Abstract
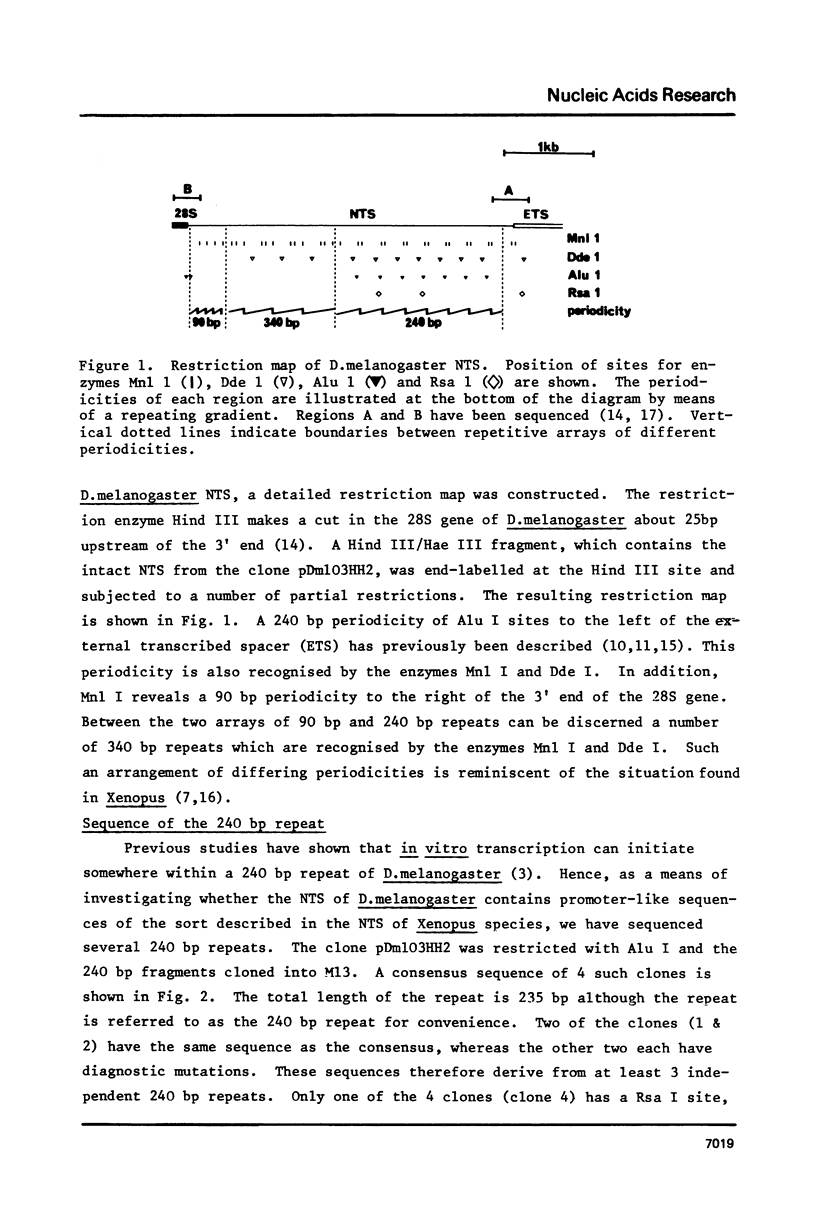
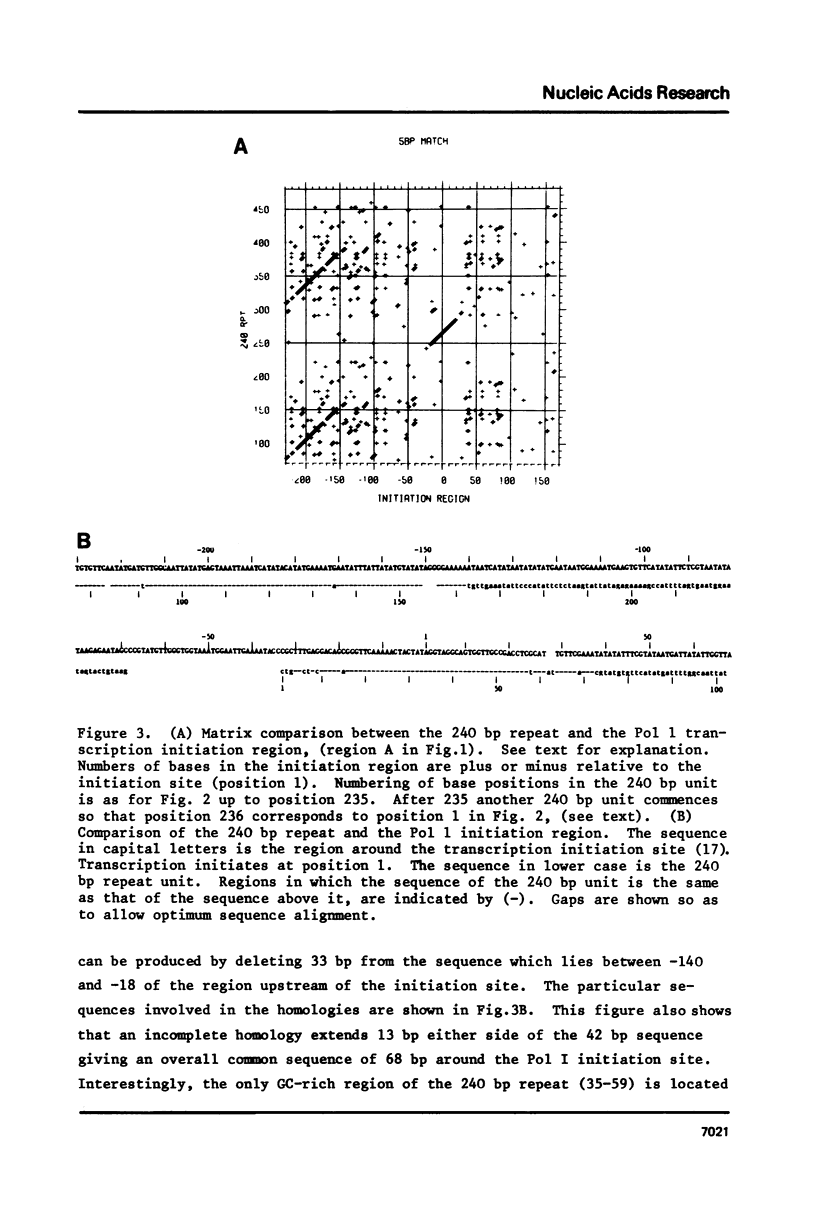
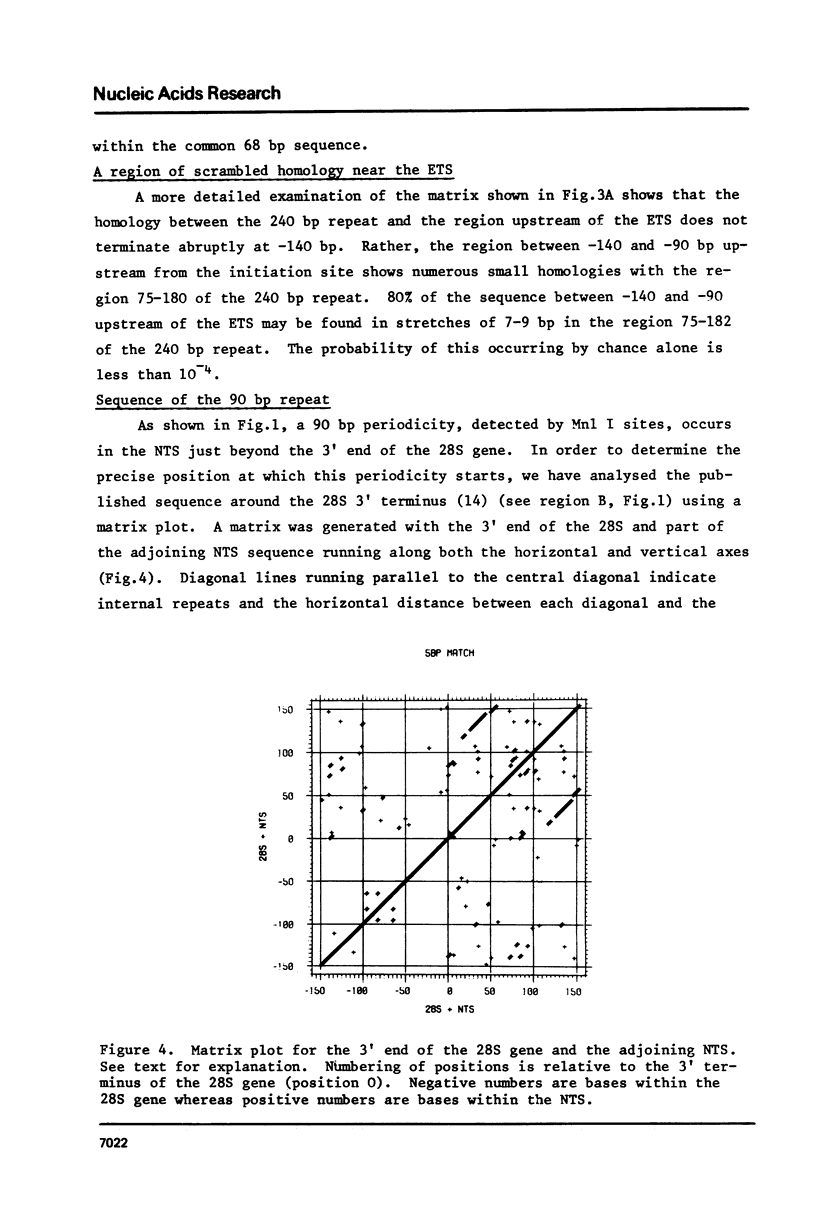
Sequence analysis of the rDNA nontranscribed spacer (NTS) of D.melanogaster shows that it contains many regions that are perfectly homologous with the Pol I transcription initiation site. Each initiation site homology is contained within a 240 base pair repeated sequence which are organised in a tandem array terminating 140 bases pairs upstream from the transcription initiation site. We discuss the significance of these regions and the maintenance of multiple homologies against the observed high rate of spacer divergence between related Drosophila species. In addition there are two other regions of repetition in the NTS: a 90 base pair periodicity commencing 16 bases downstream from the 3' end of the 28S gene and a number of 340 base pair repeats. A similar complex of different periodicities has been described in Xenopus and may be a general feature of NTS sequences.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Gait M. J., Mayol L., Young I. G. A short primer for sequencing DNA cloned in the single-stranded phage vector M13mp2. Nucleic Acids Res. 1980 Apr 25;8(8):1731–1743. doi: 10.1093/nar/8.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach R., Allet B., Crippa M. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 1981 Oct 24;9(20):5311–5330. doi: 10.1093/nar/9.20.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Thoday J. M., Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982 Feb 18;295(5850):564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., McIntyre G. A. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur J Biochem. 1970 Sep;16(1):1–11. doi: 10.1111/j.1432-1033.1970.tb01046.x. [DOI] [PubMed] [Google Scholar]
- Grummt I. Mapping of a mouse ribosomal DNA promoter by in vitro transcription. Nucleic Acids Res. 1981 Nov 25;9(22):6093–6102. doi: 10.1093/nar/9.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Kunz W., Petersen G., Renkawitz-Pohl R., Glätzer K. H., Schäfer M. Distribution of spacer length classes and the intervening sequence among different nucleolus organizers in Drosophila hydei. Chromosoma. 1981;83(2):145–158. doi: 10.1007/BF00286785. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Restriction analysis of spacers in ribosomal DNA of Drosophila melanogaster. Nucleic Acids Res. 1979 Sep 11;7(1):205–215. doi: 10.1093/nar/7.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K., Dawid I. B. The nucleotide sequence at the transcription termination site of ribosomal RNA in Drosophila melanogaster. Nucleic Acids Res. 1981 Apr 24;9(8):1801–1811. doi: 10.1093/nar/9.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Identification of the in vivo and in vitro origin of transcription in human rDNA. Nucleic Acids Res. 1982 Jul 10;10(13):3933–3949. doi: 10.1093/nar/10.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Barnett T., Murtif V. L. Nontranscribed spacers in Drosophila ribosomal DNA. Chromosoma. 1981;82(5):637–655. doi: 10.1007/BF00285773. [DOI] [PubMed] [Google Scholar]
- Reichel R., Monstein H. J., Jansen H. W., Philipson L., Benecke B. J. Small nuclear RNAs are encoded in the nontranscribed region of ribosomal spacer DNA. Proc Natl Acad Sci U S A. 1982 May;79(10):3106–3110. doi: 10.1073/pnas.79.10.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Glätzer K. H., Kunz W. Characterization of cloned ribosomal DNA from Drosophila hydei. Nucleic Acids Res. 1980 Oct 24;8(20):4593–4611. doi: 10.1093/nar/8.20.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungger D., Achermann H., Crippa M. Transcription of spacer sequences in genes coding for ribosomal RNA in Xenopus cells. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3957–3961. doi: 10.1073/pnas.76.8.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Trendelenburg M. F., Krohne G., Franke W. W. Lengths and patterns of transcriptional units in the amplified nucleoli of oocytes of Xenopus laevis. Chromosoma. 1977 Mar 16;60(2):147–167. doi: 10.1007/BF00288462. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Strachan T., Coen E., Webb D., Dover G. Modes and rates of change of complex DNA families of Drosophila. J Mol Biol. 1982 Jun 15;158(1):37–54. doi: 10.1016/0022-2836(82)90449-1. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]



