Abstract
Like all aerobic organisms, plants require molecular oxygen for respiratory energy production. In plants, hypoxic conditions can occur during natural events (e.g., flooding), during developmental processes (e.g., seed germination), and in cells of compact tissues with high metabolic rates. Plant acclimation responses to hypoxia involve a modulation of gene expression leading to various biochemical, physiological, and morphological changes that stave off eventual anoxia. In contrast with the animal kingdom, a direct oxygen-sensing mechanism in plants has been elusive so far. However, two recent independent studies show that oxygen sensing in plants operates via posttranslational regulation of key hypoxia response transcription factors by the N-end rule pathway. The N-end rule is an evolutionarily conserved pathway for protein degradation that relates the fate of a protein with the identity of its N-terminal residues. Results from these studies demonstrate that oxygen-dependent modification and targeted proteolysis of members of the ethylene response factor group VII transcription factor family regulate hypoxia-responsive gene expression in Arabidopsis thaliana. The discovery of this plant hypoxia-sensing mechanism sets the stage for further research on plant homeostatic response to oxygen, which could be relevant to understanding plant distributions in flood-prone ecosystems and improving hypoxia tolerance of crops.
INTRODUCTION
Although plants are a major source of oxygen released into the atmosphere, they are also strongly dependent on external oxygen supply. This is true not only for nonphotosynthesizing organs, such as roots, storage organs, flowers, or fruits, but also during situations when photosynthesis is absent (during the night) or limited (Drew, 1997; Bailey-Serres and Voesenek, 2008). Unlike animals, plants lack an active oxygen distribution system. This means that dense and metabolically active tissues, such as stems, meristems, and fruits, can quickly develop hypoxic cores, even when oxygen is externally available (Rolletschek et al., 2004; Armstrong et al., 2009). Oxygen deficiency can also occur in the external environment, such as during winter ice encasement or flooding. Flooding, including root waterlogging and complete plant submergence, leads to low oxygen availability because of severely restricted rates of gas diffusion under water (Armstrong, 1979).
Under conditions of limited oxygen availability, ATP production via oxidative phosphorylation is constrained, negatively affecting the cellular energy status. Acclimative responses to hypoxia include a switch to an energy-conserving mode, involving lower ATP consumption and the use of pyrophosphate as an alternative energy donor (Huang et al., 2008). The biochemical and physiological reconfiguration triggered by hypoxia is also reflected in the molecular response of plants to hypoxia, which includes the induction of genes encoding fermentative and glycolytic enzymes, many transcription factors (TFs), and several genes of unknown function (Christianson et al., 2010; Mustroph et al., 2010; Narsai et al., 2011). Despite a considerable variation in the tolerance to prolonged hypoxia, most plant species switch on this core set of hypoxia-responsive genes. Furthermore, there is a surprisingly conserved response of different cell types of Arabidopsis thaliana to low-oxygen stress (Mustroph et al., 2009), making it likely that there is a common mechanism regulating gene expression during oxygen limitation. Despite knowledge regarding the molecular response to hypoxia, the actual mechanisms leading to the induction of low-oxygen-responsive genes in plants has been elusive (Fukao and Bailey-Serres, 2004; Bailey-Serres and Chang, 2005).
THE HUNT FOR THE ELUSIVE OXYGEN SENSOR IN PLANTS
The precise and tight control of molecular and physiological responses to hypoxia indicates that plants must possess sensitive oxygen-sensing mechanisms. However, whether plants achieve this using direct sensing or via oxygen-mediated indirect changes has been less certain.
Direct Sensing Mechanisms
A direct sensor could be a transcriptional regulator or TF that is activated or repressed directly by oxygen. Such direct oxygen-sensing systems exist in higher animals and microorganisms. In mammals, under normoxic conditions, the TF hypoxia-inducible factor-1-alpha (HIF-1α) is rapidly degraded by the proteasome after hydroxylation via 2-oxoglutarate-dependent prolyl and asparaginyl hydroxylases. During hypoxia, oxygen-dependent hydroxylation is strongly inhibited, and HIF-1α induces many hypoxia-responsive genes (Bruick, 2003; Haddad, 2004; Brahimi-Horn et al., 2005). Rhizobial bacteria use a different system: A heme-binding His protein kinase, FixL, is inactivated by oxygen binding to the heme domain, but in the absence of oxygen, it phosphorylates the TF FixJ, thereby activating it (Akimoto et al., 2003; Gilles-Gonzalez and Gonzalez, 2005). A similar mechanism exists in Escherichia coli, in which the oxygen-sensitive protein E. coli direct oxygen-sensing phosphodiesterase has a phosphodiesterase activity (Green et al., 2009).
These signal transduction pathways have two things in common. First, the accumulation of the signaling protein is posttranscriptionally regulated. This means that oxygen-dependent protein turnover or activation is the major determinant of TF activity, and not the mRNA levels. Second, these regulators contain functional heme groups and belong to a family of PAS domain proteins, which can be regulated either by oxygen or redox changes (Taylor and Zhulin, 1999; Gilles-Gonzalez and Gonzalez, 2004).
Based on these observations, it was hypothesized that plants might possess a similar signal transduction system (Asif et al., 2009). The genome of plants contains many PAS domain proteins as well as prolyl hydroxylases. However, the PAS domain proteins have taken a different role in plant signal transduction, namely the perception and transduction of light signals. The well-known photoreceptors, phytochromes and phototropins, belong to the PAS protein family in plants (Taylor and Zhulin, 1999). Plants also possess many prolyl hydroxylases, some of which are even induced by low-oxygen stress (Vlad et al., 2007). However, these genes are not very close homologs of the animal hypoxia-responsive enzymes (Mustroph et al., 2010), and were recently described as being exclusively involved in cell wall modifications in root hair cells (Velasquez et al., 2011).
The hypoxia-inducible, oxygen-binding plant hemoglobins have been suggested to be involved in oxygen sensing (Hunt et al., 2002; Thiel et al., 2011). Despite evidence for a role in hypoxia tolerance (Hunt et al., 2002), the low dissociation constant of hemoglobins for oxygen precludes their possible function as an oxygen sensor or carrier (Abbruzzetti et al., 2011). Besides heme-based direct oxygen-sensing mechanisms, there could be other proteins that are sensitive to oxygen, (e.g., Fe-S proteins). The iron group, associated with Cys groups from the protein sensory domain, can be changed because of the oxygen status of the cell (Green et al., 2009). A prominent example for such a sensing system is the regulator FNR from E. coli (Spiro and Guest, 1990; Green et al., 2009). However, again no orthologs for this protein were found in higher plants.
Indirect Sensing Mechanisms
The lack of evidence for a direct oxygen sensor in plants led to the suggestion that indirect sensing could be involved. Indirect sensing methods are suggested to involve detection of changes occurring because of variation in oxygen availability, such as the sensing of redox status (NADH/ NAD+ ratios), reactive oxygen species (ROS) (H2O2, NO), or the energy status (ATP levels) of a plant cell. Nonsymbiotic hemoglobins have been suggested to be involved in the sensing of NO or other ROS (Sowa et al., 1998; Dordas et al., 2003a; Dordas et al., 2003b; Thiel et al., 2011). However, it has never been demonstrated how a possible signal from hemoglobin could be transferred to the nucleus. Recently, it has been suggested that hemoglobin has a metabolic function in scavenging NO or recycling NAD(P)H (Thiel et al., 2011).
Monitoring the energy status of plant cells under low-oxygen stress is another possible sensing mechanism, because the limitation of oxidative phosphorylation during hypoxia causes a rapid drop in ATP levels. This signaling cascade could involve many protein kinases, such as calcineurin B-like interacting protein kinase (CIPK15), which was recently shown to be involved in the anaerobic germination of rice (Oryza sativa) seedlings (Lee et al., 2009). The protein kinases KIN10/11 could also be part of this energy-sensing network (Baena-González et al., 2007).
Hypoxia also causes an increase in mitochondrial calcium efflux (Subbaiah et al., 1998). This modulation of intracellular calcium levels has been linked to hypoxia-mediated gene expression (Chung and Ferl, 1999; Tsuji et al., 2000). There are indications that phospholipase C (Reggiani and Laoreti, 2000; Reggiani, 2006), Rho-like small G proteins (Baxter-Burrell et al., 2002), and ethylene (He et al., 1996) are also part of this complex signal transduction chain involving calcium signaling.
PLANT TRANSCRIPTION FACTORS ASSOCIATED WITH THE LOW-OXYGEN RESPONSE
The low-oxygen responsive signaling cascade involves various TFs. The exposure of Arabidopsis roots to hypoxic conditions has been found to differentially regulate the expression of TFs belonging to the MYB, basic domain/Leu zipper, basic helix-loop-helix, and the ethylene response factor (ERF) families (Licausi et al., 2011b). Various other studies have demonstrated the role of specific members of these TF families in different plant species. A general involvement of MYB TFs in mediating hypoxia tolerance has been shown in Arabidopsis (Hoeren et al., 1998), rice (Mattana et al., 2007), and wheat (Triticum aestivum) (Lee et al., 2007). In Arabidopsis seeds, the TFs ANAC102 and VIN3 were involved in the seedling response to hypoxic stress (Bond et al., 2009; Christianson et al., 2009). A G-box binding factor, GBF1, was suggested to induce the ADH1 promoter in maize (Zea mays), although it has never been cloned (de Vetten and Ferl, 1995).
Members of the group VII AP2-ERF family are the most intensely studied TFs in the context of plant adaptive responses to flooding-induced hypoxia and have emerged as important regulators of the low-oxygen response. SUB1-A, a member of the rice group VII AP2-ERF family (Nakano et al., 2006), was demonstrated as being both necessary and sufficient for enhanced survival of submergence by initiating molecular responses that repressed GA-mediated shoot elongation under water. SUB1-A rice thus survives submergence stress by using an energy-saving quiescence strategy involving conservative consumption of carbohydrate reserves (Fukao et al., 2006; Xu et al., 2006). A contrasting strategy is used by deepwater rice, in which the related group VII ERF TFs, SNORKEL1 and SNORKEL2, mediate rapid elongation of submerged rice internodes. This facilitates the emergence of the shoot organs above water, which then act as “snorkels” to aerate the whole plant (Hattori et al., 2009).
Although these TFs regulate specific adaptive responses in rice varieties with different strategies to survive submergence-associated hypoxia, a more general involvement of group VII ERF TFs in hypoxia responses has also been suggested (HRE1, HRE2 [Licausi et al., 2010], RAP2.2 [Hinz et al., 2010]) (Table 1). In Arabidopsis, two of the five members of group VII ERF are strongly induced by low-oxygen stress, whereas the other three are constitutively expressed (Table 1). However, plants that lack both HRE1 and HRE2 or RAP2.2 are still responsive to hypoxia, although the level of gene induction is reduced in comparison with the wild type (Hinz et al., 2010; Licausi et al., 2010). In addition, it was shown that RAP2.12 could bind the hypoxia-responsive promoter of ADH1 (Papdi et al., 2008). However, group VII ERFs are not restricted to mediating low-oxygen responses and are also involved in responses to drought, salt, or osmotic stress (Fukao et al., 2011; Park et al., 2011) or pathogen response (Ogawa et al., 2005), indicating the presence of a complex regulatory network.
Table 1.
Characteristics of Group VII ERFs in Arabidopsis Related to the Hypoxic Response and Degradation by the N-End Rule Pathway
| Gene Name | HRE1* | HRE2* | RAP2.3 | RAP2.2** | RAP2.12** |
| ATG code | At1g72360 | At2g47520 | At3g16770 | At3g14230 | At1g53910 |
| Control expression level (root/shoot)a | 187/103 | 45/93 | 408/4464 | 507/892 | 1114/574 |
| Hypoxic expression level (root/shoot)a | 7059/301 | 2647/1239 | 498/3051 | 554/950 | 750/403 |
| Presence of MC at N terminus | Yes | Yes | Yes | Yes | Yes |
| Protein degradation in vitrob | Yes | Yes | Yes | Yes | Yes |
| Protein degradation in planta | Yesb | Yesb | Not tested | Not tested | Not tested |
| Protein accumulation under hypoxia | Nobb | Yesb | Not tested | Not tested | Yesc |
| Effect of lower gene expression | Slightly lower induction of ADH activity in T-DNA insertion lined; lower induction of hypoxic genes in RNAie | unchanged induction of ADH activity in T-DNA insertion lined; decreased salt tolerancef | T-DNA insertion is less tolerant to hypoxia, lower ADH induction under hypoxiaj | ||
| Effect of overexpression | More tolerant to hypoxiabg; higher ADH expressiondg | More tolerant to hypoxiabg; unchanged gene expressiong; increased tolerance to floodingf; increased salt tolerancef | Higher expression of defense genes (PDF1.2, GST6)i | More tolerant to hypoxia, higher ADH induction under hypoxiaj; RAP2.2 protein does not accumulate, little gene expression changes under controlk | More tolerant to submergence, higher induction of hypoxic genesc |
| Interacting partnersh | At2g38490 (CIPK22, SnRK3.19) | ACBP2l, ACBP4m, OBF4n, TTL3, RCD1 | SINAT2 (Ring finger protein, At3g58040)k | ACBP1c, ACBP2c, At2g44040 | |
| Other remarks | mRNA induction inhibited by CHXg | mRNA induction not inhibited by CHXg | Involved in defense response?i | Induced by darknessj; involved in regulation of carotenoid biosynthesisk | |
| Intracellular localization | Nucleus (N-terminal tag)c | Nucleus (N-terminal tag)cf | Nucleus (N-terminal tag)i; Nucleus and cytosolm | Membrane (nucleus under hypoxia, C-terminal tag)c |
*An HRE1/HRE2 double T-DNA insertion line was less tolerant to hypoxia and showed lower induction of hypoxic genes (late response; Licausi et al., 2010). ** RAP2.2/2.12 amiRNA-plants (knockdown for both genes) showed reduced induction of hypoxia-responsive genes (Licausi et al., 2011a). CHX, cycloheximide; MC, N-terminal Met-Cys.
Data are expression values (RMA-normalized) from roots and shoots of Arabidopsis seedlings (Mustroph et al., 2009).
Arabidopsis Interactome Mapping Consortium, 2011, http://interactome.dfci.harvard.edu/A_thaliana/.
A MOLECULAR MECHANISM FOR OXYGEN SENSING: THE N-END RULE PATHWAY
Protein degradation has emerged as a common theme in the regulation of plant responses to stress and in developmental programs (Lyzenga and Stone, 2011). The results from two independent studies now suggest that the N-end rule pathway of protein degradation serves to regulate the low-oxygen response in plants (Gibbs et al., 2011; Licausi et al., 2011a). Both studies identified members of the group VII ERFs as substrates for this pathway. The oxygen-dependent stability of these ERFs in turn dictates the switching on or off of downstream signaling networks involved in the acclimation response to hypoxia. The major finding is that members of group VII ERFs, possessing a characteristic N-terminal motif (N-degron), are posttranslationally modified in an oxygen-dependent manner via the N-end rule pathway of protein degradation.
The N-end rule pathway degrades proteins that are characterized by a specific N-terminal amino acid, the type of amino acid determining whether a protein will be degraded by the proteasome (Graciet et al., 2009; Holman et al., 2009). Lys, His, and Arg are direct controllers of this pathway, defined as primary destabilizing residues, and target the protein to the proteasome by the UBR box-containing E3 ligase PROTEOLYSIS6 (PRT6). Other destabilizing residues, such as Phe, Trp, and Tyr, are recognized by another E3-ligase, PRT1 (Stary et al., 2003). Some amino acids (Asp and Glu) are referred to as secondary destabilizing residues, because their presence makes a protein accessible to modification by arginyl-tRNA:protein arginyltransferase (ATE1/2). This enzyme transfers an Arg residue to the target protein, thereby making it a substrate for PRT6. Furthermore, three tertiary destabilizing residues are known, Asn, Gln, and Cys. N-terminal Asn and Gln residues can be enzymatically transformed by deamidases into Asp or Glu, respectively, and can then be processed further by ATE1/2 and PRT6. There are at least three ways to produce a destabilizing N-terminal residue in proteins: 1) cleavage of proteins by endopeptidases in the cytosol, 2) removal of signaling or transit peptides, and 3) removal of the first Met residue by Met aminopeptidase. The latter occurs only if the second residue of a protein is small (Ala, Cys, Gly, Pro, Ser, Thr, Val) (Sriram et al., 2011).
Interestingly, proteins with a Cys residue at the second position can be processed by the N-end rule pathway in mammals and plants. Such a protein can become accessible to ATE1/2 after oxidation of the N-terminal Cys via an unknown mechanism and subsequently can be processed by the N-end rule pathway. The oxidation of Cys was suggested to be a potential sensor of the redox or oxygen status of a cell (Graciet et al., 2009; Holman et al., 2009). Cys oxidation was also observed for the mouse heterotrimeric G proteins RGS4 and RGS5, involved in the cardiovascular signaling pathway, which naturally contain Cys-2. It was shown that both proteins were degraded by the N-end rule pathway in an oxygen-dependent (Lee et al. 2005) and/or NO-dependent (Hu et al., 2005) manner.
Considering that a similar pathway operates in animals, and the fact that TFs mediating hypoxia responses contain the characteristic N-degron, it was perhaps only a matter of time before clues were translated into convincing evidence for the existence of a similar pathway in plants. In this context, it is interesting to note that although both studies arrived at similar results, the clues that led to these independent discoveries were different.
For Gibbs et al. (2011), the first hint for these exciting findings was the analysis of microarray data of Arabidopsis seeds lacking PRT6 (prt6). Surprisingly, these seeds showed higher expression of a set of genes that are regulated by low-oxygen stress in Arabidopsis (Liu et al., 2005; Loreti et al., 2005; Branco-Price et al., 2008; Mustroph et al., 2009; van Dongen et al., 2009). After this surprising finding, the high expression of hypoxia-inducible genes in both mutants was confirmed by microarray analysis of seedlings of prt6 as well as the other component of this pathway, ate1ate2 (Figure 1). This observation led to the assessment of putative substrates for the N-end rule pathway, namely proteins starting with Met-Cys. The Arabidopsis genome contains ~200 proteins starting with this motif, including the five members of the Arabidopsis group VII ERF family, of which three had been shown to be involved in low-oxygen stress regulation (Hinz et al., 2010; Licausi et al., 2010). These two observations led the authors to hypothesize that ERF TFs, especially those of the group VII subfamily, could be hypoxia sensors in plants.
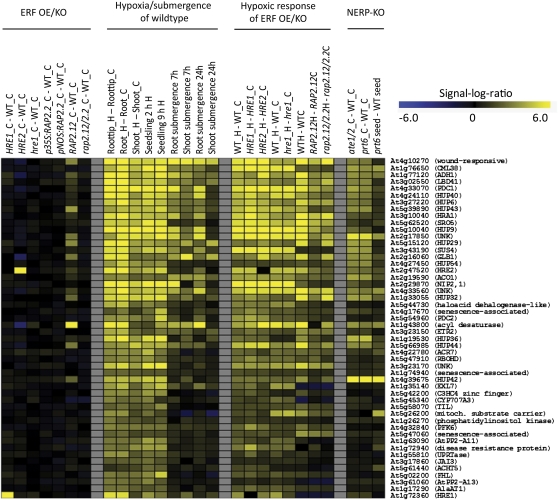
Figure 1.
Heatmap for Core Hypoxia Response Genes in Arabidopsis.
Expression data for the 49 core hypoxia response genes (Mustroph et al., 2009) and HRE1 (which is not included in the 49 core genes) under low-oxygen stress and in mutants for group VII ERF tfs or the N-end rule pathway. Data are signal-log ratios (stress versus control or mutant versus the wild type) of robust multiarray average–normalized expression data obtained from ATH1 microarray chips. Yellow color represents upregulation and blue color represents downregulation of the respective genes. Maximal color intensities refer to a signal-log ratio of ±6.0 (64-fold change). Data are from the following sources: HRE1, HRE2 overexpression (OE) (Licausi et al., 2010); hre1-RNA interference (RNAi) (Yang et al., 2011); RAP2.2 overexpression (Welsch et al., 2007); RAP2.12 overexpression and artificial micro RNA (amiRNA) (Licausi et al., 2011a); hypoxic response of root tips, roots, and shoots (Mustroph et al., 2009); hypoxic response of seedlings (Branco-Price et al., 2008); submergence response of roots and shoots (Lee et al., 2011); hypoxic response of HRE1 and HRE2 overexpressors (Licausi et al., 2010); hypoxic response of hre1-RNAi (Yang et al., 2011); hypoxic response of RAP2.12 overexpression and amiRNA (Licausi et al., 2011a); and knockouts (KOs) of N-end rule pathway genes PRT6 and ATE1/ATE2 (Gibbs et al., 2011). Abbreviations of gene names were taken from Mustroph et al. (2010). HUP, hypoxia-induced unknown protein; UNK, unknown protein not included in the list of HUP genes.
To prove this hypothesis, an extended study was performed to test the stability of group VII ERFs, namely the five Arabidopsis members of this group, HRE1, HRE2, RAP2.2, RAP2.3 (EBF), and RAP2.12. All of these TFs share a common N-terminal consensus sequence, including the Met-Cys initiating motif (MCGGAII). First, a heterologous rabbit reticulocyte lysate assay was used to test the stability of the five proteins and their modified versions containing a mutated Met-Ala motif. These in vitro experiments showed that all five proteins could be degraded via the proteasome when an N-terminal Cys was present, whereas the substitution of Cys with Ala inhibited protein degradation. Furthermore, the two proteins HRE1 and HRE2 were also tested in planta by overexpression, revealing protein degradation of the wild-type protein and stability of the mutated protein without an N-terminal Cys under aerated control conditions. In line with this finding, the wild-type protein was stabilized in the prt6 mutant, where the degradation of N-end rule pathway target proteins is inhibited. The final and most important experiment showed that hypoxic treatment reduced protein degradation of the wild-type HRE2 in ecotype Columbia-0 but not HRE1. Although HRE1 accumulation under hypoxia could not be demonstrated in planta, seedlings expressing N-terminally mutated versions of HRE1 and HRE2 had enhanced survival under hypoxic conditions, as previously shown for a transgenic line overexpressing the wild-type version of HRE1 (Licausi et al., 2010).
By contrast, the discovery of Licausi et al. (2011a) stems from observations that overexpressing the group VII ERF RAP2.12 causes a significant increase in the expression of hypoxia-responsive genes only under hypoxic conditions (Figure 1). This suggested that aerobic conditions prevented the action of RAP2.12. Interestingly, a lack of constitutive expression of ADH transcript and activity levels was observed previously in overexpression lines of HRE1, HRE2, and RAP2.2 (Hinz et al., 2010; Licausi et al., 2010). Licausi et al. (2011a) further observed that the RAP2.12 activity was no longer dependent on hypoxic conditions in plants overexpressing RAP2.12 with a modified or deleted N terminus. In these lines, a signature hypoxia-responsive gene expression profile was observed under aerobic conditions, thus linking the N terminus with the oxygen-dependent regulation of RAP2.12 activity.
The authors also convincingly demonstrated that RAP2.12 was important for survival under hypoxic conditions through its activity of inducing hypoxic-responsive gene expression. A higher percentage of RAP2.12 overexpressors survived submergence-associated hypoxia compared with the wild-type plants, explained by the faster and stronger induction of hypoxia-responsive genes. Transgenic knockdown lines of RAP2.12 and its close homolog RAP2.2 showed decreased induction of some hypoxia-responsive genes upon hypoxic treatment. In addition, the insertion of a hypoxia-responsive element from the At-HB1 promoter in front of the luciferase gene permitted its activation by RAP2.12. Combined with the observation that transcript stability of RAP2.12 was not affected by N-terminal modifications, the authors concluded that posttranslational modifications were responsible for the oxygen-dependent regulation of RAP2.12 activity.
Modification of the N-terminal region also affected the subcellular localization of RAP2.12. Wild-type RAP2.12 fused to green fluorescent protein localized at the plasma membrane under aerobic conditions, seemed to restrict itself to the nucleus under hypoxic conditions, and disappeared upon reoxygenation. Deletion of the conserved N-terminal residues of RAP2.12 resulted in both membrane and nuclear localization under aerobic conditions, thus explaining the hypoxia-responsive gene expression of these lines under aerobic conditions. Hypoxia resulted in the localization of all RAP2.12 fusion proteins to the nucleus. Interestingly, the fusion protein with a modified N terminus persisted in the nucleus, even upon reoxygenation.
Based on the conserved sequence of residues present in the N terminus of RAP2.12, the authors identified it as a possible substrate of the N-end rule pathway. This they successfully demonstrated in two ways. First, they showed increased stability of RAP2.12 in mutants lacking components of the N-end rule pathway (ate1ate2, prt6) under both aerobic and hypoxic conditions, in contrast with the oxygen-dependent lability of RAP2.12 in the wild type. Second, they found that replacing the critical N-terminal Cys with Ala, which modified the N-degron, caused RAP2.12 to be localized to the nucleus, independent of oxygen concentrations.
Considering the lack of hydrophobic domains in RAP2.12, Licausi et al. (2011a) also sought to find interacting partners that could sequester RAP2.12 at the plasma membrane. This sequestration would have an important function, because it would prevent the nuclear and cytoplasmic localization of RAP2.12 under aerobic conditions and thereby its degradation by the N-end rule pathway, which is active at these locations. Based on previous studies showing interactions between other group VII ERF members and Acyl CoA binding proteins (ACBPs) (Li and Chye, 2004; Li et al., 2008), the authors tested interactions of RAP2.12 with ACBP1 and ACBP2, the only two membrane-bound members of the Arabidopsis ACBP family. Using a combination of yeast two-hybrid and bimolecular fluorescence complementation, the authors established a positive interaction between RAP2.12 and ACBP1 and ACBP2.
Interestingly, both evaluations tested the survival of the N-end rule pathway mutants, prt6 and ate1ate2, in which levels of the oxygen-labile ERFs persist with contrasting results. Whereas Gibbs et al. (2011) found 7-d-old mutant seedlings grown on sugar-supplemented Murashige and Skoog plates to be more tolerant to hypoxia than wild-type seedlings, Licausi et al. (2011a) found a reduction in the tolerance of 5-week-old mutant plants grown on soil to submergence-associated hypoxia in darkness. Although the two experiments are not directly comparable because of differences in treatment (hypoxia versus complete submergence) and developmental stage (7-d-old seedling versus 5-week-old plants), the increased tolerance of mutant seedlings could be caused by the presence of Suc in the agar medium used. This would bypass the shortage of energy these seedlings would otherwise face. By comparison, the adult plants used in the submergence screen would have both a higher carbohydrate consumption rate and limited reserves.
The results of Licausi et al. (2011a) indicate that degradation of RAP2.12 upon reoxygenation is as important for survival as hypoxia-mediated stability of the protein. This is further corroborated by observations that transgenic lines expressing altered N-end versions of RAP2.12 that consequently persisted upon reoxygenation had significantly reduced survival compared with the wild-type plants or plants overexpressing the wild-type version of the protein. Degradation of RAP2.12 upon reoxygenation allows the molecular responses to hypoxia to be switched off. This would be critical for submerged plants to initiate molecular responses aimed at readjustment to the higher light and oxygen levels upon de-submergence and to recover growth and photosynthesis.
FUTURE PERSPECTIVES: THIS IS JUST THE BEGINNING
The identification of an oxygen-sensing mechanism in plants (Figure 2) is an important breakthrough and a very exciting finding, but it is just the beginning. The two studies have provided an excellent basis for future studies and further questions, and there is little doubt that the picture that emerges will be highly complex.
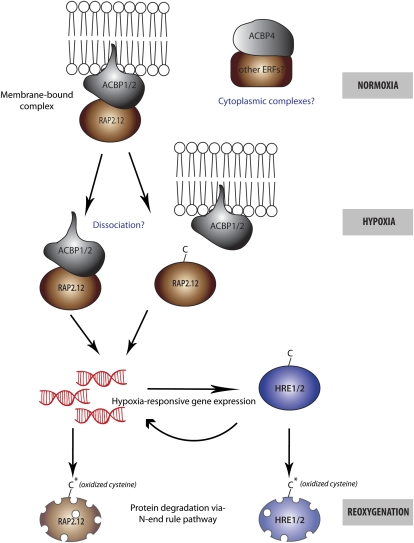
Figure 2.
An Oxygen-Sensing Mechanism in Plants Mediated by Group VII ERF TFs.
Under normoxic conditions, constitutively expressed ERFs, such as RAP2.12, form a complex with membrane-bound ACBPs. ACBP-ERF complexes could also occur in the cytoplasm. In such complexes, oxidation of the N-terminal Cys of these ERFs, which targets them for degradation by the N-end rule pathway, might be prevented. When oxygen concentrations fall, ERFs move to the nucleus to induce hypoxia-responsive gene expression. It is not clear whether hypoxia triggers ERF dissociation from ACBPs. Hypoxia also induces expression of other ERFs, such as HRE1 and HRE2, which could then induce their downstream target genes. Upon reoxygenation, the oxidation of N-terminal Cys of these ERFs would target them for degradation, and this would switch off the hypoxia response.
The Oxygen-Sensing Model
The oxidation of the terminal Cys of the N-degron could be mediated directly by oxygen-dependent enzymatic or chemical mechanisms or indirectly by changes in the cellular homeostasis that occur because of a decrease in O2 availability, such as changes in the cytosolic pH, increased calcium fluxes, changes in the redox state of the cell, and/or generation of ROS. If oxidation occurs enzymatically, the enzyme responsible (as yet unidentified) could qualify as a potential “oxygen sensor,” whereas group VII ERFs would be mediators of the oxygen sensing. To qualify as a bona fide oxygen sensor, the candidate protein must display activity that is directly proportional to oxygen availability. This would require a complete biochemical characterization of the protein mediating the oxidation of the terminal Cys, in particular, the dissociation constant for oxygen.
Both studies (Gibbs et al., 2011; Licausi et al., 2011a) used very low external O2 conditions to mimic hypoxia. However, under physiological conditions, oxygen concentrations typically vary between 1 and 21%. Furthermore, studies have shown that hypoxia-responsive gene expression is evident even upon mild decreases in oxygen availability (van Dongen et al., 2009), suggesting that activation of TFs controlling hypoxia-responsive gene expression might occur at concentrations much higher than 1%. It is therefore of interest to determine the concentrations of O2 at which ERF stabilization occurs and whether this varies for different ERF members and different organs and cell types. There is evidence that TFs associated with hypoxia-responsive gene expression are regulated in an oxygen concentration-dependent manner (Licausi et al., 2011b). Although all Arabidopsis group VII ERFs are possible substrates for the N-end rule pathway, it is likely that, depending on the oxygen concentration, different combination of TFs are active, leading to expression of specific target gene sets.
Furthermore, although HRE1 and HRE2 can be degraded via the N-end rule pathway, they are unlikely to be the primary oxygen-sensing mediators, because neither of these ERFs is constitutively expressed, and a primary sensor of ambient oxygen levels must be present before oxygen levels decrease. It can be speculated that constitutively expressed proteins, such as RAP2.12 and RAP2.2, switch on early hypoxia-response genes as oxygen concentrations decrease. Hypoxia also induces expression of secondary ERFs, such as HRE1 and HRE2. These would then induce expression of partly overlapping sets of downstream hypoxia-response genes. Upon reoxygenation, the oxygen lability of all these ERFs would ensure switching off of the hypoxia response, which has been demonstrated to negatively affect survival during hypoxic conditions (Licausi et al., 2011a) (Figure 2). Preliminary support for the hypothesis of a two-step regulatory system comes from the observation that hre1 hre2 knockout plants show decreased induction of hypoxic genes, particularly after longer durations of hypoxic stress, compared with wild-type plants (Licausi et al., 2010). Therefore, although Arabidopsis group VII ERFs are identified as potential oxygen-sensing mediators, it is unlikely that they activate hypoxia-responsive gene networks simultaneously. Extensive evidence exists in the literature establishing the differential regulation of group VII ERF family members by hormones and other factors (Büttner and Singh, 1997; Welsch et al., 2007; Hinz et al., 2010; Hess et al., 2011; Yang et al., 2011). Structural changes could also contribute to different sensitivities to oxygen availability (for example, HRE2 is shorter than the other group VII ERFs in a region downstream of the DNA binding domain) (Licausi et al., 2010). It is also possible that different oxygen concentrations invoke different group VII ERFs, depending on the responses they mediate.
The interaction of ERFs with ACBPs could be an important aspect of the oxygen-sensing mechanism. Complex formation with ACBPs in the membrane might be an important manner for the sequestration and protection of the protein from oxidation under aerobic conditions (Figure 1). In this context, the regulation of ACBP docking sites could be important for hypoxia tolerance. Furthermore, it will be interesting to note whether the interaction with membrane-bound ACBPs is a common theme among the other group VII ERFs. A complex formation with cytosolic ACBPs (ACBP4, ACBP5) cannot be ruled out, considering evidence demonstrating association of these ACBPs with another ethylene-responsive binding protein, RAP2.3/AtEBP (AT3G16770) (Li et al., 2008). Although Licausi et al. (2011a) suggested that RAP2.12 dissociates from ACBPs upon hypoxia, this was inconclusive based on the evidence presented. It is possible that RAP2.12 travels in a complex to the nucleus (Figure 2). If dissociation occurs, it will be interesting to unravel how hypoxia might trigger this process.
A further open question is the binding of the group VII TFs to hypoxia-inducible promoters. Although this has been shown for RAP2.12 binding to the ADH1 promoter (Papdi et al., 2008) and the HB1 promoter (Licausi et al., 2011a), the remaining ERFs have not been shown to bind to hypoxic promoters. Furthermore, the hypoxia-responsive element has not been identified. Although the authors suggest the motif ATCTA to be the binding site, this motif alone is unlikely to be specific for the hypoxic response for several reasons. First, the motif occurs in the −500 bp promoter region of more than 15,000 Arabidopsis genes. Second, the motif was also enriched in promoter regions of carotenogenesis genes (Welsch et al., 2007). Therefore, extensive promoter studies are needed to fully understand the complex regulatory network mediating hypoxia responses.
Variation in Hypoxia Tolerance
A long-standing question in the field of plant anaerobiosis has been to identify how sensing, signaling, and response mechanisms vary between hypoxia-tolerant and -intolerant genotypes. In the context of oxygen sensing, this is still an open question. The results presented by Gibbs et al. (2011) provide some clues in this direction. Most of the rice group VII ERFs that mediate adaptive responses to flooding contain the N-terminal Met-Cys motif (e.g., SUB1-A, SUB1-B, SK1, SK2, but not SUB1-C). However, despite the presence of this motif in SUB1-A, it was not degraded by the N-end rule pathway when assessed in the reticulocyte lysate system (Gibbs et al., 2011). The authors suggest that the resistance of SUB1-A, present only in flood-tolerant rice cultivars (Xu et al., 2006), to degradation by the N-end rule pathway could enhance hypoxia responses. It will be interesting to determine whether other rice TFs from this group, including sequences from different cultivars, are also regulated by this pathway and whether specific structural changes allow some proteins to evade degradation by the N-end rule pathway. Such studies, both within and between plant species, will also provide deeper insight into the variation in the susceptibility of group VII ERFs to oxygen-dependent degradation and the relation therein to hypoxia tolerance. In addition, the timing and the magnitude of responses to low oxygen could impart crucial distinctions in the ability to successfully survive prolonged hypoxia.
In some situations, plants can be warned of impending low-oxygen conditions before these conditions set in. For example, when plants are submerged, the gaseous plant hormone ethylene accumulates to high concentrations within the plant organs well before oxygen levels drop seriously. In the flooding-tolerant species Rumex palustris, a RAP2.12 ortholog was rapidly upregulated upon submergence before the onset of hypoxia (Licausi et al., 2011a). Furthermore, preliminary results suggest that this upregulation is mediated by ethylene. It is possible that submergence-tolerant plants, such as R. palustris, use hormonal regulators, such as ethylene, to increase levels of oxygen-sensing proteins. Sequestering more of these proteins in complexes at the membrane when oxygen conditions are not yet limiting might allow a faster and stronger response to hypoxia and increase chances of survival. Thus, signals like ethylene could act as priming mechanisms for activation of hypoxia responses. In fact, the expression of HRE1 and RAP2.2 is partially regulated by ethylene in Arabidopsis (Hinz et al., 2010; Hess et al., 2011; Yang et al., 2011).
In addition, although group VII ERFs are essential to survival and activate a core set of hypoxia response genes, the activation of this core set is conserved across species regardless of tolerance levels (Mustroph et al., 2010) and most likely does not determine the variation in hypoxia tolerance. The identification of downstream signaling networks is therefore of great importance to identify mechanisms mediating variation in hypoxia tolerance.
Acknowledgments
R.S. received funding from the Centre for Biosystems Genomics 2012, and A.M. received funding from Deutsche Forschungsgemeinschaft project MU 2755/4-1.
AUTHOR CONTRIBUTIONS
R.S. and A.M. contributed equally to the writing of this article.
References
- Abbruzzetti S., Faggiano S., Spyrakis F., Bruno S., Mozzarelli A., Astegno A., Dominici P., Viappiani C. (2011). Oxygen and nitric oxide rebinding kinetics in nonsymbiotic hemoglobin AHb1 from Arabidopsis thaliana. IUBMB Life 63: 1094–1100 [DOI] [PubMed] [Google Scholar]
- Akimoto S., Tanaka A., Nakamura K., Shiro Y., Nakamura H. (2003). O2-specific regulation of the ferrous heme-based sensor kinase FixL from Sinorhizobium meliloti and its aberrant inactivation in the ferric form. Biochem. Biophys. Res. Commun. 304: 136–142 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W. (1979). Aeration in higher plants. Adv. Bot. Res. 7: 225–332 [Google Scholar]
- Armstrong W., Webb T., Darwent M., Beckett P.M. (2009). Measuring and interpreting respiratory critical oxygen pressures in roots. Ann. Bot. (Lond.) 103: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M.H., Trivedi P.K., Misra P., Nath P. (2009). Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Funct. Integr. Genomics 9: 525–535 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Chang R. (2005). Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. (Lond.) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Voesenek L.A.C.J. (2008). Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A., Yang Z.B., Springer P.S., Bailey-Serres J. (2002). RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Bond D.M., Wilson I.W., Dennis E.S., Pogson B.J., Jean Finnegan E. (2009). VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J. 59: 576–587 [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn C., Mazure N., Pouysségur J. (2005). Signalling via the hypoxia-inducible factor-1alpha requires multiple posttranslational modifications. Cell. Signal. 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K.A., Jang C.J.H., Larive C.K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Bruick R.K. (2003). Oxygen sensing in the hypoxic response pathway: Regulation of the hypoxia-inducible transcription factor. Genes Dev. 17: 2614–2623 [DOI] [PubMed] [Google Scholar]
- Büttner M., Singh K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.A., Llewellyn D.J., Dennis E.S., Wilson I.W. (2010). Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal. Behav. 5: 1006–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.A., Wilson I.W., Llewellyn D.J., Dennis E.S. (2009). The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.J., Ferl R.J. (1999). Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121: 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten N.C., Ferl R.J. (1995). Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J. 7: 589–601 [DOI] [PubMed] [Google Scholar]
- Dordas C., Hasinoff B.B., Igamberdiev A.U., Manac’h N., Rivoal J., Hill R.D. (2003a). Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. Plant J. 35: 763–770 [DOI] [PubMed] [Google Scholar]
- Dordas C., Rivoal J., Hill R.D. (2003b). Plant haemoglobins, nitric oxide and hypoxic stress. Ann. Bot. (Lond.) 91(Spec No): 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew M.C. (1997). Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 223–250 [DOI] [PubMed] [Google Scholar]
- Fukao T., Bailey-Serres J. (2004). Plant responses to hypoxia—is survival a balancing act? Trends Plant Sci. 9: 449–456 [DOI] [PubMed] [Google Scholar]
- Fukao T., Xu K., Ronald P.C., Bailey-Serres J. (2006). A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Isa N.M., Gramuglia S., Fukao T., Bassel G.W., Correia C.S., Corbineau F., Theodoulou F.L., Bailey-Serres J., Holdsworth M.J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez M.A., Gonzalez G. (2004). Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96: 774–783 [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez M.A., Gonzalez G. (2005). Heme-based sensors: Defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99: 1–22 [DOI] [PubMed] [Google Scholar]
- Graciet E., Walter F., Maoiléidigh D.O., Pollmann S., Meyerowitz E.M., Varshavsky A., Wellmer F. (2009). The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. USA 106: 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Crack J.C., Thomson A.J., LeBrun N.E. (2009). Bacterial sensors of oxygen. Curr. Opin. Microbiol. 12: 145–151 [DOI] [PubMed] [Google Scholar]
- Haddad J.J. (2004). Oxygen sensing and oxidant/redox-related pathways. Biochem. Biophys. Res. Commun. 316: 969–977 [DOI] [PubMed] [Google Scholar]
- Hattori Y., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- He C.J., Morgan P.W., Drew M.C. (1996). Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiol. 112: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess N., Klode M., Anders M., Sauter M. (2011). The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol. Plant. 143: 41–49 [DOI] [PubMed] [Google Scholar]
- Hinz M., Wilson I.W., Yang J., Buerstenbinder K., Llewellyn D., Dennis E.S., Sauter M., Dolferus R. (2010). Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren F.U., Dolferus R., Wu Y.R., Peacock W.J., Dennis E.S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman T.J., et al. (2009). The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R.G., Sheng J., Qi X., Xu Z., Takahashi T.T., Varshavsky A. (2005). The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437: 981–986 [DOI] [PubMed] [Google Scholar]
- Huang S., Colmer T.D., Millar A.H. (2008). Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci. 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Hunt P.W., Klok E.J., Trevaskis B., Watts R.A., Ellis M.H., Peacock W.J., Dennis E.S. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 17197–17202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.W., Chen P.W., Lu C.A., Chen S., Ho T.H., Yu S.M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee M.J., Tasaki T., Moroi K., An J.Y., Kimura S., Davydov I.V., Kwon Y.T. (2005). RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 102: 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Mustroph A., Sasidharan R., Vashisht D., Pedersen O., Oosumi T., Voesenek L.A., Bailey-Serres J. (2011). Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Lee T.G., Jang C.S., Kim J.Y., Kim D.S., Park J.H., Kim D.Y., Seo Y.W. (2007). A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 129: 375–385 [Google Scholar]
- Li H.Y., Chye M.L. (2004). Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54: 233–243 [DOI] [PubMed] [Google Scholar]
- Li H.Y., Xiao S., Chye M.L. (2008). Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 59: 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F., Kosmacz M., Weits D.A., Giuntoli B., Giorgi F.M., Voesenek L.A.C.J., Perata P., van Dongen J.T. (2011a). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F., van Dongen J.T., Giuntoli B., Novi G., Santaniello A., Geigenberger P., Perata P. (2010). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Licausi F., Weits D.A., Pant B.D., Scheible W.R., Geigenberger P., van Dongen J.T. (2011b). Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190: 442–456 [DOI] [PubMed] [Google Scholar]
- Liu F.L., Vantoai T., Moy L.P., Bock G., Linford L.D., Quackenbush J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Poggi A., Novi G., Alpi A., Perata P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga W.J., Stone S.L. (October 20, 2011). Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. http://dx.doi.org/10.1093/jxb/err310 [DOI] [PubMed]
- Mattana M., Vannini C., Espen L., Bracale M., Genga A., Marsoni M., Iriti M., Bonazza V., Romagnoli F., Baldoni E., Coraggio I., Locatelli F. (2007). The rice Mybleu transcription factor increases tolerance to oxygen deprivation in Arabidopsis plants. Physiol. Plant. 131: 106–121 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Rocha M., Geigenberger P., Whelan J., van Dongen J.T. (2011). Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 190: 472–487 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pan L., Kawai-Yamada M., Yu L.H., Yamamura S., Koyama T., Kitajima S., Ohme-Takagi M., Sato F., Uchimiya H. (2005). Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 138: 1436–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Abrahám E., Joseph M.P., Popescu C., Koncz C., Szabados L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.Y., Seok H.Y., Woo D.H., Lee S.Y., Tarte V.N., Lee E.H., Lee C.H., Moon Y.H. (2011). AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem. Biophys. Res. Commun. 414: 135–141 [DOI] [PubMed] [Google Scholar]
- Reggiani R. (2006). A role for ethylene in low-oxygen signaling in rice roots. Amino Acids 30: 299–301 [DOI] [PubMed] [Google Scholar]
- Reggiani R., Laoreti P. (2000). Evidence for the involvement of phospholipase C in the anaerobic signal transduction. Plant Cell Physiol. 41: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Rolletschek H., Weschke W., Weber H., Wobus U., Borisjuk L. (2004). Energy state and its control on seed development: Starch accumulation is associated with high ATP and steep oxygen gradients within barley grains. J. Exp. Bot. 55: 1351–1359 [DOI] [PubMed] [Google Scholar]
- Sowa A.W., Duff S.M.G., Guy P.A., Hill R.D. (1998). Altering hemoglobin levels changes energy status in maize cells under hypoxia. Proc. Natl. Acad. Sci. USA 95: 10317–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Guest J.R. (1990). FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6: 399–428 [DOI] [PubMed] [Google Scholar]
- Sriram S.M., Kim B.Y., Kwon Y.T. (2011). The N-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 12: 735–747 [DOI] [PubMed] [Google Scholar]
- Stary S., Yin X.J., Potuschak T., Schlögelhofer P., Nizhynska V., Bachmair A. (2003). PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 133: 1360–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Bush D.S., Sachs M.M. (1998). Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol. 118: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.L., Zhulin I.B. (1999). PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J., Rolletschek H., Friedel S., Lunn J.E., Nguyen T.H., Feil R., Tschiersch H., Müller M., Borisjuk L. (2011). Seed-specific elevation of non-symbiotic hemoglobin AtHb1: Beneficial effects and underlying molecular networks in Arabidopsis thaliana. BMC Plant Biol. 11: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Nakazono M., Saisho D., Tsutsumi N., Hirai A. (2000). Transcript levels of the nuclear-encoded respiratory genes in rice decrease by oxygen deprivation: Evidence for involvement of calcium in expression of the alternative oxidase 1a gene. FEBS Lett. 471: 201–204 [DOI] [PubMed] [Google Scholar]
- van Dongen J.T., Fröhlich A., Ramírez-Aguilar S.J., Schauer N., Fernie A.R., Erban A., Kopka J., Clark J., Langer A., Geigenberger P. (2009). Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann. Bot. (Lond.) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S.M., et al. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Vlad F., Spano T., Vlad D., Daher F.B., Ouelhadj A., Fragkostefanakis S., Kalaitzis P. (2007). Involvement of Arabidopsis prolyl 4 hydroxylases in hypoxia, anoxia and mechanical wounding. Plant Signal. Behav. 2: 368–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Maass D., Voegel T., Dellapenna D., Beyer P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang C.Y., Hsu F.C., Li J.P., Wang N.N., Shih M.C. (2011). The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 156: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]