Abstract
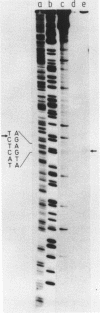
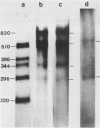
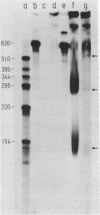
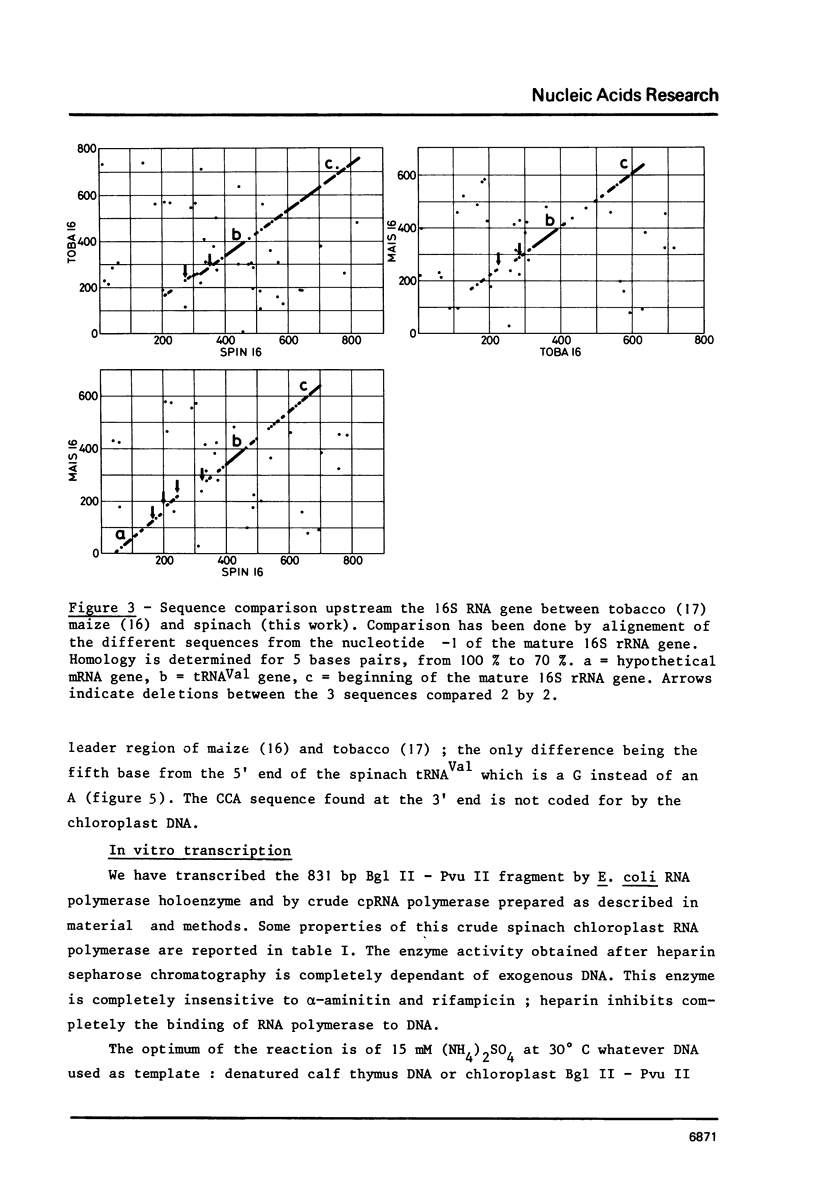
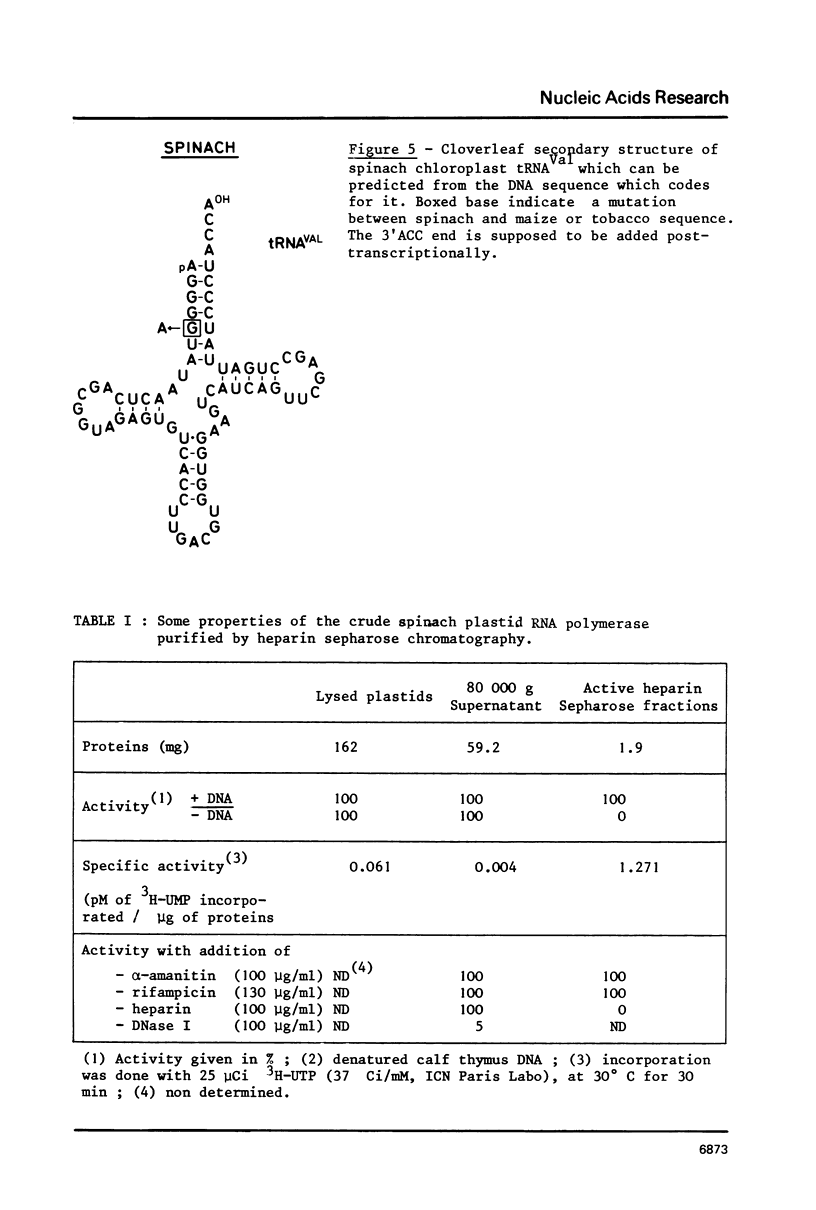
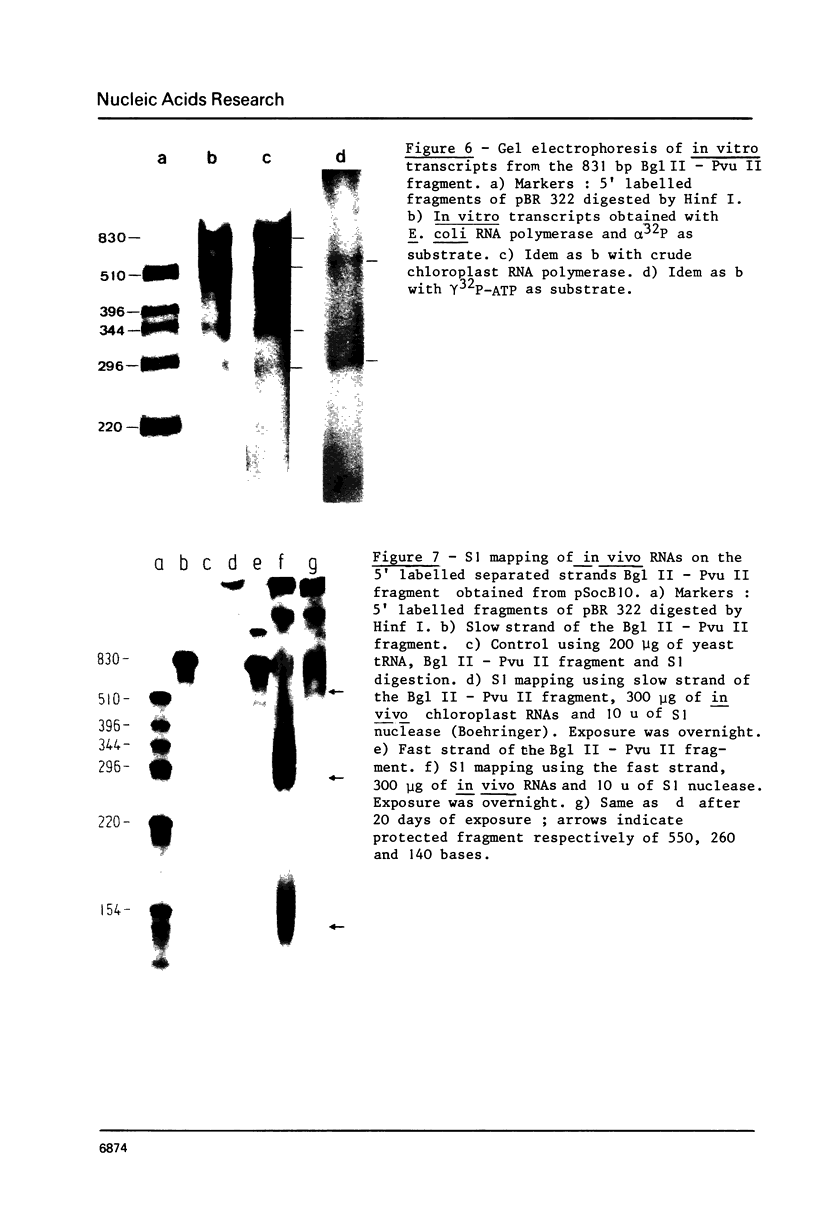
A cloned fragment of spinach chloroplast DNA carrying 140 bp of the 16S rRNA gene and 691 bp upstream this gene has been analysed by DNA sequencing, by in vitro transcription, by S1 mapping with chloroplast RNAs and purified 16S rRNA from 30S ribosomal subunits. A tRNAVal gene has been located between the position 394 and 465. Crude chloroplast RNA polymerase has been purified by heparin sepharose chromatography of a 80 000 g supernatant from pure lysed spinach plastids and used to transcribe the cloned Bg1 II-Pvu II DNA fragment. Four in vitro transcripts of about 830, 550, 350 and 260 bases were obtained whatever RNA polymerase used: the chloroplast or the E. coli enzyme. The transcripts of 550 and 260 bases are initiated by ATP. S1 mapping with in vivo chloroplasts RNAs on 5' labelled separated strands from Bg1 II-Pvu II fragments indicates 2 protected DNA fragments respectively of 140 and 260 bases on the strand which codes for rRNAs and possibly one protected DNA fragment of 550 bases on the other strand. The start site of the 260 bases transcript might correspond to the initiation site of transcription of the rRNA genes. The possibility that the 550 bases transcription of the non coding strand for rRNA genes corresponds to the beginning of a mRNA is discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Grummt I., Allet B. The nucleotide sequence of the initiation region of the ribosomal transcription unit from mouse. Nucleic Acids Res. 1981 Apr 10;9(7):1559–1569. doi: 10.1093/nar/9.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M., Briat J. F., Laulhere J. P. Influence of the ionic environment on the in vitro transcription of the spinach plastid DNA by a selectively bound RNA-polymerase DNA complex. Biochim Biophys Acta. 1981 Oct 27;655(3):374–382. doi: 10.1016/0005-2787(81)90048-4. [DOI] [PubMed] [Google Scholar]
- Bohnert H. J., Gordon K. H., Crouse E. J. Homologies among ribosomal RNA and messenger RNA genes in chloroplasts, mitochondria and E. coli. Mol Gen Genet. 1980;179(3):539–545. doi: 10.1007/BF00271743. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Gigot C., Laulhere J. P., Mache R. Visualization of a Spinach Plastid Transcriptionally Active DNA-Protein Complex in a Highly Condensed Structure. Plant Physiol. 1982 May;69(5):1205–1211. doi: 10.1104/pp.69.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J. F., Laulhere J. P., Mache R. Transcription activity of a DNA-protein complex isolated from spinach plastids. Eur J Biochem. 1979 Jul;98(1):285–292. doi: 10.1111/j.1432-1033.1979.tb13187.x. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Mache R. Properties and characterization of a spinach chloroplast RNA polymerase isolated from a tanscriptionally active DNA-protein complex. Eur J Biochem. 1980 Oct;111(2):503–509. doi: 10.1111/j.1432-1033.1980.tb04966.x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horney D. L., Webster D. A. Deoxyribonuclease: a sensitive assay using radial diffusion in agarose containing methyl green-DNA complex. Biochim Biophys Acta. 1971 Sep 30;247(1):54–61. doi: 10.1016/0005-2787(71)90806-9. [DOI] [PubMed] [Google Scholar]
- Klemenz R., Geiduschek E. P. The 5' terminus of the precursor ribosomal RNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 25;8(12):2679–2689. doi: 10.1093/nar/8.12.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Kee S. G., Efstratiadis A., Kafatos F. C. Amplification and characterization of a beta-globin gene synthesized in vitro. Cell. 1976 Jun;8(2):163–182. doi: 10.1016/0092-8674(76)90001-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. A modified pBR322 vector with improved properties for the cloning, recovery, and sequencing of blunt-ended DNA fragments. Gene. 1982 Feb;17(2):189–196. doi: 10.1016/0378-1119(82)90072-5. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Schwarz Z., Kössel H., Schwarz E., Bogorad L. A gene coding for tRNA is located near 5' terminus of 16S rRNA gene in Zea mays chloroplast genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4748–4752. doi: 10.1073/pnas.78.8.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverthorne J., Ellis R. J. Protein synthesis in chloroplasts. VIII. Differential synthesis of chloroplast proteins during spinach leaf development. Biochim Biophys Acta. 1980 Apr 30;607(2):319–330. doi: 10.1016/0005-2787(80)90084-2. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tohdoh N., Shinozaki K., Sugiura M. Sequence of a putative promoter region for the rRNA genes of tobacco chloroplast DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5399–5406. doi: 10.1093/nar/9.20.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohdoh N., Sugiura M. The complete nucleotide sequence of 16S ribosomal RNA gene from tobacco chloroplasts. Gene. 1982 Feb;17(2):213–218. doi: 10.1016/0378-1119(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Venegas A., Sewell E. T., Masiarz F. R., DeGennaro L. J., Weinberg F., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. II. Physical map and nucleotide sequence of the 5 S ribosomal RNA gene and adjacent intergenic regions. J Biol Chem. 1977 Nov 25;252(22):8126–8135. [PubMed] [Google Scholar]
- Whitfeld P. R., Herrmann R. G., Bottomley W. Mapping of the ribosomal RNA genes on spinach chloroplast DNA. Nucleic Acids Res. 1978 Jun;5(6):1741–1751. doi: 10.1093/nar/5.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienand U., Schwarz Z., Feix G. Electrophoretic elution of nucleic acids from gels adapted for subsequent biological tests. Application for analysis of mRNAs from maize endosperm. FEBS Lett. 1979 Feb 15;98(2):319–323. doi: 10.1016/0014-5793(79)80208-2. [DOI] [PubMed] [Google Scholar]
- Wu R. Nucleotide sequence analysis of DNA. I. Partial sequence of the cohesive ends of bacteriophage lambda and 186 DNA. J Mol Biol. 1970 Aug;51(3):501–521. doi: 10.1016/0022-2836(70)90004-5. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Tandem promoters direct E. coli ribosomal RNA synthesis. Cell. 1979 May;17(1):225–234. doi: 10.1016/0092-8674(79)90310-6. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Gilbert S. F., Nomura M. DNA sequences of promoter regions for rRNA operons rrnE and rrnA in E. coli. Cell. 1979 May;17(1):201–209. doi: 10.1016/0092-8674(79)90308-8. [DOI] [PubMed] [Google Scholar]