific carbohydrate structures, resulting in the regulation of various cells via glycoconjugates. Thus, they can identify the cell types and cell development stages including embryonic stem (ES) and induced pluripotent stem (iPS) cells through histochemical applications, flow cytometry, and lectin microarrays [6] because cells often display altered surface glycoproteins and glycolipids depending on the physiological and pathological conditions. Lectins are widely distributed in all taxa from microbial organisms, plant, and animal and are involved in numerous cellular processes that depend on their specific recognition of complex carbohydrates. Based on the structural similarity of carbohydrate recognition domain (CRD) and their characteristics, animal lectins are classified into several categories: C-type lectins (CTLs), galectins, I-type lectins, pentraxins, P-type lectins, tachylectins, and so forth [7].
Intensive investigations have been carried out to clarify the biochemical and physiological properties of humoral lectins of marine resources including marine cyanobacteria, algae, and invertebrates such as barnacles [8–10], sea urchins [11, 12], sea cucumbers [13–18], horseshoe crabs [19–30], tunicates [31–36], mollusks [37–39], and fishes [40–69] as shown in Table 1. For fish and sea invertebrate lectins, they could be mainly classified into CTLs, galectins, F-type lectins, and rhamnose binding lectin (RBL) families in addition to the Ricin-type, Lily-type, 6x β-propeller/Tectonin-type lectins (Table 1). Here, we review these multiple lectins from marine resources including fishes and sea invertebrate in terms of their structure-activity relationships and molecular evolution.
Table 1.
Lectins from marine organisms.
| Organism | Lectins | Binding specificity | Lectin family | References |
|---|---|---|---|---|
| [Protozoan] | ||||
| Sponge | ||||
| (Aphrocallistes vastus) | LECCI | d-Galactose | C-type | [93] |
| (Axinella polypoides) | AP-I/II/III/V | d-Galactose | Ricin type | [94] |
| AP-IV | hexuronic acid | |||
| (Geodia cydonium) | GCLT1 | LacNAc | Galectin | [95] |
| (Ptilota filicina) | PFL | d-Galactose | [96] | |
|
| ||||
| [Mollusk] | ||||
| Tridacna | ||||
| (Tridacna maxima) | tridacin | N-acetyl galactosamine | C-type | [37] |
| Abalone | ||||
| (Haliotis laevigata) | PLC | d-Galactose/d-Mannose | C-type | [38] |
| Penguin wing oyster | ||||
| (Pteria penguin) | PPL | d-Galactose | RBL | [39] |
|
| ||||
| [Arthropod] | ||||
| Barnacle | ||||
| (Megabalanus rosa) | BRA-1~ 3 | d-Galactose | C-type | [8, 9] |
| (Balanus rostratus) | BRL | d-Galactose | C-type | [10] |
| Horseshoe crab | ||||
| (Tachypleus tridentatus) | TL-1 | LPS (KDO), LTA | 6x β-propeller/Tectonin | [19] |
| TL-2 | GlcNAc/GalNAc, LTA | 5x β propeller | [20–22] | |
| TL-3 | LPS (O-antigen) | F-type | [23] | |
| TL-4 | LPS(O-antigen), LTA, | [24] | ||
| TLs-5 | Fucose | Ficolin | [25, 26] | |
| TL-P | N-acetyl group | 6x β-propeller/Tectonin | [27] | |
| GBP/PAP/LBP | GlcNAc | [28] | ||
|
| ||||
| [Echinoderm] | ||||
| Sea cucumber | ||||
| (Cucumaria echinata) | CEL-I/CEL-II | GlcNAc | C-type | [13, 14] |
| CEL-IV | GlcNAc-Galactose | C-type | [13, 15] | |
| CEL-III | d-Galactose and so forth | Ricin type | [13, 16] | |
| (Stichopus japonicus) | SPL-1 | uronic acid | C-type | [17] |
| SPL-2 | LacNAc and so forth | C-type | [17] | |
| SJL-1 | d-Galactose | C-type | [18] | |
| Urchin | ||||
| (Anthocidaris crassispina) | SUEL | d-Galactose | RBL | [11] |
| Echinoidin (ECH) | d-Galactose | C-type | [12] | |
| Starfish | ||||
| (Asterina pectinifera) | LacNAc | C-type | [97] | |
|
| ||||
| [Protochordate] | ||||
| Ascidian | ||||
| (Polyandrocarpa misakiensis) | TC14 | d-Galactose | C-type | [31] |
| (Botryllus schlosseri) | Bs RBL-1~5 | l-Rhamnose | RBL | [32, 33] |
| (Halocynthia roretzi) | P36 | d-Galucose | C-type | [34] |
| P40-P50 | ficolin | [35] | ||
| 41KD | d-Glactose | C-type (ficolin) | [36] | |
|
| ||||
| [Fishes] | ||||
| Japanese eel | ||||
| (Anguilla japonica) | AJL-1 | β-Galactoside | Galectin | [40] |
| AJL-2 | Lactose | C-type | [41] | |
| eCL-1/eCL-2 | Lactose | C-type | [42] | |
| European eel | ||||
| (Anguilla anguilla) | AAA | Fucose | F-type | [43] |
| Electric eel | ||||
| (Electrophorus electricus) | Electrolectin | Lactose and so forth | Galectin | [44] |
| Conger eel | ||||
| (Conger myriaster) | Congerin I | Lactose and so forth | Galectin | [45, 46] |
| Congerin II | Lactose and so forth | Galectin | [45, 47] | |
| Congerin P | Lactose/mannose | Galectin | unpublished | |
| conCL-s | mannose | C-type | [48] | |
| Windowpane flounder | ||||
| (Lophopsetta maculate) | d-Galactose | Galectin | [49] | |
| Zebrafish | ||||
| (Danio rerio) | Drgal1-L1–L3 | LacNAc | Galectin | [50] |
| Shishamo smelt | ||||
| (Osmerus lanceolatus) | OLABL | d-Galactose | C-type | [51] |
| OLL | l-Rhamnose | RBL | [52] | |
| Catfish | ||||
| (Arius thalassinus) | d-Galactose | Galectin | [53] | |
| (Silurus asotus) | SAL | l-Rhamnose | RBL | [54] |
| saIntL | d-Mannose | intelectin | [55] | |
| Steelhead trout | ||||
| (Oncorhynchus mykiss) | STL-1–3 | l-Rhamnose | RBL | [56, 57] |
| Chum salmon | ||||
| (Oncorhynchus keta) | CSL-1–3 | l-Rhamnose | RBL | [58] |
| White-spotted charr | ||||
| (Salvelinus leucomaenis) | WCL-1, 3 | l-Rhamnose | RBL | [59] |
| Spanish mackerel | ||||
| (Scomberomorus niphonius) | SML | l-Rhamnose | RBL | [60] |
| Sweet fish (ayu) | ||||
| (Plecoglossus altivelis) | SFL | l-Rhamnose | RBL | [61] |
| Ponyfish | ||||
| (Leiognathus nuchalis) | PFL-1,2 | l-Rhamnose | RBL | [62] |
| Far-East dace | ||||
| (Tribolodon brandti) | TBL-1–3 | l-Rhamnose | RBL | [63] |
| Striped bass | ||||
| (Morone saxatilis) | Fucose | F-type | [64] | |
| Sea bass | ||||
| (Dicentrarchus labrax) | DlFBL | Fucose | F-type | [65] |
| Japanese sea perch | ||||
| (Lateolabrax japonicus) | JspFL | Fucose | F-type | [66] |
| Pufferfish | ||||
| (Fugu rubripes) | Pufflectin | d-Mannose | Lily-type | [67] |
| Scorpionfish | ||||
| (Scorpaena plumieri) | plumieribetin | d-Mannose and so forth | Lily-type | [68] |
| Carp | ||||
| (Cyprinus carpio) | carpFEL | GlcNAc | 6x β-propeller/Tectonin | [69] |
AAA: Anguilla anguilla agglutinin; AJL: Anguilla japonica lectin; BRA: Megabalanus rosa lectin; BRL: Balanus rostratus lectin; CEL: Cucumaria echinata lectin; CSL: chum salmon lectin; GCLT1: Geodia cydonium lectin; OLABL: Osmerus lanceolatus asialofetuin-binding lectin; OLL: Osmerus lanceolatus lectin; PFL: ponyfish lectin; PLC: perlucin; PPL: Pteria penguin lectin; SAL: Silurus asotus lectin; SFL: sweet fish (ayu) lectin; SJL: Stichopus japonicus lectin; SML: Spanish mackerel lectin; SPL: sea cucumber plasma lectin; STL: Steelhead trout lectin; SUEL: sea urchin egg lectin; TBL: Tribolodon brandti lectin; WCL: white spotted charr lectin; KDO: 2-keto-3-deoxyoctonate; LPS: lipopolysaccharide; LTA: lipoteichoic acid; GalNAc: N-acetyl galactosamine; GlcNAc: N-acetyl glucosamine; LacNAc: N-acetyl lactosamine.
2. C-Type Lectin Family
C-type lectins (CTLs) are one of major animal lectin family, of which members bind in a Ca2+-dependent fashion to mono- and oligosaccharides. They adopt generally multi-domain structures and contain one or more highly conserved CRD consisting of 115–130 amino acid residues [72], which has a unique mixed α~β topology [73]. In the presence of Ca2+, CTLs initiate a broad range of biological processes such as adhesion, endocytosis, and pathogen neutralization [74]. C-type lectin domain (CTLD) superfamily is a large group of extracellular proteins with conserved CRD sequences but different function including more than a thousand identified members, most of which lacking lectin activity. Since CTLD superfamily has been first classified into 7 groups (I to VII) by Drickamer in 1993 [75]; the classification of CTLD was revised in 2002 with additional seven groups (VIII to XIV) by Drickamer and Fadden [76]; and in 2005 with three new groups (XV to XVII) by Zelensky and Gready [77]. Group I contains four members, versican, aggrecan, neurocan, and brevican, which contain a proteoglycan core peptide and a single CTLD in the vicinity of the C-terminus and for which the name “lectican” has been proposed [78]. Group II CTLs have a single transmembrane domain, an extracellular carboxyl terminus, and a cytoplasmic amino terminus; typical examples are the hepatocyte asialoglycoprotein receptor (ASGR) [79], dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN), and macrophage receptors. Group III CTLs are collectins, which consist of an amino terminal collagen domain and a carboxyl terminal CTLD. They participate in the host defense mechanism through complement activation and include serum mannose binding protein (MBP) and pulmonary surfactant proteins [80]. Group IV CTLs, or selectins, are involved in the adhesive interaction between leukocytes and vascular endothelial cells such as L-selectin (leucocytes), E-selectin (endothelial cells), and P-selectin (platelets) [81]. Group V CTLs are involved in signal transduction; typical examples are the natural killer cell receptors [82] and the low-affinity IgE receptor (CD23) [83]. Group VI CTLs are type I transmembrane proteins consisting of a cysteine-rich domain, a fibronectin type II domain, and tandem CRDs. This group includes the macrophage cell surface mannose receptor and a dendritic cell surface molecule DEC-205 [84]. Group VII includes soluble single-CRD proteins. This group includes the pancreatic stone protein (PSP) isolated from pancreatic stones, lithostathine, which considered an inhibitor of calcite crystal growth [85–87]. Interestingly, CTLs isolated from marine invertebrates such as acorn barnacle hemolymph [9, 88] and pearl shells [38] have also been reported to have unique multiple functions in their biomineralization, that is, the inhibitory activity toward the crystal growth of calcium carbonate based on their high association constants to calcium ions. Furthermore, fish-specific CTLD proteins have been identified as the type II antifreeze proteins (AFPs), which inhibit the freezing by binding to ice in plasma of cold-water-living fish species including herring (Clupea harengus), rainbow smelt (Osmerus mordax), Japanese smelt (Hypomesus nipponensis), sea raven (Hemitripterus americanus), and longsnout poacher (Brachyopsis rostratus) [89–91].
Zelensky and Gready (2004) have reported the genome-level analysis on CTLD superfamily in Fugu rubripes genome that demonstrate the divergence of all CTLD superfamily proteins except for two groups V and VII present in mammals [92]. They also identified fish-specific CTLDs, AFPs, and dual-CTLD proteins, in Fugu genome. CTLs are diversified and widely distributed in animal kingdom including fishes as unique structural motifs and functional domains, regardless of whether they possess the sugar-binding properties or not.
3. Galectin Family
Galectins are family of carbohydrate-binding proteins defined by their Ca2+-independent affinity for β-galactoside sugar, sharing a conserved sequence motif within their CRD of about 130 amino acid residues, the lack of a signal peptide and attached carbohydrate, and predominantly cytoplasmic location [99]. Based on the structural features, galectins can be classified into three types: prototype (monomer or homodimer of single carbohydrate-binding domain), tandem-repeat type (two carbohydrate-binding domains on a single chain), and chimera type (carbohydrate-binding domain and an extra N-terminal domain on a single chain) (Figure 1). Galectins have proposed to participate in diverse physiological phenomena such as development, differentiation, morphogenesis, immunity, apoptosis, metastasis of malignant cell, and so forth. To date, several galectins were identified from fishes including Japanese eel Anguilla japonica [40], electric eel Electrophorus electricus [44], channel catfish Ictalurus punctuates [53], windowpane flounder Lophopsetta maculate [49], zebrafish Danio rerio [50], and conger eel Conger myriaster [45–47].
Figure 1.
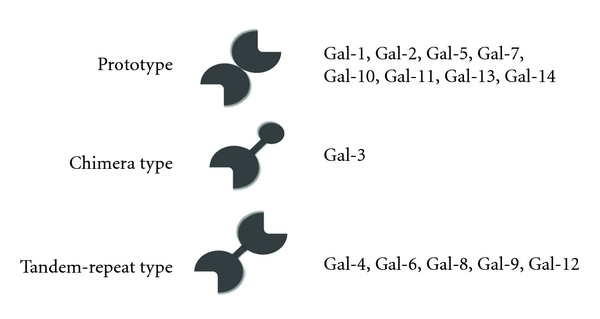
Classification of galectins.
Conger eel contains two prototype galectins, congerins I (Con I) and II (Con II). They are components of the biological defense system: these proteins mainly exist in the frontier organs and tissues that delineate the body from the outer environment, such as the epidermal club cells of the skin, wall of the oral cavity, pharynx, esophagus, and gills [46, 100], and agglutinate marine pathogen bacteria such as Vibrio anguillarum [101], and have opsonic and cytotoxic activities against cells [102–104]. It was reported that other fish galectin isolated from Japanese eel, AJL-1, also showed agglutinating activity against pathogenic Gram-positive bacteria, S. difficile [40]. Con I and Con II consist of 136 and 135 amino acid residues, respectively, and both contain several common structural characteristics: acetylated N-termini and no cysteine residue that is related to oxidizing inactivation found in mammalian galectins. Each subunit of Con I and Con II has one carbohydrate-binding site, and they form a homodimer to exhibit divalent crosslinking activity. Since a gene duplication event, these genes have been evolving at a high dN/dS (nonsynonymous/synonymous substitution) rate (2.6) under selection pressure, causing amino acid changes in Con I and Con II [46]. Usually, nonsynonymous substitutions in coding regions are restrained, by purifying selection, to maintain protein structure or function, whereas synonymous substitutions and nucleotide substitution for noncoding regions accumulate constantly by random genetic drift. Therefore, dN/dS ratios are normally smaller than 1.0 (∼0.2 in various genes). In other words, Con I and Con II have evolved via accelerated amino acid substitutions under positive selection. Similar evolutionary behavior has been observed only in several gene families including that of biological offense and defense systems such as snake venom isozymes and conus peptides and reproduction systems [105]. The structure of Con I demonstrated a protein-fold evolution by swapping β-strands between subunits, which altered the β-sheet topology at the dimer interface from entirely antiparallel to partially parallel to entangle two subunits, although Con I and Con II adopt the similar subunit structure with “jelly-roll” motif consisting of five-stranded and six-stranded β-sheets (Figure 2) [70, 71]. Domain swapping has been hypothesized as a mechanism in quaternary structure formation in protein evolution and reported several proteins [106, 107]. One of the important features of strand swapping seems to be increasing the stability of the quaternary structure, which is required for essential divalent cross-linking activity in agglutinating pathogenic bacteria (Figure 2). On the other hands, crystal structure analysis of Con II revealed that a MES (2-(N-morpholino) ethane sulfonic acid) molecule that corresponding to sulfonosugar bound to the cleft near the known CRD [71]. The similar extension of binding site toward the nonreducing end of lactose was observed in the fungal prototype galectins, ACG and CGL2 [108, 109]. Crystal structure of Con II complex with lacto-N-fucopentaose III at 2.2 Å resolution supported the extension of carbohydrate-binding site to the position where the MES was observed, indicating that the potent natural ligands of Con II include the additional moieties at the nonreducing end of lactose [110]. Actually, the differences in the thermostability and carbohydrate specificities between Con I and Con II were also observed [47]. Furthermore, to identify the determinants of selection pressures in the evolutionary process and the structural elements associated with the unique carbohydrate-binding activities of Con I and Con II, we recently reconstructed a probable ancestral form of congerin (Con-anc) that corresponded to the putative amino acid sequence at the divergence of Con I and Con II in the phylogenic tree [111, 112]. It was found that Con-anc showed the properties similar to those of Con II in terms of thermostability and carbohydrate-recognition specificity, although Con-anc shares the higher sequence similarity with Con I than Con II. From the differences between Con-anc and Con II on the sugar-binding specificities and mutation analysis of Con II, Con II has been considered to acquire the binding ability to the α2,3-sialyl galactose moieties such as GM3 and GD1a during the accelerated evolutionary event from Con-anc with the replacement of Arg3 and Tyr123 residues of Con II. The α2,3-sialyltransferases have been recently isolated and cloned from the pathogenic marine bacteria Vibrio sp. and Photobacterium phosphoreum [113, 114], suggesting that α2,3-sialyl galactose-containing sugars, which are presumed to be targets for Con II, may specifically be present in pathogenic marine bacteria. On the other hand, Con I has evolved from the ancestral congerin Con-anc to increase the binding activity against α1,4-fucosylated N-acetyl glucosamine [114]. These findings emphasize that the carbohydrate-binding ability and the specificities of congerins are diversified via accelerated evolution.
Figure 2.
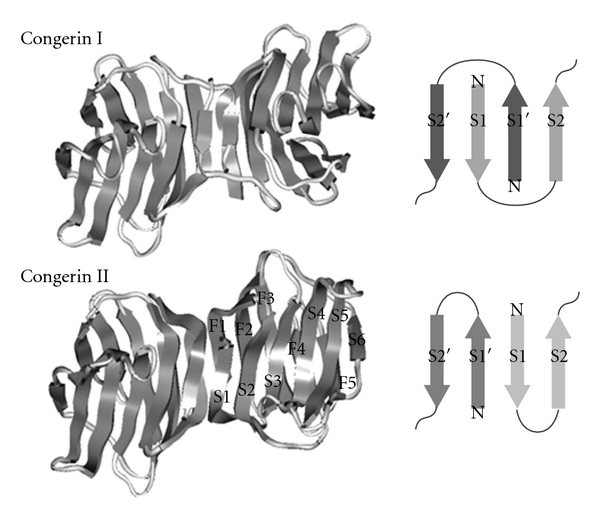
Schematic presentation of the wild-type congerin I (top: PDB code 1c1f) and congerin II (bottom: PDB code 1is5) and the topologies of β-sheet at subunit interface [70, 71].
Cooper (2002) reviewed the genome-wide screening of galectin gene families (galectinomics) based on the genomic sequence database including Arabidopsis, Drosophila, Caenorhabditis, Xenopus, human, and zebrafish Danio [115]. In the zebrafish genome, homologues of mammalian galectins-1, 3, 4, 9, and HSPC159 were reported. Table 2 summarized the results of searching for the homologues genes encoding galectins and related proteins in the eighth integrated assembly of the zebrafish genome, Zv8 (release 59, August 2010). In the zebrafish embryo, four proto-type galectin-1-like proteins, Drgal1-L1, Drgal1-L2, Drgal1-L3, and splice variant of Drgal1-L2, one chimera-type Drgal3, and two tandem-repeat galectins, Drgal9-L1 and Drgal9-L2, have been identified and characterized [50, 116]. They exhibited distinct phase-specific expression patterns during embryo development; for example, Drgal1-L1 is maternal; Drgal1-L2 is zygotic and expressed at postbud stage in notochord, while Drgal1-L3, Drgal3, Drgal9-L1, and Drgal9-L2 are both maternal and zygotic, and ubiquitously in adult tissues [50]. Furthermore, knock-down experiments in zebrafish embryo showed that Drgal1-L2 plays a key role in somatic cell differentiation through the skeletal muscle formation [117]. Zebrafish genes encoding proto-type galectins contain four exons with highly conserved exon-intron boundaries to that of mammalian galectin-1. Recently, the homologue of galectin-related interfiber protein (Grifin), which is a lens crystallin protein and one of galectin-related proteins (GRPs), has been also identified in zebrafish (DrGrifin) (Table 2), especially in the lens, particularly in the fiber cells of 2 days post fertilization embryos, and adult zebrafish tissues such as oocytes, brain, and intestine [118].
Table 2.
Zebrafish galectin-related genes.
| Length | Chromosome | Exons | Gene ID* | ||
|---|---|---|---|---|---|
| Transcripts (bp) | Protein (AA) | ||||
| Drgal1 (proto) | |||||
| lgals1 (Gal1-L1) | 883 | 134 | 3 | 4 | 326706 |
| lgals2a (Gal1-L2) | 850 | 134 | 3 | 4 | 405830 |
| lgals2b (Gal1-L3) | 545 | 118 | 6 | 4 | 393486 |
| si:ch211-10a23.2 | 1951 | 149 | 13 | 4 | 567812 |
|
| |||||
| Drgal3 (chimera) | |||||
| zgc112492 (Gal3) | 1993 | 561 | 3 | 5 | 550351 |
| lgals3l (Gal3-like) | 958 | 228 | 17 | 6 | 325599 |
| lgals3bpa (Gal3-like) | 2005 | 567 | 3 | 5 | 677742 |
| lgals3bpb (Gal3-like) | 1983 | 572 | 3 | 5 | 405809 |
| si:ch211-67f24.5 | 1925 | 369 | 3 | 5 | 125833577 |
|
| |||||
| Drgal4 (tandem) | |||||
| LOC567193 | 3981 | — | 18 | 25 | 567193 |
|
| |||||
| Drgal8 (tandem) | |||||
| LOC100334749 | 864 | 287 | 17 | 7 | 100334749 |
| si:ch211-199I3.2 | 537/402 | 153/133 | 20 | 9 | 368889 |
|
| |||||
| Drgal9 (tandem) | |||||
| zgc92326 (Gal9-L1) | 2591 | 321 | 15 | 11 | 327284 |
| lgals9l1 | 1075 | 310 | 15 | 10 | 337597 |
| LOC100148547 | 1017 | 282 | 15 | 7 | 100148547 |
| zgc171951 | 1453 | 280 | 15 | 7 | 100124603 |
|
| |||||
| Others | |||||
| Grifin (zgc92897) | 684 | 139 | 3 | 4 | 445036 |
| GRP (zgc136758) | 3518 | 164 | 1 | 5 | 723995 |
| GRP (si:ch211-101n13.9) | 438 | 145 | 13 | 2 | 563573 |
| LOC100535066 | 3984 | 1327 | 18 | 29 | 100535066 |
| c7orf23 (zgc:112101) | 1011 | 154 | 18 | 3 | 550443 |
*Gene ID: ID numbers in NCBI's database for gene-specific information [98].
Furthermore, novel galectin-related protein named CvGal, which contains four canonical galectin CRDs, has been discovered from the hemocytes of the eastern oyster, Crassostrea virginica [119]. CvGal can recognize both endogenous and exogenous ligands including bacteria, algae, and Perkinsus sp. as a soluble opsonin for pathogens or as a hemocyte surface receptor for both microbial pathogens and algae food ingested into digestive ducts and as a modulator for up-regulation of CvGal itself [119]. Unique domain architecture for genes/proteins consist of galectin CRDs, nematogalectin, was also found in freshwater hydrozoan Hydra and marine hydrozoan Clytia [120]. Nematogalectin, a 28 kDa protein with an N-terminal GlyXY domain that can form a collagen triple helix followed by galectin CRD, is a major component of the nematocyst tubule and is transcribed by nematocyte-specific alternative splicing [120]. Thus, the galectin family proteins also diversified by unique evolutionary process including tandem duplication and accelerated evolution.
4. F-Type Lectin Family
F-type lectins (fucolectin), which bind fucose and share characteristic sequence motif, have been identified as immuno-recognition molecules in invertebrates and vertebrates such as horseshoe crab (Tachypleus tridentatus) [23] and Japanese eel (Anguilla japonica) [43]. The crystal structures of single CRD and tandem CRDs of F-type lectins with a jellyroll β-barrel topology have been reported for Anguilla japonica agglutinin (AAA) and MsaFBP32 from striped bass (Morone saxatilis), respectively, [121, 122]. Bianchet et al. described that the fold structure of AAA, F-type lectin motifs, is widely distributed in other proteins even with lower sequence similarities, for example, C1 and C2 repeats of blood coagulation factor V, C-terminal domain of sialidase, N-terminal domain of galactose oxidase, APC10/DOC1 ubiquitin ligase and XRCC1 [121]. Furthermore, it has been reported that the several proteins are homologous to or contained with F-type lectin CRDs, of which examples include Streptococcus pneumoniae TIGR4, furrowed receptor and CG9095 of Drosophila melanogaster, Xenopus laevis pentraxin 1 fusion protein, Microbulbifer degradans ZP_00065873.1, and yeast allantoises [121, 123] in addition to the tandem-repeated types of F-type lectins found in modern teleosts [64–66, 122], while F-type lectin CRD motifs are absent in genomes of higher vertebrates such as reptiles, birds, and mammals.
5. Rhamnose-Binding Lectin Family
The rhamnose-binding lectins (RBLs) are a family of animal lectins that show the specific binding activities to l-rhamnose or d-galactose and mainly isolated from eggs and ovary cells of fishes and invertebrates [39, 56, 57, 126]. Sea urchin egg lectin (SUEL) is the first example of isolated and sequenced RBL family [11]. SUEL forms a homodimer composed of two identical subunits, which consist of 105 amino acid residues including single CRD, via intersubunit disulfide bond, resulting in the hemagglutinating activity with bivalent binding properties. To date, the RBL family has been found in over 20 species of fish, which located specifically in oocytes, ovaries, and skin mucus [52, 54, 56–63]. RBLs have been also found in the mantle of penguin wing oyster [39], and ascidians [32, 33]. Except for the reproductive cells including oocyte and egg, RBLs are mainly located in the tissue related to the immune system such as mucous cells of gill, goblet cells of intestine, spleen, thrombocyte, lymphocyte, monocyte, and neutrophil [127, 128]. Moreover, RBLs were isolated from spores of the microsporidian fish parasite, Loma salmonae, which was located in gill tissue [129] and Glugea plecoglossi from ayu eggs [61], respectively. Thus, it is possible that RBLs participate in the self-defense mechanisms. In fact, the receptor of RBL from amago (Oncorhynchus rhodurus) was expressed on the peritoneal macrophage after inflammatory stimulation [130], and RBLs from grass carp (Ctenopharyngodon idellus) roe induced a dose-dependent increase in phagocytic activity of seabream macrophage [131].
6. Structural Characterization of RBLs: Primary Structures and Classification
Three RBLs, named CSL1, CSL2, and CSL3, have been isolated as rhamnose-binding lectins from chum salmon (Oncorhynchus keta) eggs [41]. The amino acid sequences among CSLs show the 42–52% identities, while CSLs show the 94 to 97% sequence identities compared to corresponding three RBLs, STL1, STL2, and STL3, from steelhead trout (Oncorhynchus mykiss) eggs, respectively. Moreover, CSL1, CSL2 and CSL3 are composed of 4, 18, and 2 subunits via noncovalent binding, respectively.
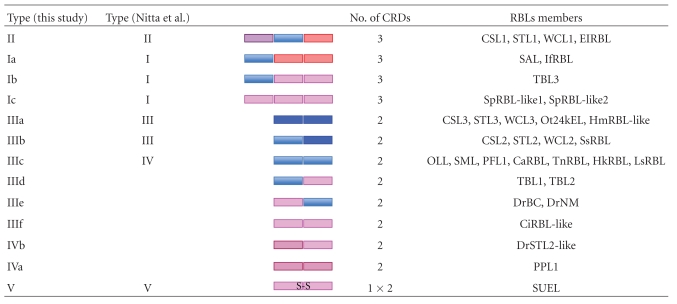
Most RBLs are composed of two or three tandem-repeated CRDs, which consist of about 95 amino acid residues, and share the conserved topology of four disulfide bonds. Figure 3 shows the aligned amino acid sequences of various RBL-CRDs. The disulfide bond pairings of RBLs have been determined for Spanish mackerel lectin (SML) by protein sequencing combined with peptide mapping [59] and for SEL24K from Chinook salmon by matrix-assisted laser desorption/ionization (MALDI) mass-spectrometry [132], respectively. Each RBL-CRD had the same disulfide bonding patterns: Cys(1)–Cys(3), Cys(2)–Cys(8), Cys(4)–Cys(7), and Cys(5)-Cys(6) (Figure 3). Furthermore, two characteristic peptide motifs, -(AN)YGR(TD)-(YGR-motif) and -DPCX(G)T(Y)KY(L)-(DPC-motif), which are located at the N- and C-terminal regions in each domain, respectively, are conserved in almost RBL-CRDs. However, the structural variations for S–S bonds and motifs are observed in PPL and N-terminal CRDs of CSL1, STL1, WCL1, and ElRBL (Figure 3). Previously, RBLs have been classified into five groups (Types I to V) based on their domain structures and the hemagglutination activity against human erythrocytes and sugar specificity against lactose (Table 3) [133]. Type I is composed of three tandemly repeated domains. Type II has two tandem-repeated domains with an extra domain. Types III and IV have two tandem-repeated domains, but they have different hemagglutination activity and sugar specificity. Type V has only one RBL domain and exits in a homodimer with a disulfide linkage between subunits. On the other hand, the phylogenetic tree constructed from the amino acid sequences of CRDs derived from several RBLs revealed that RBL-CRD can be classified into seven groups, RBL-CRD1 to RBL-CRD7, as shown in Figure 4. Based on their structural features of RBL-CRDs compositions, RBLs can be classified into 13 subgroups (Ia to V) (Table 3). The subunit of CSL1 is composed of 286 amino acid residues with three tandemly repeated domains (Type II), while the subunits of CSL2 and CSL3 are composed of 195 amino acid residues with two tandem-repeated domains (Type III).
Figure 3.
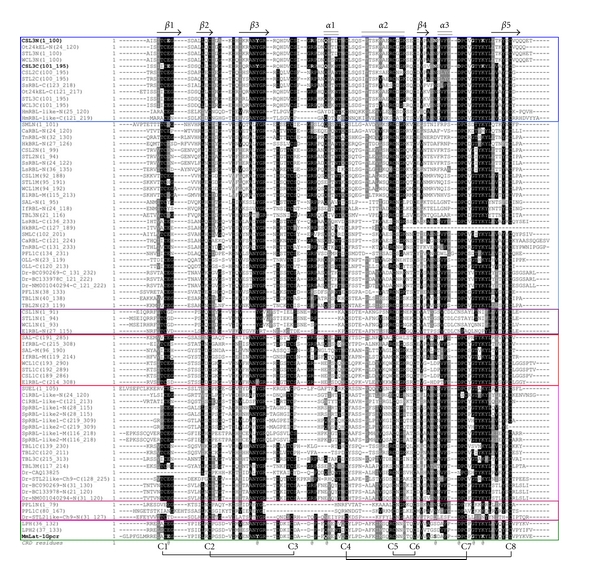
Aligned amino acid sequences of RBL-CRDs. Multiple alignment was achieved using the CLUSTAL X program. Secondary structural elements are shown as cylinders (α helix, α1–α3) and arrows (β strand, β1–β5). Disulfide-bond paring of eight half-cystine residues (C1–C8) is indicated at the bottom. Bold-faced amino acids indicate the residues involved in the carbohydrate binding. The sequence data were obtained from the Entrez protein sequence database including SwissProt, PIR, PRF, and PDB. The CRDs located in the N-terminal, middle, and C-terminal regions are represented by N, M, and C, respectively. CSL: chum salmon (Oncorhynchus keta) egg lectin, OLL: shishamo smelt (Osmerus lanceolatus) egg lectin, PFL: ponyfish (Leiognathus nuchalis) egg lectin, PPL: penguin wing oyster (Pteria penguin) lectin, SAL: catfish (Silurus asotus) egg lectin, SFL: ayu (Pleacoglossus altivelis) egg lectin, SML: Spanish mackerel (Scomberomorous niphonius) egg lectin, STL: steelhead trout (Oncorhynchus mykiss) egg lectin, SUEL: sea urchin (Anthocidaris crassispina) egg lectin, TBL: far east dace (Tribolodon brandti) egg lectin, WCL: white spotted charr (Salvelinus leucomaenis) egg lectin. Bs: Botryllus schlosseri, Ca: snakehead (Channa argus), Ci: Ciona intestinalis, Dr: zebrafish (Danio rerio), El: northern pike (Esox lucius), Hk: spotted seahorse (Hippocampus kuda), Hm: Hydra magnipapillata, If: blue catfish (Ictalurus furcatus), Ls: humphead snapper (Lutjanus sanguineus), Mm: house mouse (Mus musculus), Ot: small-eared galago (Otolemur garnettii), Sp: Strongylocentrotus purpuratus, Ss: Atlantic salmon (Salmo salar), Tn: green pufferfish (Tetraodon nigroviridis).
Table 3.
Classification of RBL family lectins.
|
Figure 4.
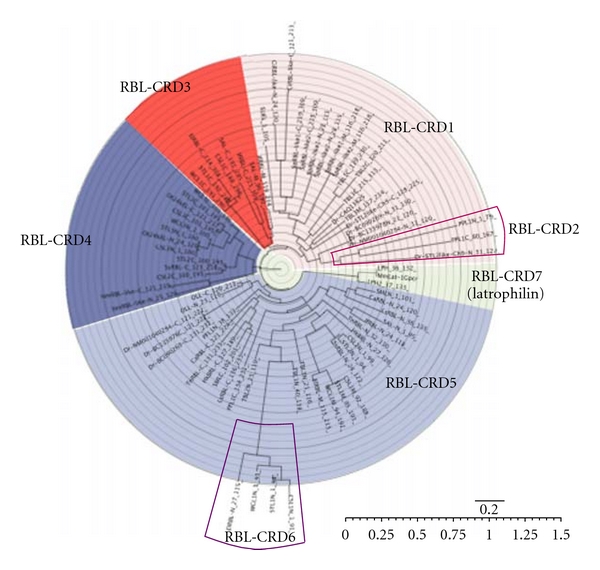
Phylogenetic tree of CRDs of RBL family lectins. A phylogenetic tree was constructed by the neighbor-joining algorithm based on an evolutionary distance matrix constructed by Kimura's method.
Furthermore, recent studies including genome-wide screening revealed several variations in the RBL families. It was found that the genes containing the distinctive structural motif of RBL-CRDs broadly distributed in almost all the animals including invertebrate (Hydra magnipapillata, Hydractinia echinata, Strongylocentrotus purpuratus, Nematostella vectensis, Caenorhabditis remanei, Triatoma dimidiata), Chordates (Ciona intestinalis, Botryllus schlosseri, Branchiostoma floridae), and vertebrate including bony fish such as Danio rerio, Oncorhynchus mykiss, and so forth, amphibian Xenopus tropicalis and mammalians such as Mus musculus, Rattus norvegicus, Homo sapiens, and so forth, and also in the bacterium (Flavobacterium) and plants (Arabidopsis thaliana, Medicago truncatula) when the genomic database was retrieved. For example, the RBL homologues have been reported as integrated domains involved in the ligand binding in membrane receptors such as polycystic kidney disdase-1-like (PKD-1) [134], axon guidance receptor EVA-1 [135], HuC21orf63 [136], and the adhesion-class G-protein-coupled receptor latrophilin (LPHN) [137, 138]. Figure 5 summarized the domain architectures of RBL superfamily proteins that contained RBL-CRDs in their sequences as a domain structure. (TSP1: thrombospondin-type 1 repeats, OLF: olfactomedin-like domain, HormR: hormone receptor domain, GPS: G-protein-coupled receptor proteolytic site domain, 7tm_2: 7 transmembrane receptor, latrophili: latrophilin cytoplasmic C-terminal region, CTLTD: C-type lectin domain. PLAT: polycystic kidney disease protein-1-like 2, PKD_channel: polycystin cation channel, FAS8C: coagulation factor 5/8 C-Terminal domain, discoidin domain, LCCL: limulus-clotting factor C, Coch-5b2, and Lgl1-lectin domain, IPPc: inositol polyphosphate phosphatase, catalytic domain homologues, RhoGAP_OCR: GTPase-activator protein for Rho-like Small GTPases in oculocerebrorenal syndrome of Lowe-1-like protein, Prp1: proline-rich protein 1, PurA: adenylosuccinate synthase, AMN1: antagonist of mitotic exit network protein 1, VWD: von Willebrand factor domain, NHL: Ncl-1, HT2A, and Lin-41 proteins, PAN module: plasminogen/hepatocyte growth factor-Apple domains of the plasma prekallikrein/coagulation factor XI-Nematode proteins module, *1: AAH90269/AAI22308/50383/CAM56745/56747/CAX13501/NP_001035384/001038891/XP_692814/001922851/003199967/003200629/706941/003200654/003200655, *2: AAI22302/50374/51864/51941/54461/54558/55629/62646/62650/CAK11496-11501/11504/11506/11509/11515/CAM56419-56422/56424/56426/56637/56466/56467/56470-56473CAP09587/09516/CAQ13825/14198-14199/NP_001038882/001082844/001082910/001082869/001093874/001093910/001096104/001098618/001103190/001103311/001103315/001103334/001103858/001104195/001108359/001103581/001103589/001095862/001103856/001138280/001128340/001153845/002663370/002663371/002666350/XP_003199229/003199230/002663369/692138/003200629/706941/003200654-655/003200654-655/003201119/003201121/003201123/003201136/002666347/003201138-39/002667144/002666349/001333550. Beside the RBLs (Types I–V), several groups are classifiable as an RBL superfamily: the immune recognition molecules, rhamnospondins (Rsps), LPHNs, Arabidopsis galactosidases, and others as shown in Figure 5. Rsp gene has been identified in colonial hydroid Hydractinia symbiolongicarpus and was found to encode a secreted modular protein of 726 amino acids composed of N-terminal serine-rich domain, eight tandem-repeated thrombospondin type 1 repeats (TSRs), and C-terminal RBL-CRD [139]. Rsps have diversified by gene duplication and predicted to act as immune recognition molecules from the evidence of gene structure and their expression profiles in the polyp's hypostome, which face to the external environment and pathogen. On the other hand, Caenorhabditis elegans EVA-1 forms a complex with SAX-3/roundabout (Robo) receptor and functions as a coreceptor for shiga-like toxin 1 (SLT-1)/slit proteins in guiding cell and axon migrations [135]. Furthermore, human C21orf63, an EVA-1 ortholog, has been identified from the Down's syndrome project [136] and reported to have specific affinity to heparin.
Figure 5.
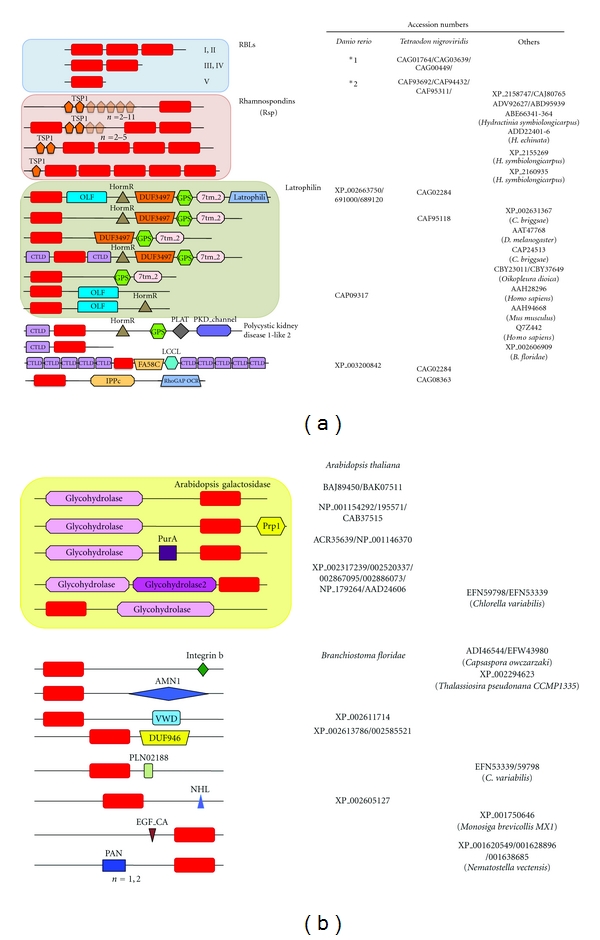
Domain architecture of RBL superfamily. The proteins with similar domain architectures have been identified using the Conserved Domain Architecture Retrieval Tool (CDART) [124].
LPHNs are synaptic Ca2+-independent α-latrotoxin (LTX) receptor, a novel member of the secretin family of G-protein-coupled receptors containing seven transmembrane regions as well as long N-terminal extracellular sequences containing a 19-amino acid signal peptide, and a serine/threonine-rich glycosylation region (Figure 5) [137]. LTX is a component of the venom of the black widow spider (latrodectus mactans) and stimulates exocytosis of γ-aminobutyric acid- (GABA-) containing presynaptic vesicles via interaction with LPHN. Recently, LPHN3, which is the most brain-specific LPHN, has been reported to be involved in the pathogenesis of attention-deficit/hyperactivity disorder [138]. Thus, the RBL-CRDs are diversified and widely distributed in the functional proteins as unique structural motifs. Similar examples for the domain architecture of proteins including membrane receptors have been reported in other lectin families such as CTLD [74–79] and F-type lectin superfamilies [122, 123].
7. Gene Structure of RBL Family
More recently, cloning and characterization of a gene for snakehead lectin (SHL) from Channa argus and its promoter region have been reported (Genebank accession nos.: EU693900) [140]. SHL gene, which consists of 2,382 bp from the transcription initiation site to the end of 3′ untranslated region (UTR) and includes two tandem RBL-CRDs with 35% identity, contains nine exons and eight introns. The first 40 bp of exon 1 is 5′UTR, and the signal peptide is encoded by exons 1 and 2. The N-terminal CRD is encoded by exons 3, 4, and 5, and C-terminal CRD is encoded by exons 6, 7, and 8. Exon 9 includes the C-terminal region of SHL and 3′UTR. These suggest that RBL-CRDs are located in three exons; respectively, and RBL may be diverged and evolved by gene duplication and/or exon shuffling. The 5′ flanking regions contained some unique consensus sequence for the nuclear factor of interleukin 6 (NF-IL6) and IFN-γ activation sites.
On the other hand, Rsp gene was predicted to encode a secreted protein of 726 amino acids composed of a signal peptide, an N-terminal serine-rich domain (SRD), eight TSRs, and a RBL-CRD at C-terminal region, consisting of 13 exons and 12 introns [139, 141]. However, RBL-CRD of Rsp was located in only single exon (Exon 12), suggesting that the gene structure of Rsp RBL-CRD is different from that of SHL RBL-CRD. Whole genome sequences have been determined for several living organisms including marine organisms such as zebrafish, Danio rerio, of which data can be available and allow us to establish the full inventory of any particular gene family in the genome. Thus, searching the zebrafish database with RBL-CRD sequences revealed that the genes encoding RBLs with tandem-repeated CRDs (Types I to IV) were located in chromosomes 9, 19, and 21 (Table 4). Furthermore, it was found that several genes encoding single RBL-CRD proteins, almost all of which physiological functions are largely unknown, are located in chromosomes 2, 9, 23, and especially in chromosome 22 (Table 4), although the genes for latrophilins and C21orf63 homolog were located in chromosomes 22 and 14, respectively.
Table 4.
Zebrafish RBL-related genes.
| Length | Chromosome | Exons | Gene ID* | ||
|---|---|---|---|---|---|
| Transcripts (bp) | Protein (AA) | ||||
| Type I/II (3CRDs) | |||||
| LOC100536087 | 1063 | 274 | 19 | 10 | 100536087 |
| LOC555549 | 1144 | 297 | 19 | 10 | 555549 |
|
| |||||
| Type III/IV (2CRDs) | |||||
| Zgc136410 | 878 | 222 | 21 | 9 | 449685 |
| LOC100536640 | 978 | 233 | 19 | 9 | 100536640 |
| LOC100535118 | 715/868 | 167/209 | 9 | 8/9 | 100535118 |
| LOC100536038 | 1050 | 272 | 19 | 9 | 100536038 |
| LOC100536135 | — (pseudo) | — | 19 | 9 | 798039 |
|
| |||||
| 1CRD | |||||
| LOC100333730 | 654/618 | 119/137 | 9 | 6 | 100333730 |
| LOC100301575 | 665 | 128 | 22 | 6 | 100301575 |
| LOC798111 | 668 | 128 | 22 | 6 | 798111 |
| LOC100535660 | 469 | 129 | 22 | 7 | 100535660 |
| MGC171851 | 677 | 128 | 22 | 6 | 798039 |
| LOC100331690 | 815 | 186 | 22 | 7 | 100331690 |
| LOC100537146 | 837 | 188 | 22 | 7 | 100537146 |
| LOC100537102 | 804 | 179 | 22 | 7 | 100537102 |
| LOC100536958 | 821 | 188 | 22 | 7 | 100536958 |
| LOC794480 | 750 | 137 | 23 | 9 | 794480 |
|
| |||||
| Others | |||||
| Latrophilin-3-like | 609 | 131 | 22 | 6 | 445036 |
| Latrophilin 2 | 669 | 128 | 22 | 7 | 94733430 |
| DKEYP-98A7.10 | 620 | 168 | 22 | 7 | 100126138 |
| LOC792919 | 652 | 190 | 2 | 6 | 792919 |
| C21orf63 homolog | 1189 | 380 | 14 | 7 | 100537109 |
*Gene ID: ID numbers in NCBI's database for gene-specific information [98].
8. Sugar-Binding Specificities and Physiological Functions of RBLs
Sugar-binding specificities of CSLs were investigated thoroughly by frontal affinity chromatography (FAC) using 100 kinds of sugar chains including N-linked and glycolipid-type glycans [142]. Interestingly, all of CSL1, CSL2, and CSL3 showed the high specific binding activity against globotriaosyl ceramide (Gb3; Galα1-4Galβ1-4Galβ1-Cer also known as CD77), which is located in lipid raft and upregulated through immune responses [143] and is also known as the functional receptor for various toxins such as Shiga toxin (Stx) [144], regardless of their low sequence homologies (42–52%) and different oligomeric structures.
CSLs induced proinflammatory cytokines, including IL-1β1, IL-1 β2, TNF-α1, TNF-α2, and IL-8, by recognizing Gb3 on the surface of the peritoneal macrophage cell line (RTM5) from rainbow trout and an established fibroblastic-like cell line (RTG-2) from gonadal tissue of the fish [142]. RBL from catfish, SAL, has also induced the alterations of gene expression in Burkitt's lymphoma cells [145]. Furthermore, CSLs showed the cytotoxicity against Gb3-displaying Caco-2 and Lovo cells via an apoptotic pathway through the recognizing of Gb3 on the cell surfaces in a dose-dependent manner, while it was not observed with DLD-1 and HCT-15 human colonic tumor cell lines lacking Gb3 [125].
RBLs from fish eggs such as STLs and CSLs also interacted and agglutinate Gram-negative and Gram-positive bacteria by recognizing the cell-surface lipopolysaccharides and lipoteichoic acid, respectively, [17, 146]. On the other hand, PPL, an RBL from Pteria penguin pearl shell, also showed the strong agglutinating activity against some Gram-negative bacteria such as Escherichia coli by recognizing lipopolysaccharides in the presence of the high concentration (500 mM) of NaCl, in which condition the oligomerization of PPL was induced, although its carbohydrate-binding specificity was quite different from that of CSLs; PPL binds to d-galactose but not l-rhamnose [39]. RBLs can recognize the O-antigen, which is the immunodominant structure exposed to the environment and is highly variable among bacterial strains, via diverse carbohydrate recognition ability. RBLs bind to glycolipids and glycoproteins of the microsporidian fish pathogens [61, 129], and the RBL receptor was expressed on peritoneal macrophages of fishes after an inflammatory stimulation [130, 147]. More recently, it was found that CSLs induced the production of radical oxygen species (ROS) in RTM5 cells in a dose-dependent manner. This effect was not inhibited by l-rhamnose or dl-threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP), an inhibitor of glucosyl ceramide synthesis, suggesting that carbohydrate recognition domains of CSLs were not involved in the respiratory burst of RTM5 cells.
Thus, CSLs are multifunctional lectins through binding to the carbohydrate such as Gb3 on cells and l-rhamnose/d-galactose residue of O-antigen of lipopolysaccharides. However, this carbohydrate-binding specificity of RBL leads to the interesting questions of how CSL can strongly recognize the different sugars such as Gb3, l-rhamnose, d-galactose, all of which common structural features are the orientation of anomeric hydroxy groups at C2 and C4.
9. Structural Characterization of RBL-CRDs
More recently, the highly ordered structure of CSL3 composed of two subunits of 20 kDa has been determined at 1.8 Å resolution [146]. The homodimer of CSL3 revealed a kinked dumbbell shape, in which two lobes are connected through linkers composed of two 5-residue peptides (-QQQET-) (Figure 6(a)). Each lobe seems to be a single globular protein with a pseudo-twofold axis and includes two antiparallel β sheets with two (β2 and β4) and three (β1, β3, and β5) strands and three helices (α1-3) (Figure 6(a)). The N- and C-terminal domains, both of which share 35% sequence identity with the RBL-CRD for mouse latrophilin-1 (LPHN-1), their folds were similar each other and superimposed on the RBL domain of LPHN-1, which has been recently reported [148], with rmsds of 1.4 and 1.5 Å for 94 Ca atoms, respectively. These RBL domains adopt a unique α/β fold with long structured loops involved in monosaccharide recognition.
Figure 6.
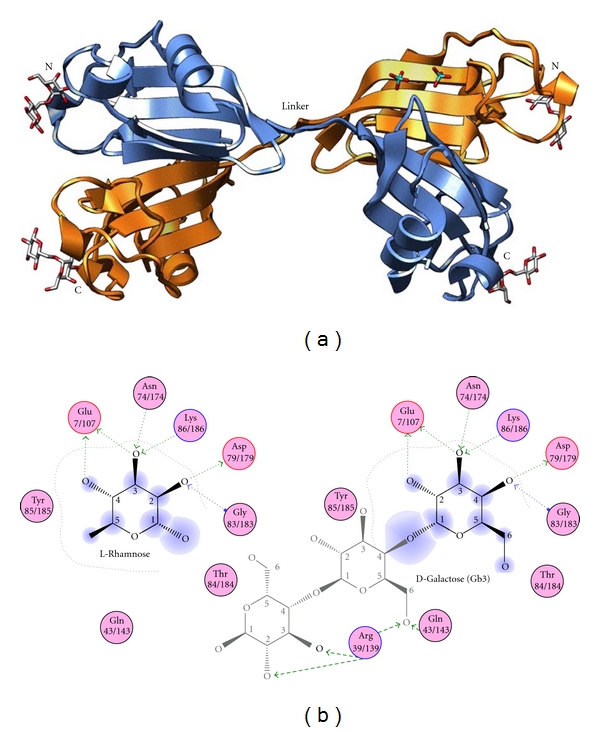
Crystal structure of CSL3. (a) Pseudotetrameric structure of CSL3 dimer (PDB code: 2z×3) [125]. Each carbohydrate-binding domain is differently colored. The bound melibioses and phosphates are shown in stick models. (b) Comparison of the carbohydrate-binding manners of CSL3 complex with l-rhamnose (left) and Gb3 (right).
It was found that the monosaccharide (rhamnose) or nonreducing end residues (Gal1 of melibiose and Gb3) share the same conserved primary binding sites in CSL3 (Glu7/107, Tyr27/127, Lys86/186, and Gly83/183) and LPHN-1 (Glu42, Tyr63, Lys120, and Gly117), respectively, (Figure 6(b)). Asn74/174 and Asp79/179 are additionally used in the primary site in CSL3. The melibiose and Gb3 complex structures revealed the oligosaccharide recognition mechanism of RBL. The Arg39/139 and Gln43/143 residues of CSL3, which bind to the carbohydrate in the 2nd and 3rd sites, are the key residues in determining specificity for oligosaccharides, Gb3. The total numbers of hydrogen bonds between CSL3 and rhamnose, melibiose, and Gb3 are 7, 8, and 10, respectively, which are consistent with the observed high affinity (K d = 2.6 × 10−5 M) of CSL3 to Gb3.
Interestingly, RBLs can bind to l-rhamnose and nonreducing d-galactose moiety of melibiose and Gb3 at the same binding site. These specific and characteristic binding abilities can be explained by the recognizing mechanisms involved in the hydrogen bonds between O2, O3, and O4 atoms of monosaccharide and the side chains of Glu7/107, Asn74/174, Asp79/179, Lys86/186, and the main chain of Gly83/183 of CSL3; that is, Glu7/107 forms hydrogen bond with O4 atom of l-rhamnose, which correspond to O2 of inverted d-galactose, while Gly83/183 forms hydrogen bond with O2 atom of l-rhamnose and O4 of inverted d-galactose form, respectively, (Figure 6(b)). These carbohydrate recognition mechanisms of lectins for the inverted carbohydrates were also found in the case for F-type lectins, which bind to both α-l-fructose and 3-O-methyl-d-galactose [122].
10. Perspectives
Since lectins isolated from marine resources are highly diversified in terms of not only structure but also functional aspects including specific and unique carbohydrate specificities as reviewed in this paper, they can be used for biomedical application as drug delivery system or diagnostic markers. For example, RBL family lectins are useful for diagnosis of pathological condition involved in Gb3 ceramide such as Burkitt's lymphoma having high malignancy. Furthermore, RBLs showed the physiological functions independent of carbohydrate-recognition ability such as ROS-inducing activities although its molecular mechanism has not yet been clarified. More recently, novel calcium-dependent mannose-binding lectin, intelectin, which is structurally identical to the intestinal receptor for lactoferrin and contained fibrinogen-related domain, has been identified from the skin mucus of catfish Silurus asotus [55]. Thus, there are a large and growing number of diversified lectins in marine resources. Further study will be necessary to elucidate the detailed structure-activity relationships of diversified marine lectins and to develop the potent therapeutic drugs.
Abbreviations
- AAA:
Anguilla japonica agglutinin
- AFPs:
Antifreeze proteins
- ASGR:
Asialoglycoprotein receptor
- CN-V:
Cyanovirin
- Con I:
Congerin I
- Con II:
Congerin II
- CRD:
Carbohydrate-recognition domain
- CSL:
Chum salmon lectin
- CTLs:
C-type lectins
- CTLD:
C-type lectin domain
- DC-SIGN:
Dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin
- EC50:
Half maximal effective concentration
- ES:
Embryonic stem
- FACT:
Frontal affinity chromatography technique
- GABA:
γ-Aminobutyric acid
- GRFT:
Griffithsin
- GRPs:
Galectin-related proteins
- HIV:
Human immunodeficiency virus
- iPS:
Induced pluripotent stem
- LPHN:
Latrophilin
- LTX:
α-Latrotoxin
- MALDI:
Matrix-assisted laser desorption/ionization
- MBP:
Mannose-binding protein
- MES:
2-(N-Morpholino) ethane sulfonic acid
- PKD-1:
Polycystic kidney disdase-1 like
- PPL:
Pteria penguin lectin
- PSP:
Pancreatic stone protein
- RBL:
Rhamnose-binding lectin
- Rsp:
Rhamnospondins
- SAL:
Silurus asotus lectin
- SHL:
Snakehead lectin
- SLT-1:
Shiga-like toxin 1
- SML:
Spanish mackerel lectin
- STL:
Steelhead trout lectin
- SUEL:
Sea urchin egg lectin
- TSRs:
Thrombospondin-type-1 repeats.
References
- 1.Vo TS, Kim SK. Potential anti-HIV agents from marine resources: an overview. Marine Drugs. 2010;8(12):2871–2892. doi: 10.3390/md8122871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith VJ, Desbois AP, Dyrynda EA. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Marine Drugs. 2010;8(4):1213–1262. doi: 10.3390/md8041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burja AM, Banaigs B, Abou-Mansour E, Grant Burgess J, Wright PC. Marine cyanobacteria—a prolific source of natural products. Tetrahedron. 2001;57(46):9347–9377. [Google Scholar]
- 4.Mori T, O’Keefe BR, Sowder RC, et al. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. Journal of Biological Chemistry. 2005;280(10):9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- 5.O’Keefe BR, Beutler JA, Cardellina JH, et al. Isolation and characterization of niphatevirin, it human-immunodeficiency-virus-inhibitor glycoprotein from the marine sponge Niphates erecta . European Journal of Biochemistry. 1997;245(1):47–53. doi: 10.1111/j.1432-1033.1997.t01-1-00047.x. [DOI] [PubMed] [Google Scholar]
- 6.Toyoda M, Yamazaki-Inoue M, Itakura Y, et al. Lectin microarray analysis of pluripotent and multipotent stem cells. Genes to Cells. 2011;16(1):1–11. doi: 10.1111/j.1365-2443.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- 7.Gabius HJ. Animal lectins. European Journal of Biochemistry. 1997;243(3):543–576. doi: 10.1111/j.1432-1033.1997.t01-1-00543.x. [DOI] [PubMed] [Google Scholar]
- 8.Muramoto K, Kamiya H. The amino-acid sequence of multiple lectins of the acorn barnacle Megabalanus rosa and its homology with animal lectins. Biochimica et Biophysica Acta. 1990;1039(1):42–51. doi: 10.1016/0167-4838(90)90224-4. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya H, Jimbo M, Yako H, et al. Participation of the C-type hemolymph lectin in mineralization of the acorn barnacle Megabalanus rosa . Marine Biology. 2002;140(6):1235–1240. [Google Scholar]
- 10.Muramoto K, Kamiya H. The amino-acid sequence of a lectin of the acorn barnacle Megabalanus rosa . Biochimica et Biophysica Acta. 1986;874(3):285–295. doi: 10.1016/0167-4838(90)90224-4. [DOI] [PubMed] [Google Scholar]
- 11.Ozeki Y, Matsui T, Suzuki M, Titani K. Amino acid sequence and molecular characterization of a D-galactoside-specific lectin purified from sea urchin (Anthocidaris crassispina) eggs. Biochemistry. 1991;30(9):2391–2394. doi: 10.1021/bi00223a014. [DOI] [PubMed] [Google Scholar]
- 12.Giga Y, Ikai A, Takahashi K. The complete amino acid sequence of echinoidin, a lectin from the coelomic fluid of the sea urchin Anthocidaris crassispina. Homologies with mammalian and insect lectins. Journal of Biological Chemistry. 1987;262(13):6197–6203. [PubMed] [Google Scholar]
- 13.Hatakeyama T, Kohzaki H, Nagatomo H, Yamasaki N. Purification and characterization of four Ca2+-dependent lectins from the marine invertebrate, Cucumaria echinata . Journal of Biochemistry. 1994;116(1):209–214. doi: 10.1093/oxfordjournals.jbchem.a124495. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama T, Matsuo N, Shiba K, et al. Amino acid sequence and carbohydrate-binding analysis of the N-acetyl-D-galactosamine-specific C-type lectin, CEL-I, from the Holothuroidea, Cucumaria echinata . Bioscience, Biotechnology and Biochemistry. 2002;66(1):157–163. doi: 10.1271/bbb.66.157. [DOI] [PubMed] [Google Scholar]
- 15.Hatakeyama T, Ohuchi K, Kuroki M, Yamasaki N. Amino acid sequence of a C-type lectin CEL-IV from the marine invertebrate Cucumaria echinata . Bioscience, Biotechnology and Biochemistry. 1995;59(7):1314–1317. doi: 10.1271/bbb.59.1314. [DOI] [PubMed] [Google Scholar]
- 16.Hatakeyama T, Nagatomo H, Yamasaki N. Interaction of the hemolytic lectin CEL-III from the marine invertebrate Cucumaria echinata with the erythrocyte membrane. Journal of Biological Chemistry. 1995;270(8):3560–3564. doi: 10.1074/jbc.270.8.3560. [DOI] [PubMed] [Google Scholar]
- 17.Matsui T, Ozeki Y, Suzuki M, Hino A, Titani K. Purification and characterization of two Ca2+-dependent lectins from coelomic plasma of sea cucumber, Stichopus japonicus . Journal of Biochemistry. 1994;116(5):1127–1133. doi: 10.1093/oxfordjournals.jbchem.a124638. [DOI] [PubMed] [Google Scholar]
- 18.Himeshima T, Hatakeyama T, Yamasaki N. Amino acid sequence of a lectin from the sea cucumber, Stichopus japonicus, and its structural relationship to the C-Type animal lectin family. Journal of Biochemistry. 1994;115(4):689–692. doi: 10.1093/oxfordjournals.jbchem.a124397. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, Kawabata SI, Hirata M, Iwanaga S. A novel type of limulus lectin-L6. Purification, primary structure, and antibacterial activity. Journal of Biological Chemistry. 1995;270(24):14493–14499. doi: 10.1074/jbc.270.24.14493. [DOI] [PubMed] [Google Scholar]
- 20.Shigenaga T, Takayenoki Y, Kawasaki S, et al. Separation of large and small granules from horseshoe crab (Tachypleus tridentatus) hemocytes and characterization of their components. Journal of Biochemistry. 1993;114(3):307–316. doi: 10.1093/oxfordjournals.jbchem.a124173. [DOI] [PubMed] [Google Scholar]
- 21.Okino N, Kawabata SI, Saito T, Hirata M, Takagi T, Iwanaga S. Purification, characterization, and cDNA cloning of a 27-kDa lectin (L10) from horseshoe crab hemocytes. Journal of Biological Chemistry. 1995;270(52):31008–31015. doi: 10.1074/jbc.270.52.31008. [DOI] [PubMed] [Google Scholar]
- 22.Beisel HG, Kawabata SI, Iwanaga S, Huber R, Bode W. Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus . The EMBO Journal. 1999;18(9):2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito T, Hatada M, Iwanaga S, Kawabata SI. A newly identified horseshoe crab lectin with binding specificity to O-antigen of bacterial lipopolysaccharides. Journal of Biological Chemistry. 1997;272(49):30703–30708. doi: 10.1074/jbc.272.49.30703. [DOI] [PubMed] [Google Scholar]
- 24.Inamori KI, Saito T, Iwaki D, et al. A newly identified horseshoe crab lectin with specificity for blood group A antigen recognizes specific O-antigens of bacterial lipopolysaccharides. Journal of Biological Chemistry. 1999;274(6):3272–3278. doi: 10.1074/jbc.274.6.3272. [DOI] [PubMed] [Google Scholar]
- 25.Gokudan S, Muta T, Tsuda R, et al. Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kairies N, Beisel HG, Fuentes-Prior P, et al. The 2.0-Å crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13519–13524. doi: 10.1073/pnas.201523798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai T, Kawabata SI, Shishikura F, Sugita H. Purification, characterization, and amino acid sequence of an embryonic lectin in perivitelline fluid of the horseshoe crab. Journal of Biological Chemistry. 1999;274(53):37673–37678. doi: 10.1074/jbc.274.53.37673. [DOI] [PubMed] [Google Scholar]
- 28.Chiou ST, Chen YW, Chen SC, Chao CF, Teh-Yung L. Isolation and characterization of proteins that bind to galactose, lipopolysaccharide of Escherichia coli, and protein A of Staphylococcus aureus from the hemolymph of Tachypleus tridentatus . Journal of Biological Chemistry. 2000;275(3):1630–1634. doi: 10.1074/jbc.275.3.1630. [DOI] [PubMed] [Google Scholar]
- 29.Kawabata SI, Iwanaga S. Role of lectins in the innate immunity of horseshoe crab. Developmental & Comparative Immunology. 1999;23(4-5):391–400. doi: 10.1016/s0145-305x(99)00019-1. [DOI] [PubMed] [Google Scholar]
- 30.Iwanaga S. The molecular basis of innate immunity in the horseshoe crab. Current Opinion in Immunology. 2002;14(1):87–95. doi: 10.1016/s0952-7915(01)00302-8. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Takagi T, Furukohri T, Kawamura K, Nakauchi M. A calcium-dependent galactose-binding lectin from the tunicate Polyandrocarpa misakiensis . Journal of Biological Chemistry. 1990;265(3):1274–1281. [PubMed] [Google Scholar]
- 32.Gasparini F, Franchi N, Spolaore B, Ballarin L. Novel rhamnose-binding lectins from the colonial ascidian Botryllus schlosseri. Developmental & Comparative Immunology. 2008;32(10):1177–1191. doi: 10.1016/j.dci.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Franchi N, Schiavon F, Carletto M, et al. Immune roles of a rhamnose-binding lectin in the colonial ascidian Botryllus schlosseri . Immunobiology. 2011;216(6):725–736. doi: 10.1016/j.imbio.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Sekine H, Kenjo A, Azumi K, et al. An ancient lectin-dependent complement system in an ascidian: novel lectin isolated from the plasma of the solitary ascidian, Halocynthia roretzi . Journal of Immunology. 2001;167(8):4504–4510. doi: 10.4049/jimmunol.167.8.4504. [DOI] [PubMed] [Google Scholar]
- 35.Kenjo A, Takahashi M, Matsushita M, et al. Cloning and characterization of novel ficolins from the solitary ascidian, Halocynthia roretzi . Journal of Biological Chemistry. 2001;276(23):19959–19965. doi: 10.1074/jbc.M011723200. [DOI] [PubMed] [Google Scholar]
- 36.Abe Y, Tokuda M, Ishimoto R, Azumi K, Yokosawa H. A unique primary structure, cDNA cloning and function of a galactose-specific lectin from ascidian plasma. European Journal of Biochemistry. 1999;261(1):33–39. doi: 10.1046/j.1432-1327.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- 37.Baldo BA, Sawyer WH, Stick RV, Uhlenbruck G. Purification and characterization of a galactan-reactive agglutinin from the clam Tridacna maxima (Röding) and a study of its combining site. Biochemical Journal. 1978;175(2):467–477. doi: 10.1042/bj1750467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss IM, Kaufmann S, Mann K, Fritz M. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata . Biochemical and Biophysical Research Communications. 2000;267(1):17–21. doi: 10.1006/bbrc.1999.1907. [DOI] [PubMed] [Google Scholar]
- 39.Naganuma T, Ogawa T, Hirabayashi J, Kasai K, Kamiya H, Muramoto K. Isolation, characterization and molecular evolution of a novel pearl shell lectin from a marine bivalve, Pteria penguin . Molecular Diversity. 2006;10(4):607–618. doi: 10.1007/s11030-006-9051-3. [DOI] [PubMed] [Google Scholar]
- 40.Tasumi S, Ohira T, Kawazoe I, Suetake H, Suzuki Y, Aida K. Primary structure and characteristics of a lectin from skin mucus of the Japanese eel Anguilla japonica . Journal of Biological Chemistry. 2002;277(30):27305–27311. doi: 10.1074/jbc.M202648200. [DOI] [PubMed] [Google Scholar]
- 41.Tasumi S, Yang WJ, Usami T, et al. Characteristics and primary structure of a galectin in the skin mucus of the Japanese eel, Anguilla japonica . Developmental & Comparative Immunology. 2004;28(4):325–335. doi: 10.1016/j.dci.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Mistry AC, Honda S, Hirose S. Structure, properties and enhanced expression of galactose-binding C-type lectins in mucous cells of gills from freshwater Japanese eels (Anguilla japonica) Biochemical Journal. 2001;360(1):107–115. doi: 10.1042/0264-6021:3600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda S, Kashiwagi M, Miyamoto K, Takei Y, Hirose S. Multiplicity, structures, and endocrine and exocrine natures of eel fucose-binding lectins. Journal of Biological Chemistry. 2000;275(42):33151–33157. doi: 10.1074/jbc.M002337200. [DOI] [PubMed] [Google Scholar]
- 44.Levi G, Teichberg VI. Isolation and physicochemical characterization of electrolectin, a beta-D-galactoside binding lectin from the electric organ of Electrophorus electricus . Journal of Biological Chemistry. 1981;256(11):5735–5740. [PubMed] [Google Scholar]
- 45.Muramoto K, Kamiya H. The amino-acid sequence of a lectin from conger eel, Conger myriaster, skin mucus. Biochimica et Biophysica Acta. 1992;1116(2):129–136. doi: 10.1016/0304-4165(92)90109-8. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa T, Ishii C, Kagawa D, Muramoto K, Kamiya H. Accelerated evolution in the protein-coding region of galectin cDNAs, congerin I and congerin II, from skin mucus of conger eel (Conger myriaster) Bioscience, Biotechnology and Biochemistry. 1999;63(7):1203–1208. doi: 10.1271/bbb.63.1203. [DOI] [PubMed] [Google Scholar]
- 47.Muramoto K, Kagawa D, Sato T, Ogawa T, Nishida Y, Kamiya H. Functional and structural characterization of multiple galectins from the skin mucus of conger eel, Conger myriaster . Comparative Biochemistry and Physiology Part B. 1999;123(1):33–45. doi: 10.1016/s0305-0491(99)00037-1. [DOI] [PubMed] [Google Scholar]
- 48.Tsutsui S, Iwamoto K, Nakamura O, Watanabe T. Yeast-binding C-type lectin with opsonic activity from conger eel (Conger myriaster) skin mucus. Molecular Immunology. 2007;44(5):691–702. doi: 10.1016/j.molimm.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Kamiya H, Shimizu Y. Marine biopolymers with cell specificity. II. Purification and characterization of agglutinins from mucus of windowpane flounder Lophopsetta maculata . Biochimica et Biophysica Acta. 1980;622(2):171–178. doi: 10.1016/0005-2795(80)90028-8. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed H, Du SJ, O’Leary N, Vasta GR. Biochemical and molecular characterization of galectins from zebrafish (Danio rerio): notochord-specific expression of a prototype galectin during early embryogenesis. Glycobiology. 2004;14(3):219–232. doi: 10.1093/glycob/cwh032. [DOI] [PubMed] [Google Scholar]
- 51.Hosono M, Sugawara S, Ogawa Y, Kohno T, Takayanagi M, Nitta K. Purification, characterization, cDNA cloning, and expression of asialofetuin-binding C-type lectin from eggs of shishamo smelt (Osmerus [Spirinchus] lanceolatus) Biochimica et Biophysica Acta. 2005;1725(2):160–173. doi: 10.1016/j.bbagen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Hosono M, Matsuda K, Kawauchi H, et al. Comparison of N-terminal amino acid sequence of fish roe rhamnose-binding lectins. Biomedical Research. 1992;13(6):443–449. [Google Scholar]
- 53.Al-Hassan JM, Thomson M, Summers B, Criddle RS. Purification and properties of a hemagglutination factor from Arabian Gulf catfish (Arius thalassinus) epidermal secretion. Comparative Biochemistry and Physiology Part B. 1986;85(1):31–39. doi: 10.1016/0305-0491(86)90217-8. [DOI] [PubMed] [Google Scholar]
- 54.Hosono M, Ishikawa K, Mineki R, et al. Tandem repeat structure of rhamnose-binding lectin from catfish (Silurus asotus) eggs. Biochimica et Biophysica Acta. 1999;1472(3):668–675. doi: 10.1016/s0304-4165(99)00185-3. [DOI] [PubMed] [Google Scholar]
- 55.Tsutsui S, Komatsu Y, Sugiura T, Araki K, Nakamura O. A unique epidermal mucus lectin identified from catfish (Silurus asotus): first evidence of intelectin in fish skin slime. Journal of Biochemistry. 2011;150(5):501–514. doi: 10.1093/jb/mvr085. [DOI] [PubMed] [Google Scholar]
- 56.Tateno H, Saneyoshi A, Ogawa T, Muramoto K, Kamiya H, Saneyoshi M. Isolation and characterization of rhamnose-binding lectins from eggs of steelhead trout (Oncorhynchus mykiss) homologous to low density lipoprotein receptor superfamily. Journal of Biological Chemistry. 1998;273(30):19190–19197. doi: 10.1074/jbc.273.30.19190. [DOI] [PubMed] [Google Scholar]
- 57.Tateno H, Ogawa T, Muramoto K, Kamiya H, Hirai T, Saneyoshi M. A novel rhamnose-binding lectin family from eggs of steelhead trout (Oncorhynchus mykiss) with different structures and tissue distribution. Bioscience, Biotechnology and Biochemistry. 2001;65(6):1328–1338. doi: 10.1271/bbb.65.1328. [DOI] [PubMed] [Google Scholar]
- 58.Shiina N, Tateno H, Ogawa T, Muramoto K, Saneyoshi M, Kamiya H. Isolation and characterization of L-rhamnose-binding lectins from chum salmon (Oncorhynchus keta) eggs. Fisheries Science. 2002;68(6):1352–1366. [Google Scholar]
- 59.Tateno H, Ogawa T, Muramoto K, Kamiya H, Saneyoshi M. Distribution and molecular evolution of rhamnose-binding lectins in Salmonidae: isolation and characterization of two lectins from white-spotted charr (Salvelinus leucomaenis) eggs. Bioscience, Biotechnology and Biochemistry. 2002;66(6):1356–1365. doi: 10.1271/bbb.66.1356. [DOI] [PubMed] [Google Scholar]
- 60.Terada T, Watanabe Y, Tateno H, et al. Structural characterization of a rhamnose-binding glycoprotein (lectin) from Spanish mackerel (Scomberomorous niphonius) eggs. Biochimica et Biophysica Acta. 2007;1770(4):617–629. doi: 10.1016/j.bbagen.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe Y, Shiina N, Shinozaki F, et al. Isolation and characterization of L-rhamnose-binding lectin, which binds to microsporidian Glugea plecoglossi, from ayu (Plecoglossus altivelis) eggs. Developmental & Comparative Immunology. 2008;32(5):487–499. doi: 10.1016/j.dci.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 62.Okamoto M, Tsutsui S, Tasumi S, Suetake H, Kikuchi K, Suzuki Y. Tandem repeat L-rhamnose-binding lectin from the skin mucus of ponyfish, Leiognathus nuchalis . Biochemical and Biophysical Research Communications. 2005;333(2):463–469. doi: 10.1016/j.bbrc.2005.05.118. [DOI] [PubMed] [Google Scholar]
- 63.Jimbo M, Usui R, Sakai R, Muramoto K, Kamiya H. Purification, cloning and characterization of egg lectins from the teleost Tribolodon brandti . Comparative Biochemistry and Physiology Part B. 2007;147(2):164–171. doi: 10.1016/j.cbpb.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 64.Odom EW, Vasta GR. Characterization of a binary tandem domain F-type lectin from striped bass (Morone saxatilis) Journal of Biological Chemistry. 2006;281(3):1698–1713. doi: 10.1074/jbc.M507652200. [DOI] [PubMed] [Google Scholar]
- 65.Salerno G, Parisi MG, Parrinello D, et al. F-type lectin from the sea bass (Dicentrarchus labrax): purification, cDNA cloning, tissue expression and localization, and opsonic activity. Fish and Shellfish Immunology. 2009;27(2):143–153. doi: 10.1016/j.fsi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Qiu L, Lin L, Yang K, et al. Molecular cloning and expression analysis of a F-type lectin gene from Japanese sea perch (Lateolabrax japonicus) Molecular Biology Reports. 2011;38(6):3751–3756. doi: 10.1007/s11033-010-0490-7. [DOI] [PubMed] [Google Scholar]
- 67.Tsutsui S, Tasumi S, Suetake H, Suzuki Y. Lectins homologous to those of monocotyledonous plants in the skin mucus and intestine of pufferfish, Fugu rubripes . Journal of Biological Chemistry. 2003;278(23):20882–20889. doi: 10.1074/jbc.M301038200. [DOI] [PubMed] [Google Scholar]
- 68.de Santana Evangelista K, Andrich F, de Rezende FF, et al. Plumieribetin, a fish lectin homologous to mannose-binding B-type lectins, inhibits the collagen-binding α1β1 integrin. Journal of Biological Chemistry. 2009;284(50):34747–34759. doi: 10.1074/jbc.M109.002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galliano M, Minchiotti L, Campagnoli M, et al. Structural and biochemical characterization of a new type of lectin isolated from carp eggs. Biochemical Journal. 2003;376(2):433–440. doi: 10.1042/BJ20030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shirai T, Mitsuyama C, Niwa Y, et al. High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution. Structure. 1999;7(10):1223–1233. doi: 10.1016/s0969-2126(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 71.Shirai T, Matsui Y, Shionyu-Mitsuyama C, et al. Crystal structure of a conger eel galectin (Congerin II) at 1.45 Å resolution: implication for the accelerated evolution of a new ligand-binding site following gene duplication. Journal of Molecular Biology. 2002;321(5):879–889. doi: 10.1016/s0022-2836(02)00700-3. [DOI] [PubMed] [Google Scholar]
- 72.Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. Journal of Biological Chemistry. 1988;263(20):9557–9560. [PubMed] [Google Scholar]
- 73.Drickamer K, Taylor ME. Biology of animal lectins. Annual Review of Cell Biology. 1993;9:237–264. doi: 10.1146/annurev.cb.09.110193.001321. [DOI] [PubMed] [Google Scholar]
- 74.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunological Reviews. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 75.Drickamer K. Ca2+-dependent carbohydrate-recognition domains in animal proteins. Current Opinion in Structural Biology. 1993;3(3):393–400. [Google Scholar]
- 76.Drickamer K, Fadden AJ. Genomic analysis of C-type lectins. Biochemical Society Symposium. 2002;69:59–72. doi: 10.1042/bss0690059. [DOI] [PubMed] [Google Scholar]
- 77.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. The FEBS Journal. 2005;272(24):6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cellular and Molecular Life Sciences. 2000;57(2):276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stockert RJ. The asialoglycoprotein receptor: relationships between structure, function, and expression. Physiological Reviews. 1995;75(3):591–609. doi: 10.1152/physrev.1995.75.3.591. [DOI] [PubMed] [Google Scholar]
- 80.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway—its role in innate immunity and evolution. Immunological Reviews. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 81.Tedder TF, Steeber DA, Chen A, Engel P. The selectins: vascular adhesion molecules. FASEB Journal. 1995;9(10):866–873. [PubMed] [Google Scholar]
- 82.Lanier LL. NK cell receptors. Annual Review of Immunology. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 83.Kijimoto-Ochiai S. CD23 (the low-affinity IgE receptor) as a C-type lectin: a multidomain and multifunctional molecule. Cellular and Molecular Life Sciences. 2002;59(4):648–664. doi: 10.1007/s00018-002-8455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annual Review of Immunology. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 85.De Caro AM, Bonicel JJ, Rouimi P, De Caro JD, Sarles H, Rovery M. Complete amino acid sequence of an immunoreactive form of human pancreatic stone protein isolated from pancreatic juice. European Journal of Biochemistry. 1987;168(1):201–207. doi: 10.1111/j.1432-1033.1987.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 86.Petersen TE. The amino-terminal domain of thrombomodulin and pancreatic stone protein are homologous with lectins. FEBS Letters. 1988;231(1):51–53. doi: 10.1016/0014-5793(88)80700-2. [DOI] [PubMed] [Google Scholar]
- 87.Geider S, Baronnet A, Cerini C, et al. Pancreatic lithostathine as a calcite habit modifier. Journal of Biological Chemistry. 1996;271(42):26302–26306. doi: 10.1074/jbc.271.42.26302. [DOI] [PubMed] [Google Scholar]
- 88.Muramoto K, Jin DH, Niino Y, et al. Comparison of the amino acid sequences of acorn barnacle lectins showing different inhibitory activities toward the crystal growth of calcium carbonate. Fisheries Science. 2001;67(4):703–709. [Google Scholar]
- 89.Fletcher GL, Hew CL, Davies PL. Antifreeze proteins of teleost fishes. Annual Review of Physiology. 2001;63:359–390. doi: 10.1146/annurev.physiol.63.1.359. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Li Z, Lin Q, et al. Structure and evolutionary origin of Ca2+-dependent herring type II antifreeze protein. PLoS One. 2007;2(6, article e548) doi: 10.1371/journal.pone.0000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishimiya Y, Kondo H, Takamichi M, et al. Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus . Journal of Molecular Biology. 2008;382(3):734–746. doi: 10.1016/j.jmb.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 92.Zelensky AN, Gready JE. C-type lectin-like domains in Fugu rubripes . BMC Genomics. 2004;5, article 51 doi: 10.1186/1471-2164-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gundacker D, Leys SP, Schröder HC, Müller IM, Müller WEG. Isolation and cloning of a C-type lectin from the hexactinellid sponge Aphrocallistes vastus: a putative aggregation factor. Glycobiology. 2001;11(1):21–29. doi: 10.1093/glycob/11.1.21. [DOI] [PubMed] [Google Scholar]
- 94.Buck F, Luth C, Strupat K, Bretting H. Comparative investigations on the amino-acid sequences of different isolectins from the sponge Axinella polypoides (Schmidt) Biochimica et Biophysica Acta. 1992;1159(1):1–8. doi: 10.1016/0167-4838(92)90067-n. [DOI] [PubMed] [Google Scholar]
- 95.Wagner-Hülsmann C, Bachinski N, Diehl-Seifert B, et al. A galectin links the aggregation factor to cells in the sponge (Geodia cydonium) system. Glycobiology. 1996;6(8):785–793. doi: 10.1093/glycob/6.8.785-d. [DOI] [PubMed] [Google Scholar]
- 96.Sampaio AH, Rogers DJ, Barwell CJ. A galactose-specific lectin from the red marine alga Ptilota filicina . Phytochemistry. 1998;48(5):765–769. doi: 10.1016/s0031-9422(97)00966-7. [DOI] [PubMed] [Google Scholar]
- 97.Kakiuchi M, Okino N, Sueyoshi N, et al. Purification, characterization, and cDNA cloning of α-N-acethylgalactosamine-specific lectin from starfish Patiria pectinifera. Glycobiology. 2008;12(2):85–94. doi: 10.1093/glycob/12.2.85. [DOI] [PubMed] [Google Scholar]
- 98. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene.
- 99.Kilpatrick DC. Animal lectins: a historical introduction and overview. Biochimica et Biophysica Acta. 2002;1572(2-3):187–197. doi: 10.1016/s0304-4165(02)00308-2. [DOI] [PubMed] [Google Scholar]
- 100.Nakamura O, Watanabe T, Kamiya H, Muramoto K. Galectin containing cells in the skin and mucosal tissues in Japanese conger eel, Conger myriaster: an immunohistochemical study. Developmental & Comparative Immunology. 2001;25(5-6):431–437. doi: 10.1016/s0145-305x(01)00012-x. [DOI] [PubMed] [Google Scholar]
- 101.Kamiya H, Muramoto K, Goto R. Purification and properties of agglutinins from conger eel, Conger myriaster (Brevoort), skin mucus. Developmental & Comparative Immunology. 1988;12(2):309–318. doi: 10.1016/0145-305x(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 102.Nakamura O, Matsuoka H, Ogawa T, Muramoto K, Kamiya H, Watanabe T. Opsonic effect of congerin, a mucosal galectin of the Japanese conger, Conger myriaster (Brevoort) Fish and Shellfish Immunology. 2006;20(3):433–435. doi: 10.1016/j.fsi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 103.Nakamura O, Inaga Y, Suzuki S, et al. Possible immune functions of congerin, a mucosal galectin, in the intestinal lumen of Japanese conger eel. Fish and Shellfish Immunology. 2007;23(3):683–692. doi: 10.1016/j.fsi.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 104.Ogawa T, Yonemaru S, Naganuma T, Hirabayashi J, Kasai K, Muramoto K. Biomolecular Chemistry: A Bridge for the Future, Chaptre 2. Cell Function of Macromolecules. Tokyo, Japan: Maruzen; 2003. Strong induction of apoptosis of T cell lines by conger eel galectins; pp. 134–137. [Google Scholar]
- 105.Ogawa T. Molecular diversity of proteins in biological offense and defense systems. Molecular Diversity. 2006;10(4):511–514. doi: 10.1007/s11030-006-9048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Science. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Advances in Protein Chemistry. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 108.Ban M, Yoon HJ, Demirkan E, Utsumi S, Mikami B, Yagi F. Structural basis of a fungal galectin from Agrocybe cylindracea for recognizing sialoconjugate. Journal of Molecular Biology. 2005;351(4):695–706. doi: 10.1016/j.jmb.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 109.Wälti MA, Walser PJ, Thore S, et al. Structural basis for chitotetraose coordination by CGL3, a novel galectin-related protein from Coprinopsis cinerea . Journal of Molecular Biology. 2008;379(1):146–159. doi: 10.1016/j.jmb.2008.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shirai T, Shionyu-Mitsuyama C, Ogawa T, Muramoto K. Structure based studies of the adaptive diversification process of congerins. Molecular Diversity. 2006;10(4):567–573. doi: 10.1007/s11030-006-9030-8. [DOI] [PubMed] [Google Scholar]
- 111.Konno A, Ogawa T, Shirai T, Muramoto K. Reconstruction of a probable ancestral form of conger eel galectins revealed their rapid adaptive evolution process for specific carbohydrate recognition. Molecular Biology and Evolution. 2007;24(11):2504–2514. doi: 10.1093/molbev/msm185. [DOI] [PubMed] [Google Scholar]
- 112.Konno A, Yonemaru S, Kitagawa A, Muramoto K, Shirai T, Ogawa T. Protein engineering of conger eel galectins by tracing of molecular evolution using probable ancestral mutants. BMC Evolutionary Biology. 2010;10(1, article 43) doi: 10.1186/1471-2148-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takakura Y, Tsukamoto H, Yamamoto T. Molecular cloning, expression and properties of an α/β-galactoside α2,3-sialyltransferase from vibrio sp. JT-FAJ-16. Journal of Biochemistry. 2007;142(3):403–412. doi: 10.1093/jb/mvm147. [DOI] [PubMed] [Google Scholar]
- 114.Tsukamoto H, Takakura Y, Yamamoto T. Purification, cloning, and expression of an α/β-galactoside α-2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. Journal of Biological Chemistry. 2007;282(41):29794–29802. doi: 10.1074/jbc.M701907200. [DOI] [PubMed] [Google Scholar]
- 115.Cooper DNW. Galectinomics: finding themes in complexity. Biochimica et Biophysica Acta. 2002;1572(2-3):209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 116.Vasta GR, Ahmed H, Du SJ, Henrikson D. Galectins in teleost fish: Zebrafish (Danio rerio) as a model species to address their biological roles in development and innate immunity. Glycoconjugate Journal. 2004;21(8-9):503–521. doi: 10.1007/s10719-004-5541-7. [DOI] [PubMed] [Google Scholar]
- 117.Ahmed H, Du SJ, Vasta GR. Knockdown of a galectin-1-like protein in zebrafish (Danio rerio) causes defects in skeletal muscle development. Glycoconjugate Journal. 2009;26(3):277–283. doi: 10.1007/s10719-008-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahmed H, Vasta GR. Unlike mammalian GRIFIN, the zebrafish homologue (DrGRIFIN) represents a functional carbohydrate-binding galectin. Biochemical and Biophysical Research Communications. 2008;371(3):350–355. doi: 10.1016/j.bbrc.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tasumi S, Vasta GR. A galectin of unique domain organization from hemocytes of the eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus . Journal of Immunology. 2007;179(5):3086–3098. doi: 10.4049/jimmunol.179.5.3086. [DOI] [PubMed] [Google Scholar]
- 120.Hwang JS, Takaku Y, Momose T, et al. Nematogalectin, a nematocyst protein with GlyXY and galectin domains, demonstrates nematocyte-specific alternative splicing in Hydra. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(43):18539–18544. doi: 10.1073/pnas.1003256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nature Structural Biology. 2002;9(8):628–634. doi: 10.1038/nsb817. [DOI] [PubMed] [Google Scholar]
- 122.Bianchet MA, Odom EW, Vasta GR, Amzel LM. Structure and specificity of a binary tandem domain F-Lectin from striped bass (Morone saxatilis) Journal of Molecular Biology. 2010;401(2):239–252. doi: 10.1016/j.jmb.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vasta GR, Ahmed H, Odom EW. Structural and functional diversity of lectin repertoires in invertebrates, protochordates and ectothermic vertebrates. Current Opinion in Structural Biology. 2004;14(5):617–630. doi: 10.1016/j.sbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 124. http://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_how_to_domain_architecture.html.
- 125.Shirai T, Watanabe Y, Lee MS, Ogawa T, Muramoto K. Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein. Journal of Molecular Biology. 2009;391(2):390–403. doi: 10.1016/j.jmb.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 126.Tateno H. SUEL-related lectins, a lectin family widely distributed throughout organisms. Bioscience, Biotechnology and Biochemistry. 2010;74(6):1141–1144. doi: 10.1271/bbb.100086. [DOI] [PubMed] [Google Scholar]
- 127.Tateno H, Shibata Y, Nagahama Y, et al. Tissue-specific expression of rhamnose-binding lectins in the steelhead trout (Oncorhynchus mykiss) Bioscience, Biotechnology and Biochemistry. 2002;66(6):1427–1430. doi: 10.1271/bbb.66.1427. [DOI] [PubMed] [Google Scholar]
- 128.Tateno H, Yamaguchi T, Ogawa T, et al. Immunohistochemical localization of rhamnose-binding lectins in the steelhead trout (Oncorhynchus mykiss) Developmental & Comparative Immunology. 2002;26(6):543–550. doi: 10.1016/s0145-305x(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 129.Booy A, Haddow JD, Olafson RW. Isolation of the salmonid rhamnose-binding lectin STL2 from spores of the microsporidian fish parasite Loma salmonae. Journal of Fish Diseases. 2005;28(8):455–462. doi: 10.1111/j.1365-2761.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 130.Ozaki H, Ohwaki M, Fukada T. Studies on lectins of amago (Oncorhyncus rhodurus). I. Amago ova lectin and its receptor on homologous macrophages. Developmental & Comparative Immunology. 1983;7(1):77–87. doi: 10.1016/0145-305x(83)90057-5. [DOI] [PubMed] [Google Scholar]
- 131.Ng TB, Lam YW, Woo NYS. The immunostimulatory activity and stability of grass carp (Ctenopharyngodon idellus) roe lectin. Veterinary Immunology and Immunopathology. 2003;94(3-4):105–112. doi: 10.1016/s0165-2427(03)00067-9. [DOI] [PubMed] [Google Scholar]
- 132.Zhao L, Almaraz RT, Xiang F, Hedrick JL, Franz AH. Gas-phase scrambling of disulfide bonds during matrix-assisted laser desorption/ionization mass spectrometry analysis. Journal of the American Society for Mass Spectrometry. 2009;20(9):1603–1616. doi: 10.1016/j.jasms.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 133.Nitta K, Kawano T, Sugawara S, Hosono M. Regulation of globotriaosylceramide (Gb3)-mediated signal transduction by rhamnose-binding lectin. Yakugaku Zasshi. 2007;127(4):553–561. doi: 10.1248/yakushi.127.553. [DOI] [PubMed] [Google Scholar]
- 134.Weston BS, Malhas AN, Price RG. Structure-function relationships of the extracellular domain of the autosomal dominant polycystic kidney disease-associated protein, polycystin-1. FEBS Letters. 2003;538(1–3):8–13. doi: 10.1016/s0014-5793(03)00130-3. [DOI] [PubMed] [Google Scholar]
- 135.Fujisawa K, Wrana JL, Culotti JG. The slit receptor EVA-1 coactivates a SAX-3/robo-mediated guidance signal in C. elegans . Science. 2007;317(5846):1934–1938. doi: 10.1126/science.1144874. [DOI] [PubMed] [Google Scholar]
- 136.Mitsunaga K, Harada-Itadani J, Shikanai T, et al. Human C21orf63 is a heparin-binding protein. Journal of Biochemistry. 2009;146(3):369–373. doi: 10.1093/jb/mvp079. [DOI] [PubMed] [Google Scholar]
- 137.Lelianova VG, Davletov BA, Sterling A, et al. α-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. Journal of Biological Chemistry. 1997;272(34):21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- 138.Arcos-Burgos M, Jain M, Acosta MT, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Molecular Psychiatry. 2010;15(11):1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 139.Schwarz RS, Hodes-Villamar L, Fitzpatrick KA, Fain MG, Hughes AL, Cadavid LF. A gene family of putative immune recognition molecules in the hydroid Hydractinia. Immunogenetics. 2007;59(3):233–246. doi: 10.1007/s00251-006-0179-1. [DOI] [PubMed] [Google Scholar]
- 140.Jia WZ, Shang N, Guo QL. Molecular cloning of rhamnose-binding lectin gene and its promoter region from snakehead Channa argus . Fish Physiology and Biochemistry. 2010;36(3):451–459. doi: 10.1007/s10695-009-9315-9. [DOI] [PubMed] [Google Scholar]
- 141.López JA, Fain MG, Cadavid LF. The evolution of the immune-type gene family Rhamnospondin in cnidarians. Gene. 2011;473(2):119–124. doi: 10.1016/j.gene.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 142.Watanabe Y, Tateno H, Nakamura-Tsuruta S, et al. The function of rhamnose-binding lectin in innate immunity by restricted binding to Gb3. Developmental & Comparative Immunology. 2009;33(2):187–197. doi: 10.1016/j.dci.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 143.Katagiri YU, Kiyokawa N, Fujimoto J. A role for lipid rafts in immune cell signaling. Microbiology and Immunology. 2001;45(1):1–8. doi: 10.1111/j.1348-0421.2001.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 144.Lingwood CA. Role of verotoxin receptors in pathogenesis. Trends in Microbiology. 1996;4(4):147–153. doi: 10.1016/0966-842x(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 145.Kawano T, Sugawara S, Hosono M, Tatsuta T, Nitta K. Alteration of gene expression induced by Silurus asotus lectin in Burkitt’s lymphoma cells. Biological and Pharmaceutical Bulletin. 2008;31(5):998–1002. doi: 10.1248/bpb.31.998. [DOI] [PubMed] [Google Scholar]
- 146.Tateno H, Ogawa T, Muramoto K, Kamiya H, Saneyoshi M. Rhamnose-binding lectins from steelhead trout (Oncorhynchus mykiss) eggs recognize bacterial lipopolysaccharides and lipoteichoic acid. Bioscience, Biotechnology and Biochemistry. 2002;66(3):604–612. doi: 10.1271/bbb.66.604. [DOI] [PubMed] [Google Scholar]
- 147.Sugawara S, Hosono M, Ogawa Y, Takayanagi M, Nitta K. Catfish egg lectin causes rapid activation of multidrug resistance 1 P-glycoprotein as a lipid translocase. Biological and Pharmaceutical Bulletin. 2005;28(3):434–441. doi: 10.1248/bpb.28.434. [DOI] [PubMed] [Google Scholar]
- 148.Vakonakis I, Langenhan T, Prömel S, Russ A, Campbell ID. Solution structure and sugar-binding mechanism of mouse latrophilin-1 RBL: a 7TM receptor-attached lectin-like domain. Structure. 2008;16(6):944–953. doi: 10.1016/j.str.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


