This massive analysis of rice small RNAs evaluated annotated microRNAs (miRNAs) and identified new miRNAs and miRNAs regulated by environmental stresses. Of particular interest are miRNA family members with distinct sequences and organ-preferential expression; some of these guide differential target cleavage and provide new insight about how an agriculturally significant phenotype may be regulated.
Abstract
Small RNAs have a variety of important roles in plant development, stress responses, and other processes. They exert their influence by guiding mRNA cleavage, translational repression, and chromatin modification. To identify previously unknown rice (Oryza sativa) microRNAs (miRNAs) and those regulated by environmental stress, 62 small RNA libraries were constructed from rice plants and used for deep sequencing with Illumina technology. The libraries represent several tissues from control plants and plants subjected to different environmental stress treatments. More than 94 million genome-matched reads were obtained, resulting in more than 16 million distinct small RNA sequences. This allowed an evaluation of ~400 annotated miRNAs with current criteria and the finding that among these, ~150 had small interfering RNA–like characteristics. Seventy-six new miRNAs were found, and miRNAs regulated in response to water stress, nutrient stress, or temperature stress were identified. Among the new examples of miRNA regulation were members of the same miRNA family that were differentially regulated in different organs and had distinct sequences Some of these distinct family members result in differential target cleavage and provide new insight about how an agriculturally important rice phenotype could be regulated in the panicle. This high-resolution analysis of rice miRNAs should be relevant to plant miRNAs in general, particularly in the Poaceae.
INTRODUCTION
Plants have evolved sophisticated ways to adapt to various environmental stresses, such as drought, salt, extreme temperatures, and nutrient starvation. To sense, respond, and acclimate to these stressful environments, plants developed cascades of molecular networks. A large proportion of plant genes are regulated in response to stressful environments, indicating that the gene regulation mechanisms involved represent a major component of these molecular networks. Although the regulation of gene expression is largely studied at the transcriptional level, the importance of posttranscriptional regulation has been emphasized with the discovery of small RNAs.
Small RNAs are noncoding RNAs that regulate gene expression by both transcriptional and posttranscriptional silencing mechanisms in a wide variety of eukaryotic organisms (Carrington and Ambros, 2003; Mallory and Vaucheret, 2004; Carthew and Sontheimer, 2009). MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are two major types of these small RNAs. Although miRNAs and siRNAs are similar in size (20 to 24 nucleotides), their biogenesis and modes of action are markedly different. Generally, siRNAs, such as heterochromatic siRNAs, are derived from double-stranded RNAs generated by RNA-dependent RNA polymerases and are involved mainly in the modulation of transcriptional activity by directing DNA methylation and chromatin remodeling (Lippman and Martienssen, 2004; Malone and Hannon, 2009). miRNAs originate from distinct genomic loci in which miRNA genes are transcribed by RNA polymerase II into single-stranded precursor transcripts that form stem-loop secondary structures that often have an imperfectly double-stranded characteristic. These are processed to generate an miRNA-miRNA* duplex by DICER-LIKE (DCL) proteins (Park et al., 2002; Kurihara and Watanabe, 2004). The functional strand of this duplex, the miRNA, becomes associated with an Argonaute (AGO) protein in an RNA-induced silencing complex (Bohmert et al., 1998; Vaucheret, 2008). The other passenger strand, the miRNA*, is usually nonfunctional and released or degraded. The RNA-induced silencing complex recognizes mRNAs that contain sequences complementary to the miRNA and targets them for posttranscriptional regulation by cleavage and/or translational inhibition (Llave et al., 2002; Brodersen et al., 2008).
Plant miRNAs play important roles in development, responses to abiotic and biotic stress, and other processes. The functional analysis of miRNAs has become particularly prominent for the study of development, such as in organ boundary formation, organ polarity/radial patterning, and development of root, stem, leaf, and floral organs (Jones-Rhoades et al., 2006). Differential accumulation of miRNAs in different tissues is common, and many miRNAs target development-associated mRNAs encoding transcription factors (Rhoades et al., 2002; Chen, 2009). Moreover, developmental defects are associated with miRNA biogenesis mutants (Jacobsen et al., 1999; Vaucheret et al., 2004). Association of miRNAs with environmental stresses was also shown using miRNA metabolism mutants. For example, an Arabidopsis thaliana mutant with defects in HYPONASTIC LEAVES1 (HYL1), a double-stranded RNA binding protein involved in miRNA biogenesis, has increased sensitivity to the stress hormone abscisic acid (ABA) (Lu and Fedoroff, 2000). In addition, dcl1 and hua enhancer 1 (hen1) mutants are also more susceptible to ABA, salt, and osmotic stresses (Zhang et al., 2008). HEN1 is a methyltransferase that adds a 2′-O-methyl group on the 3′-terminal nucleotide of plant miRNAs and siRNAs to protect them against degradation (Chen et al., 2002; Huang et al., 2009b). The phenotypic effect of these mutants suggests that some miRNAs processed by DCL1, HYL1, and HEN1 have important roles in environmental stresses. miRNA gene expression studies also have provided the evidence of association with stress responses. They reported specific miRNAs showing altered expression patterns under environmental stresses such as cold, drought, salt, UV-B radiation, oxidative stress, or mechanical stress (Sunkar and Zhu, 2004; Sunkar et al., 2007; Zhou et al., 2007; Lu et al., 2008b; Zhou et al., 2008; Li et al., 2011). In addition, it has also been documented that several miRNAs are associated with nutrient homeostasis under phosphate or sulfate depletion conditions (Jones-Rhoades and Bartel, 2004; Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006; Hsieh et al., 2009; Lundmark et al., 2010; Nilsson et al., 2010). Although most of these observations were made in Arabidopsis, a few studies also reported stress-related miRNAs in rice (Oryza sativa; Bari et al., 2006; Zhao et al., 2007, 2009a; Sanan-Mishra et al., 2009; Jian et al., 2010; Li et al., 2011). However, genome-wide analysis of rice small RNA data has not been successful for the identification of new abiotic stress–regulated miRNAs. This may be because such miRNAs are not abundant and require deeper data sets.
At the time of this study, 1687 genes encoding miRNAs had been registered in miRBase (release 14) from 22 species of plants, including Chlamydomonas reinhardtii, moss (Physcomitrella patens), loblolly pine (Pinus taeda), Arabidopsis, Medicago truncatula, poplar (Populus spp), maize (Zea mays), and rice (Griffiths-Jones et al., 2008). Of these, 417 rice miRNA genes have been reported on the basis of cloning experiments and/or computational prediction. The computational prediction was mostly based on the conservation with other plants, especially with Arabidopsis (Reinhart et al., 2002). Traditional cloning and sequencing of small RNAs confirmed the conserved miRNAs as well as prompted the annotation of new miRNAs (Liu et al., 2005; Sunkar et al., 2005; Luo et al., 2006). More recently, next-generation deep sequencing technologies have been used to provide quantitative information and enrich the set of annotated rice miRNAs (Lu et al., 2008a; Sunkar et al., 2008; Zhu et al., 2008; Xue et al., 2009). However, given that the approaches used to identify rice miRNAs range from prediction to validation of target cleavage, it is not surprising that some miRNAs annotated in miRBase do not fit currently accepted criteria (Meyers et al., 2008). Distinguishing annotated miRNAs with poor fit to accepted annotation criteria from those that are likely bona fide would help prioritize miRNAs for more detailed studies.
A number of plant miRNA genes produce mature miRNAs with the same or similar sequences, and they are grouped into miRNA families. In rice, the 417 miRNA genes mentioned above generate 274 distinct mature miRNAs and fall into 144 miRNA families. Since miRNAs in the same family are often predicted to target the same or overlapping sets of genes, it has been assumed that these may act in a functionally redundant manner (Jones-Rhoades et al., 2006). Recently, however, deep sequencing and miRNA precursor expression studies have helped reveal that distinct family members may have sequence variations and differential expression patterns to facilitate functional specialization. Small RNA profiling with deep sequencing can distinguish members of miRNA families at single-nucleotide resolution, a difficult task with other methods, such as RNA gel blot or microarray analysis. The sequence variations include nucleotide sequence polymorphisms as well as differences in the length of mature miRNAs.
One example arising from next-generation sequencing approaches is the case of Arabidopsis miR159 and miR319, considered members of a single family since they share sequence identity at 17 of 21 nucleotides and have 5′ ends that are offset by a single nucleotide. Their sequence variations are sufficient to cause their regulation of different targets: miR159 regulates MYB mRNAs, whereas miR319 predominantly acts on TCP mRNAs (Palatnik et al., 2007). Interestingly, the identity of the first 5′ nucleotide of the miRNAs was proposed as a major determinant for AGO protein association (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008; Wu et al., 2009; Havecker et al., 2010). This implies that a one nucleotide difference at this position among miRNA family members is enough to make them functionally distinct because of their interaction with different AGO complexes. Recently, miRNA sequence length variation was also reported to have dramatic effects on miRNA functions. Whereas canonical 21-nucleotide miRNAs play a major role in posttranscriptional regulation by guiding target RNA cleavage, 22-nucleotide miRNAs have the additional function of triggering secondary siRNA production from target RNAs (Chen et al., 2010; Cuperus et al., 2010b). In addition, miRNAs 24 nucleotides in length have been proposed to direct DNA methylation at target loci (Wu et al., 2010). In plants, most of what we know about miRNA sequence variants beyond sequence analysis comes from work in Arabidopsis. Although regulation of targets by rice miRNAs was recently investigated at the genomic level with a technique called parallel analysis of RNA ends (German et al., 2008) by three groups (Wu et al., 2009; Li et al., 2010; Zhou et al., 2010), these studies did not address the cleavage specificity of rice miRNA family members with different sequences.
In Arabidopsis, differential expression patterns of miRNA family members have been reported at the precursor level in limited examples. For instance, expression patterns of MIR319a, MIR319b, and MIR319c were analyzed using promoter:β-glucuronidase fusions and revealed that MIR319 genes exhibit largely nonoverlapping spatial expression patterns in seedlings and inflorescences (Nag et al., 2009). Although there are no significant sequence differences among the mature miRNAs produced from the three miR319 loci in Arabidopsis, the distinct expression patterns of their genes suggest that they may have unique developmental functions. Consistent with this idea, a single mutation of miR319a impaired petal growth and development. In addition, analysis of the expression of miR169 precursors showed that out of 10 detectable precursors, only two were substantially downregulated by drought stress in Arabidopsis (Li et al., 2008). Although these examples are intriguing, differential expression patterns of mature miRNA family members has not been studied well, especially in rice.
In this study, we applied high-throughput sequencing to profile small RNAs and to annotate miRNA genes from rice. Our data provide insight into the authenticity of previously reported rice miRNAs and identify new miRNAs that are associated with diverse roles in rice development and environmental stress responses. Moreover, among the new cases of miRNA regulation were miRNA family members exhibiting differential accumulation in different organs that impact target selection and cleavage. Our findings related to miR529 are particularly intriguing and add new insight about how an agriculturally significant phenotype may be regulated.
RESULTS
Construction of Small RNA Libraries from Environmental Stress–Treated Rice Plants
To obtain a genome-wide comprehensive survey of miRNAs in rice, small RNA libraries were constructed from seedling, root, shoot, and panicle tissues and from those tissues treated with various environmental and nutrient stresses. In addition, seedlings and panicles from DCL1 RNA interference (RNAi) plants were also used to construct small RNA libraries (Liu et al., 2005). In total, 33 types of bar-coded libraries were constructed and grouped into five pools containing six to eight libraries each, and then each pool was subjected to high-throughput sequencing. The sequencing reads were sorted into each library using a 2-nucleotide barcode located at the 3′ end of the 5′ adaptor. Moreover, 31 additional libraries were constructed from selected biological replicates and then sequenced individually. In total, a set of 62 libraries was sequenced.
As shown in Supplemental Table 1 online, after trimming the adaptor sequences, sequencing reads of the multiplexed, bar-coded libraries ranged from 257,241 to 923,759, and those of individually sequenced libraries varied from 1,680,275 up to 5,747,232. In all, 131,121,802 reads were obtained from the 62 libraries and corresponded to 37,633,754 distinct sequences (see Supplemental Table 1 online). Out of these, 94,774,538 reads (72.3%), represented by 16,574,511 distinct sequences, perfectly matched the rice genome. Reads matching structural noncoding RNAs, such as rRNA, tRNA, snRNA (small nuclear RNA), and snoRNA (small nucleolar RNA), corresponded to 12.8% of genome matches (12,092,369 reads). After removal of sequences that did not map to the rice genome or that matched to noncoding structural RNAs, the abundance of each sequence in a library was normalized to transcripts per 2 million (TP2M) to represent the relative cloning frequency in each library.
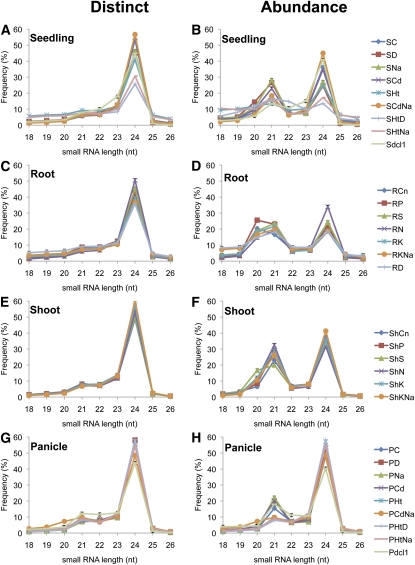
The length of the small RNA sequences ranged from 18 to 26 nucleotides as shown in Figure 1. When the distinct sequences were examined, the patterns of all the libraries were nearly identical. The 24-nucleotide small RNAs were dominant in all cases, indicating that this size class is rich in sequence diversity. In terms of total sequence abundance, there were two major peaks at 21 and 24 nucleotides, consistent with DCL cleavage products. The size distribution of small RNAs was determined more by the tissue than the stress treatment. Whereas the proportion of 21-nucleotide small RNAs among libraries was similar for different tissues, consisting of 18% in panicles, 19% in seedlings, 20% in roots, and 27% in shoots, the proportion of 24-nucleotide small RNAs varied more substantially among the different tissues, comprising 23% in roots, 32% in seedlings, 35% in shoots, and 48% in panicles. In Arabidopsis, the distribution of 24-nucleotide small RNAs is ~43% in leaves and 61% in inflorescences (Kasschau et al., 2007). In plants, the 24-nucleotide small RNAs consist mainly of siRNAs that are associated with repeats and transposons. The high level of 24-nucleotide small RNAs in both Arabidopsis inflorescences and rice panicles compared with those in vegetative tissues may suggest that reproductive tissues need more diverse repression of these elements.
Figure 1.
Small RNA Size Distributions from the Libraries of Different Tissues.
Small RNA size profiles are grouped by tissue as indicated. The size of small RNAs was plotted versus frequency (percentage) among distinct sequences ([A], [C], [E], and [G]) or total sequences ([B], [D], [F], and [H]). Seedling, root, and shoot tissues were harvested at the 2-week-old stage. Panicle was harvested at a mature stage when the panicles were ~15 cm in length. Library codes and detailed information about each library are indicated in Supplemental Table 1 online.
Unexpectedly, an additional peak at 20 nucleotides was observed in all the root libraries. This peak has not been reported previously, probably because root-derived small RNAs have not been examined in detail. Further analysis revealed that the majority of these 20-nucleotide sequences originated from several highly abundant 20-nucleotide versions of root-preferential miRNAs, such as miR156, 159, 395.2, 435, and miR3979 (see Supplemental Figure 1 online). In Arabidopsis, the dcl1-7 mutant exhibited a lower proportion of 21-nucleotide small RNAs compared with the wild type, and most of this difference can be attributed to a substantial reduction in known miRNAs (Lu et al., 2006). However, RNAi lines suppressing the function of Os-DCL1 did not show a dramatic decrease in 21-nucleotide small RNAs. When the expression level of individual miRNAs was compared between Os-DCL1 RNAi lines and the wild type, most miRNAs were decreased in the RNAi lines (see Supplemental Figure 2 online). This implies that considerable amounts of 21-nucleotide small RNAs in rice are from DCL1-independent siRNAs.
Evaluation of Annotated miRNAs
Previous computational prediction and cloning studies reported 417 rice miRNA loci that produce 274 distinct mature miRNAs, as registered in miRBase release 14.0 (Reinhart et al., 2002; Wang et al., 2004; Arazi et al., 2005; Liu et al., 2005; Sunkar et al., 2005, 2008; Luo et al., 2006; Lu et al., 2008a; Morin et al., 2008; Zhu et al., 2008; Huang et al., 2009a; Johnson et al., 2009; Xue et al., 2009). After removing 11 double-registered miRNA genes in the same loci, 406 miRNA loci were analyzed. Of these, 140 loci generate miRNAs that are conserved between Arabidopsis and rice. Since some rice miRNAs are registered solely based on computational prediction or with little evidence of expression, their authenticity is not certain (Lu et al., 2008a; Zhu et al., 2008; Wu et al., 2009).
To evaluate reported rice miRNAs, we adopted recently proposed criteria for plant miRNAs (Meyers et al., 2008). Briefly, it was proposed that processing of a miRNA stem-loop precursor by DCL enzymes is expected to give rise predominantly to an miRNA with a lower frequency of the miRNA* and possibly a low frequency of small RNAs from elsewhere in the precursor. In addition, due to the single-stranded precursor, the small RNAs from a miRNA locus should have a strong strand bias.
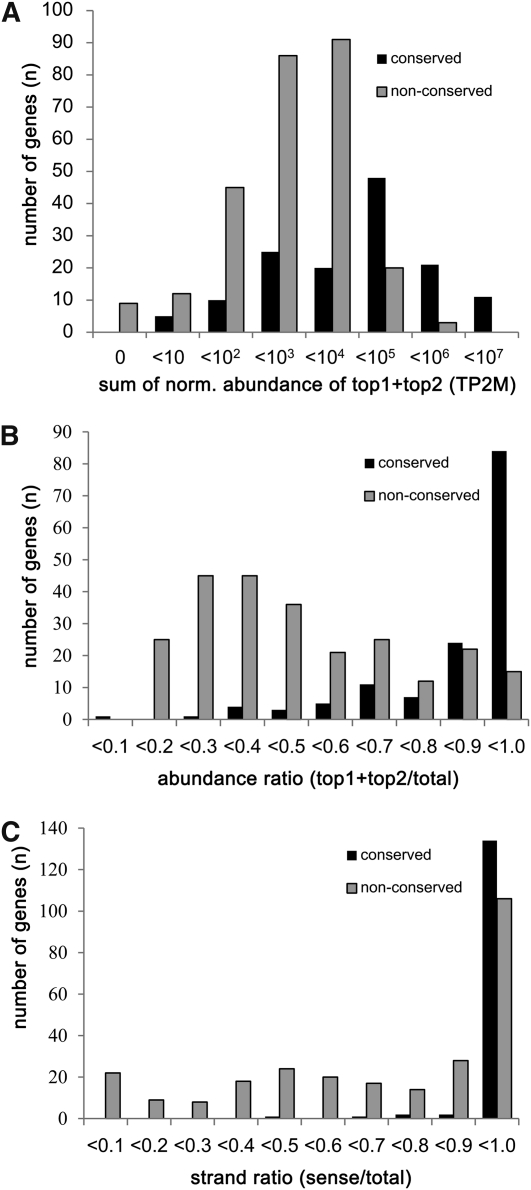
We first examined the sequencing frequency of the two most abundant small RNA sequences matching each miRNA locus (Figure 2A). Sequencing frequency was calculated as the sum of the normalized abundance from all 62 libraries. Whereas nine miRNA precursors, from miR413, miR418, miR426, miR445e, miR445g, miR1424, miR1435, miR2099, and miR2105, did not have any matching small RNAs, the others showed a range of abundance levels. The highest was miR168a, for which the sum of normalized abundance was ~16 M for the two most abundant sequences. Since 95% (133/140) of the conserved miRNA loci had sequencing frequencies of >20 for the two most abundant matching small RNAs (see Supplemental Table 2 online), we designated 41 miRNA precursors with abundances ≤20 as lowly expressed loci. Some of these had been previously annotated based solely on computational predictions or were identified from developing seeds that are not included in this study.
Figure 2.
Evaluation of 414 of Known miRNA Genes from Rice.
Known rice miRNA genes were grouped based on the conservation between rice and Arabidopsis. Sum of normalized abundance is from all the libraries. Histograms represent number of genes in each abundance range.
(A) The sum of normalized abundance is represented for the two most abundant small RNAs from each miRNA gene.
(B) The ratio of sum of the normalized abundance of the two most abundant small RNAs and all the small RNAs matching each miRNA gene was calculated.
(C) The sum of the abundance of small RNAs matching sense strands was divided by that matching both strands to calculate strand ratio for each miRNA gene.
Next, we sought to examine whether a few abundant small RNAs predominate in each miRNA locus or whether the abundance is distributed among a larger number of small RNAs (Figure 2B). To this end, the ratio of the abundance between two most abundant small RNAs and all the small RNAs matching each miRNA locus was assessed. Among conserved miRNA precursors, 95% had an abundance ratio of 0.556 or greater, whereas among nonconserved miRNAs, 31% (71/229) had ratios <0.556. Based on this analysis, we considered miRNA loci with abundance ratio of <0.4 as siRNA-like miRNA loci, and those with ratios between 0.4 and 0.5 as marginal miRNA loci rather than typical miRNA loci (see Supplemental Table 2 online).
Finally, we investigated whether most small RNAs originated from the same strand of an annotated stem-loop. For this, and the analysis below, we used the annotated stem-loop structures as defined in miRBase. Strand ratio was calculated by dividing the sequencing frequency of small RNAs from the sense strand by those from both strands of the stem-loop. Ninety-five percent of conserved miRNA stem-loops had strand ratios of 0.932 or greater (Figure 2C). However, only 39% (89/229) of nonconserved miRNAs fit this value. Accordingly, miRNA stem-loops with strand ratios <0.8 were considered siRNA-like miRNAs, and those with ratios between 0.8 and 0.9 were considered marginal miRNAs. In addition, two other annotated stem-loops were also grouped as siRNA-like miRNAs because, in these cases, the most abundant small RNAs were found in the loop region rather than in the stem of the precursor or they matched the rice genome more than 20 times. Based on these analyses, 40 stem-loops were considered as low expression loci, 198 as typical miRNA loci, 19 as marginal miRNA loci, and 149 as siRNA-like miRNA loci.
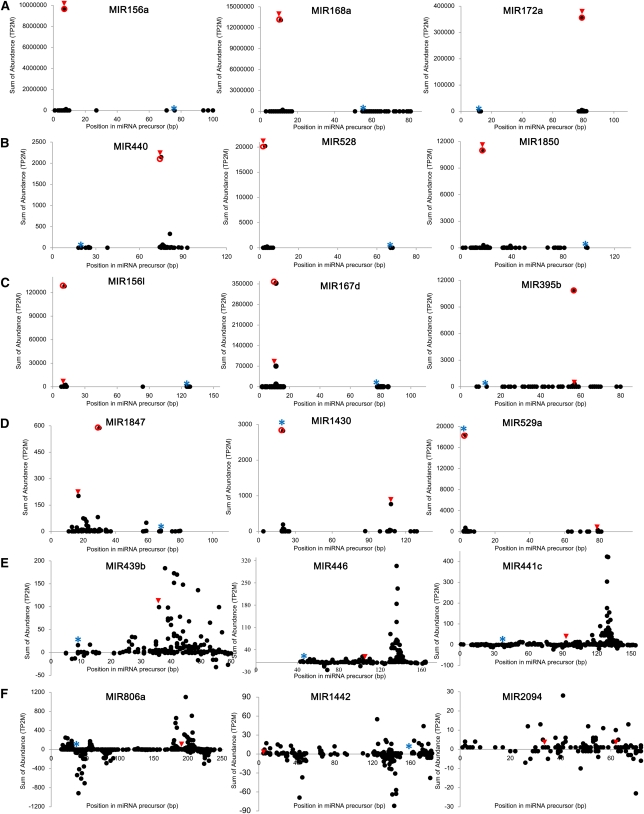
The 217 miRNA genes corresponding to the typical miRNA and marginal miRNA loci were the focus of our more detailed analysis. To determine if annotated mature miRNAs could be identified based on the distribution of small RNAs along their stem-loops, we plotted the sum of the abundance of each small RNA versus its position in the miRNA stem-loops and called these graphs S-plots (for small RNA plots). Out of 217 validated miRNA loci, in 160 loci (74%) the most abundant sequence from all the small RNAs in a locus was the annotated miRNA (Figures 3A and 3B; see Supplemental Table 3 online). However, in the remaining 57 miRNA loci, the most abundant sequence did not correspond exactly to the current miRBase reference sequence. In 52 of these cases, the most abundant sequence was a variant of the annotated miRNA with a small difference in length or start position (Figure 3C). For example, the reported miR395b (21 nucleotides) was sequenced with only a sum of the normalized abundance of 96, whereas its 20-nucleotide variant had an abundance of 10,850. We also observed miRNA loci where the miRNA* was sequenced more often than the miRNA (Figure 3D). Interestingly, miR529a* showed sequence similarity with the Os-miR156 families and shared target genes in the SPL gene family. Moreover, miR1430* and miR1433* are similar to miR169 in sequence and target the same genes of the NF-YA family (see Supplemental Table 3 online). These results indicate that relatively highly expressed annotated miRNA*s of miR529a, miR1430, and miR1433 are likely the true miRNAs and not miRNA*s. We also identified some cases where more than two distinct miRNAs are produced from a single precursor. It was shown previously that the natural antisense–miR444 loci generate more than three distinct miRNAs targeting the same MADS box genes (Lu et al., 2008a). All of these were validated by our new sequencing data, and three more new miRNAs from natural antisense–miR444 loci were found in our data (see Supplemental Table 3 online). Precursors of the miR159/319 family are relatively long, and it has been proposed that miR159a generates an additional miRNA, miR159a.2, from same arm of a precursor (Lacombe et al., 2008). We found that miR159c, miR159f, miR319a, and miR319b loci also produce other miRNAs that are similar to miR159a.2 in sequence, and we designated them as miR159cf.2, miR319a.2, and miR319b.2.
Figure 3.
Small RNA Plots of miRNA Precursors.
Small RNAs matching the indicated miRNA precursors were plotted versus the sum of their normalized abundance from all the libraries. Red arrowhead, miRNA reported in miRBase; blue asterisk, reported miRNA*; red circle, most abundant small RNA.
(A) The known and conserved miRNA is the most abundant small RNA.
(B) The known and nonconserved miRNA is the most abundant small RNA.
(C) An overlapping sequence variant is more abundant than the known miRNA.
(D) Another small RNA, unrelated to the known miRNA or same as miRNA*, is more abundant than the known miRNA.
(E) The known miRNA is poorly expressed and small RNAs matching the miRNA precursors are highly clustered.
(F) Known miRNAs are poorly expressed and small RNAs are found on the both sides of the miRNA precursor.
The plotting approach in Figure 3 also provided additional evidence of siRNA-like miRNA features found in the 149 rice miRNA loci referred to above. As shown in Figure 3E, some miRNA loci showed evenly distributed small RNAs without any prominent small RNAs of high abundance. Moreover, the other loci generated small RNAs from both strands of the annotated miRNA precursors, which is a typical characteristic of siRNA loci (Figure 3F). These results imply that plotting the high-throughput sequencing data in this way is a powerful approach to evaluate miRNA loci and to find the most reliable miRNAs within miRNA precursors.
Identification of Previously Undiscovered miRNAs and Their Targets
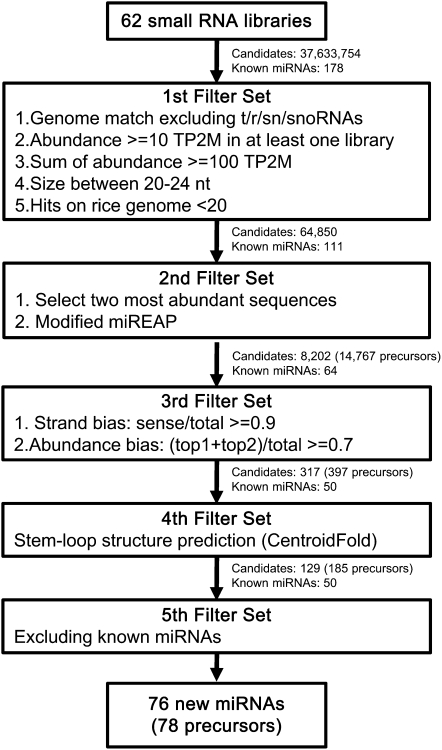
To identify previously undiscovered miRNAs in our small RNA data set, we developed a computational pipeline; this pipeline included five filter sets diagrammed in Figure 4. The filters were designed based on the consensus properties of the set of validated miRNAs fitting the new criteria for plants (Meyers et al., 2008). In the first filter, we removed small RNAs matching rRNAs, tRNAs, and other structural noncoding RNAs, such as snRNAs and snoRNAs. Small RNAs that were expressed at very low levels were excluded by requiring at least 10 TP2M in at least one library or the sum of the normalized abundance in all libraries to be at least 100. Additionally, small RNAs matching to more than 20 genomic locations were removed, and then small RNAs in the size range of 20 to 24 nucleotides were retained. Out of 32,633,754 distinct sequences, only 64,850 sequences passed this first filter set, including 111 reported miRNAs. The remaining 163 annotated miRNAs were filtered out due to low expression.
Figure 4.
Pipeline for the Identification of New miRNAs from Small RNA Libraries.
New miRNAs were identified by a series of filters shown in the diagram and described in the text. Numbers of candidate small RNAs and known miRNAs after each filter are indicated.
In the second filter, we used miREAP, an algorithm to detect and score Dicer hairpin products (https://sourceforge.net/projects/mireap/). Briefly, the small RNA sequences within 400-nucleotide segments of the rice genome were selected to predict their miRNA:miRNA* pairings, in which no more than four positions were unpaired. Consequently, 8202 sequences corresponding to 14,767 loci were obtained. Among the reported miRNAs, only 64 sequences passed this filter. The remaining 47 sequences were filtered out because they were not the most abundant small RNAs in the loci. In the third filter, we asked for two features of miRNAs: single-strand bias and an abundance bias showing one or two most prominent small RNAs from a given precursor. We obtained 317 candidates including 50 annotated miRNAs; these 317 candidates mapped to 397 loci in the rice genome.
The candidates representing 397 loci were subjected to a manual analysis of secondary structure, predicted using the CentroidFold program (Sato et al., 2009). We found that 185 loci (representing 129 distinct, mature miRNA sequences) potentially form miRNA precursor-like stem-loop structures. After removing 50 known miRNAs, 76 new miRNAs from 78 precursors were obtained. A search for sequence similarity against all the annotated miRNAs in miRBase revealed that none of these new miRNAs had been found in other plants. Most of these miRNAs were among those underrepresented in the RNAi lines of DCL1 (see Supplemental Figure 2 online).
To understand better the functions of the newly identified miRNAs, putative targets of these miRNAs were predicted using the computational algorithm psRNATarget (Dai and Zhao, 2011). A highly stringent penalty score (≤2.5, lower scores are better matches) was applied to maintain high specificity and low noise. The potentially novel miRNAs, including 76 newly identified miRNAs and 19 novel miRNAs from known miRNA precursors, were subjected to target mRNA prediction. Of the 95 miRNAs, 71 have at least one predicted target gene with a score not higher than 2.5 (see Supplemental Table 4 online). The remaining 24 miRNAs may have no target, their target mRNAs may not be annotated in the rice genome, or their targets may have more mismatches to the miRNA sequence than our cutoff permitted. In total, 356 potential target genes were predicted; 32 (9.0%) encode transcription factors; 33 (9.3%) encode proteins involved in signal transduction, such as several protein kinases and Leu-rich repeat family proteins; 72 (20.2%) encode proteins engaged in metabolic processes; 103 (28.9%) encode other proteins, and 116 (32.6%) encode proteins without annotated function (see Supplemental Table 5 online). This is in contrast with the targets of conserved miRNAs, approximately two-thirds of which encode transcription factors. Some of highest scoring potential targets were experimentally evaluated using a modified 5′-rapid amplification of cDNA ends (RACE) approach to detect cleavage events at predicted sites opposite to nucleotide 10 from the 5′ end of the miRNA (Figure 5). This approach validated the cleavage of six targets of six miRNAs, including two previously undiscovered miRNAs. Target cleavage in l-ascorbate oxidase indicated that the cleavage position is guided by miR397b.2, which is an abundant variant in the precursor, rather than by the poorly expressed miR397b that is annotated.
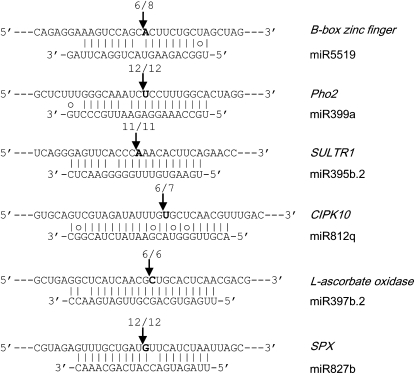
Figure 5.
Validation of Predicted Target Genes.
The arrows indicate sites verified by modified RLM 5′-RACE. Predicted cleavage sites are indicated by a bold nucleotide at position 10 relative to the 5′ end of the miRNA. The number of cloned RACE products sequenced is shown above each sequence.
Environmental Stress–Associated miRNAs and Their Targets
To find known or previously unknown miRNAs associated with environmental stress responses, we next analyzed a total of 222 validated miRNAs from the bona fide loci above, including 146 miRNA variants from annotated precursors and 76 newly identified miRNAs. Since deep-sequencing reads reflect the expression level of individual small RNAs, we compared the relative abundance of each miRNA between control and stress libraries. Whereas most miRNAs were expressed at similar levels in each stress library, we identified several miRNAs whose expression was significantly regulated by environmental stress treatment (see Supplemental Figure 3 online). To minimize noise from technical bias, we selected miRNAs showing the greatest difference between libraries and reproducible patterns of regulation in biological or technical replicate libraries. To validate these cases of regulation, we performed RNA gel blots or another gel assay called splinted-ligation-mediated miRNA detection.
Specifically, miR1425 was underrepresented in the libraries of salt- or cold-treated panicles. This expression pattern was validated, as shown in Figure 6C. RF-1, a target of miR1425, was induced under cold stress conditions when the miRNA decreased. We also found that in both the sequencing data and RNA gel blots, miR820.2 is downregulated after an 8-h drought stress treatment (Figure 6D). DRM2, a target of miR820.2 (Lu et al., 2008a), was decreased when the miRNA decreased, which may indicate that the miRNA is fine-tuning another, stronger mechanism that serves as the major repressor of the target.
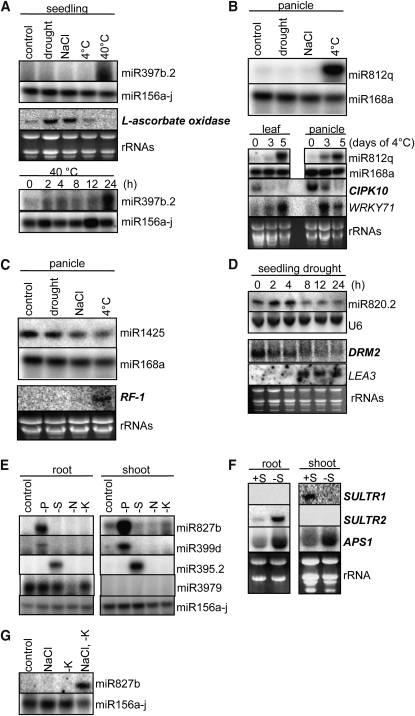
Figure 6.
Expression Patterns of Environmental Stress–Responsive miRNAs and Their Target Genes.
Splinted-ligation-based miRNA detection ([A] to [C] and [E] to [G]) or RNA gel blots (D) were used for miRNA expression analysis. U6, miR156a-j, and miR168a were used for loading controls. RNA gel blots were used for mRNA expression analysis. Genes in bold are targets of miRNAs. The other genes were used for controls of stress treatments. rRNAs served as loading controls. Seedling, root, and shoot tissues were harvested at the 2-week-old stage. Panicle was harvested at a mature stage when the panicles were ~15 cm in length.
We confirmed that the well-studied miR399 and miR827 were both upregulated by phosphate starvation, as reported previously in Arabidopsis (Fujii et al., 2005; Aung et al., 2006; Chiou et al., 2006; Hsieh et al., 2009; Pant et al., 2009; Lundmark et al., 2010) and in rice (Bari et al., 2006; Lin et al., 2010). Intriguingly, miR827 was also strongly induced by the combined stress of salt stress under conditions of potassium starvation (Figure 6G). This is not an additive response of the two stresses because imposing either single stress, high salt, or potassium starvation did not detectably elevate the levels of miR827. The double stress response could imply a link between phosphate starvation and high salt plus potassium starvation or that miR827 has a dual function under both the phosphate stress and the double stress regimes.
Under nitrogen starvation conditions, a newly discovered miRNA, miR3979, was downregulated in roots. Since miR3979 is a root-preferential miRNA, this regulation was found only in roots, not in shoots. It is well known that miR395 is upregulated by sulfate starvation stress in Arabidopsis. We also found that miR395.2 was specifically induced by sulfate starvation stress in rice but not by the other nutrient starvations (Figure 6E). SULTR1, which is a target of miR395.2, was cleaved in the exact site expected for miR395.2 (Figure 5) and was expressed at a low level under sulfate starvation conditions, when the miR395.2 levels are increased (Figure 6F). However, the expression levels of SULTR2 and APS1, which are also targets of miR395.2, were not obviously inversely correlated with the accumulation levels of miR395.2. This might be due to differences in spatial expression patterns between miR395.2 and its target genes. In Arabidopsis, whereas miR395 is expressed in phloem companion cells, SULTR2;1 mRNA accumulates in the xylem parenchyma cells (Kawashima et al., 2009).
Although most of the miRNAs shown to be regulated by environmental stress in Figure 6 correlate with their deep sequencing data, we also tested others because their target genes were known as stress-associated genes. This led to the identification of miR397b.2 and miR812q, neither of which showed significant differential expression in sequencing frequency. The expression of miR397b.2 is induced by heat stress, and its target, l-ascorbate oxidase, is cleaved as expected (Figure 5) and shows downregulation when heat stressed (Figure 6A). A newly identified miRNA, miR812q, is specifically upregulated by cold stress in both leaf and panicle (Figure 6B). Its target RNA is inversely regulated and encodes a CBL-interacting protein kinase (CIPK10'; Kolukisaoglu et al., 2004). Cleavage of this new target was confirmed by 5′-RACE (Figure 5). This implies that miR812q may play a major role in CIPK10 expression.
Differentially Expressed miRNAs among Tissues
To analyze the extent of differential miRNA expression, the sequencing data for the 222 validated newly discovered and known miRNAs were subjected to hierarchical clustering in an unsupervised manner (Eisen et al., 1998). Notably, hierarchical clustering showed that miRNA expression was mostly separated by tissue types rather than by stress (see Supplemental Figure 4 online). In other words, the miRNA expression patterns of different stresses within root, shoot, seedling, or panicle were more similar to each other than to the patterns of expression among different tissues.
The abundance of each miRNA within seedling root, seedling shoot, or panicle was examined and compared by averaging the normalized read values from libraries of the same tissue. Since differential expression of miRNAs detected at a low level (<10 TP2M) may represent the limit of detection rather than biological variability, we focused on the highly abundant miRNAs for differential abundance analyses. As expected, miRNA expression varied from highly specific to ubiquitous. Tissue-preferential miRNAs were selected that exhibited at least fivefold higher expression in one tissue compared with the others. Sixty (27%) of 222 validated miRNAs exhibited at least this high degree of tissue specificity (Figure 7). The expression of selected differentially expressed miRNAs was verified with the splinted-ligation-mediated miRNA detection method. Consistently, all 10 miRNAs tested correlated with the differential expression detected by sequencing. Compared with other tissues, tissue preferential miRNAs were found more often for panicle. Notably, 18 of the newly found miRNAs with panicle-preferential expression were not found at all among the sequences from the other tissue libraries. This implies that flowers may require a more complex set of miRNAs to regulate gene expression than do stems, leaves, or roots.
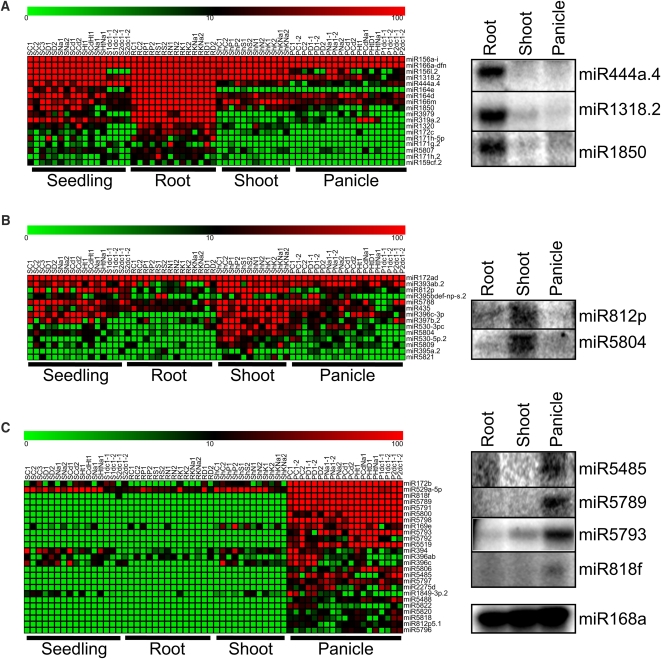
Figure 7.
Expression Profiling Analysis and Validation of Tissue-Preferential Expression.
miRNAs that are expressed at least 5 times higher in one tissue than the other tissues were identified, clustered by average linkage hierarchical clustering, and depicted in a heat map representation. Red represents high expression and green represents low expression. Seedling, root, and shoot tissues were harvested at the 2-week-old stage. Panicle was harvested at a mature stage when the panicles were ~15 cm in length. Detailed information about each library is indicated in Supplemental Table 1 online. Expression of selected miRNA was validated with splinted-ligation-based miRNA detection. Root-preferential miRNAs (A), shoot-preferential miRNAs (B), and panicle-preferential miRNAs (C).
Differential Expression of Members in the Same Family of miRNAs
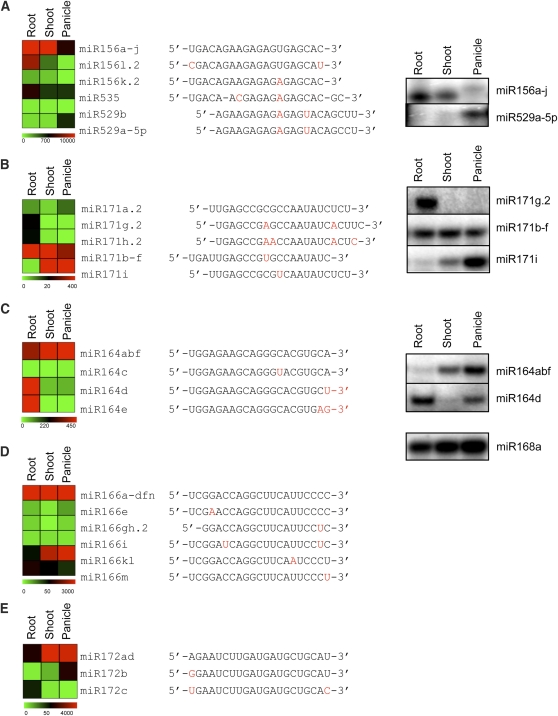
Many plant miRNAs belong to closely related families whose members differ by only a few nucleotides in sequence. These miRNAs may be redundant or diverse in their targets and function. To investigate this further, we examined the expression pattern of distinct miRNAs within families in different tissues. A total of 222 validated miRNAs were grouped into 148 families based on the sequence similarity (see Supplemental Table 3 online). Out of these, 24 miRNA families have more than two distinct mature miRNA sequences. Expression patterns of different tissues revealed that miRNA members in six families, including miR156, miR171, miR164, miR166, and miR172, were differentially expressed (Figure 8). To validate the expression patterns, the splinted-ligation-mediated miRNA detection method was used. Since this method is sensitive enough to discriminate closely related sequences especially with different length or nucleotides at 3′ ends of small RNAs, we were able to validate the expression pattern of miRNA members that differ only in a few nucleotides. For example, miR156a-j is preferentially expressed in root and shoot of seedlings but less expressed in the panicle. However, miR529-5p, which shares 14 nucleotides with miR156a-j, showed the opposite expression pattern (i.e., panicle-preferential expression; Figure 8A). In the case of miR171 family members, miR171g.2 is root preferential, whereas miR171b-f is ubiquitous and miR171i is less expressed in roots (Figure 8B). Interestingly, in both the miR156 and miR171 families, sequence differences are located within the functionally critical 5′-proximal sequences, suggesting that they may target genes different from those targeted by the other members. However, despite their differential expression patterns, the miR164 family members miR164abf and miR164d showed sequence differences in their 3′ ends, which are not critical in target selection (Figure 8C; Mallory et al., 2004).
Figure 8.
Differential Expression of Sequence Variants in the Same miRNA Family.
miRNA expression data from 2-week-old root, shoot, and 15-cm panicle were mean centered and represented by a heat map. The sequences of miRNA family members were aligned, and nucleotides that differ are shown in red. Validation of the expression patterns was accomplished by splinted-ligation-based miRNA detection. miR168a was used as constitutive expression control. miR156 family (A), miR171 family (B), miR164 family (C), miR166 family (D), and miR172 family (E).
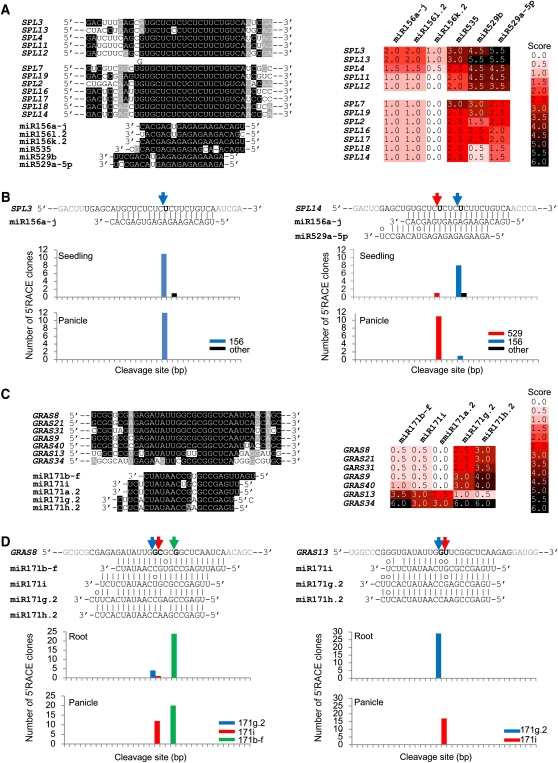
To investigate whether differences in sequence as well as differential expression patterns of miRNA family members would affect the selection of their targets, we predicted targets of each sequence variant and examined specific target cleavage sites in relevant tissues. For this purpose, the miR156 and miR171 families were selected for detailed analysis. Among members of the miR156 family, miR156a-j, which is preferentially expressed in seedlings, has 12 potential targets with a score of <2.5 that encode members of the SPL (for Squamosa Promoter binding protein Like) gene family. By contrast, panicle-preferential miR529a-5p is predicted to target the transcripts from only seven SPL genes among the 12 miR156a-j targets (Figure 9A). Seedling-preferential miR156l.2 and ubiquitously expressed miR156k.2 and miR535 share the same targets as miR156a-j. However, miR529b, which is similar to miR529a-5p in a sequence but ubiquitously expressed, is predicted to target the same genes as miR529a-5p. To validate specific target cleavages by those miRNA family members, cleavage sites were mapped from RNAs isolated from seedlings or panicles using the modified 5′-RACE procedure. Given that the 5′ end of miR529a-5p is offset by four nucleotides relative to miR156a-j and AGO invariably catalyzes cleavage of targets opposite of the bond between nucleotides 10 and 11 from the 5′ end of the miRNA, the cleavage product will differ by four nucleotides depending on which miRNA guides the processing event. As shown in Figure 9B, cleavage site mapping indicated that SPL3 is predominantly targeted by miR156a-j both in seedlings and panicle. However, SPL14, which is a common target of miR156a-j and miR529a-5p, is targeted by both miRNAs in seedlings, whereas it is predominantly targeted by miR529a-5p in panicle (Figure 9B). Although our 5′-RACE experiment does not provide quantitative information about cleavages between tissues, detection of preferential cleavage by a specific miRNA compared with one or more of the others in the same tissue suggests the effect is due to the particularly high level of the corresponding miRNA. This implies that a group of SPL genes including SPL14 has additional posttranscriptional regulation by miR529a-5p in the panicle compared with another group of SPL genes including SPL3.
Figure 9.
Differential Selection of Target Genes by Differentially Expressed miRNAs.
(A) Sequence alignment of miR156 family members and their target genes. The table next to the target genes represents target score for each member of the miR156 family.
(B) Validation of target cleavage by RLM 5′-RACE. Predicted cleavage sites are indicated by a bold nucleotide at position 10 relative to the 5′ end of the miRNA. The number of cloned RACE products sequenced is shown in the histogram.
(C) Sequence alignment of the miR171 family and their target genes.
(D) Validation of target cleavage by the miR171 family. The cleavage of GRAS13 shown formiR171g.2 could be equally the result of miR171 h.2. Names of target genes were from a previous publication (Tian et al., 2004), and their Michigan State University Rice Genome Annotation Project locus names are shown in Supplemental Table 8 online.
miR171 sequence variants were implicated in the targeting of different sets of genes that encode GRAS transcription factors (Figure 9C). Whereas both the ubiquitously expressed miR171b-f and the panicle-preferential miR171i have five potential targets, including GRAS8, 21, 31, 9, and 40 with a score of ≤ 2.5 score, root-preferential miR171g.2 and miR171 h.2 can target not only GRAS8, 21, and 31 but also GRAS13. The latter has less chance of being a target of miR171b-f and miR171i due to a penalty score exceeding 2.5. Three major cleavage sites detected in GRAS8 support it as a common target of miR171b-f, miR171i, miR171g.2, and miR171 h.2 (Figure 9D). In roots, two predominant cleavage sites correspond to miR171b-f and miR171g.2, whereas cleavage at the miR171i site was minor. However, in the panicle, instead of the miR171g.2-mediated cleavage of GRAS8, cleavage guided by miR171i was predominant as well as that mediated by miR171b-f. GRAS13 was predicted to be targeted by root-preferential miR171g.2 and miR171 h.2 but not by miR171b-f or miR171i (Figure 9C). Consistent with this, there is only one major cleavage site mediated by miR171g.2 and miR171 h.2 in root. However, in the panicle, one cleavage site corresponding to miR171i was found. Even though this site has a target score of 3.0, it seems that miR171 can mediate its cleavage in panicle. These data suggest that differentially expressed miRNA sequence variants may function as dynamic regulators of plant development.
DISCUSSION
The work in this study furthered our understanding of rice miRNAs in several ways. The deep sequencing of 62 small RNA libraries not only allowed effective small RNA profiling but also the confident annotation of miRNA genes. Specifically, this facilitated evaluation and categorization of annotated rice miRNAs, many of which have siRNA characteristics, and the identification of ~100 new miRNAs, using recent miRNA criteria. New cases of miRNA regulation in response to temperature, drought, nutrient, and multiple stresses were identified along with their target RNAs. Additional examples of tissue-preferential expression were also found. Moreover, we showed that differential regulation of sequence variants within miRNA families occurs. Our detailed analysis demonstrated that this can lead to the cleavage of different targets in rice and presumably in other species that may contribute to important agronomic traits.
Evaluation of Annotated miRNAs
Considering the huge complexity of the small RNA population in plants, it is a challenge to distinguish a relatively small number of miRNAs from a vast excess of siRNAs, some of which closely resemble miRNAs in their structural and biochemical characteristics. New criteria for confident annotation of a small RNA as an authentic miRNA were proposed based on its predominant processing from a miRNA stem-loop precursor by a DCL enzyme (Meyers et al., 2008). Compared with siRNA loci where small RNAs are highly distributed with low-abundance distinct siRNAs on both strands, typical miRNA loci generate one unique and highly abundant mature miRNAs with a limited number of low-abundance small RNAs on the same strand of the precursor.
We found that the S-plots are suitable for visualizing the distribution and abundance of small RNAs on an miRNA locus and for qualifying typical miRNAs based on the aforementioned criteria. Using this method, we were able to confirm only half (198 of 406) of the annotated rice miRNA loci with characteristics of typical miRNAs. However, the other loci were considered either as marginal miRNAs (19, 4.7%), low expression loci (40, 9.9%), or as siRNA-like miRNAs (149, 36.7%). This observation is similar to what was reported in another study with a much smaller sample size (Zhu et al., 2008). They classified 164 (54.5%) of 301 annotated rice miRNA as miRNA loci, 52 (17.3%) as low expression, and 85 (28.2%) as siRNA-like miRNA loci. In addition, most of their designations overlapped with ours. With the aid of deeper sequencing data and more diverse RNA sources than the earlier study, our results clearly expand the list of typical rice miRNAs as evidenced by the greater proportion of miRNAs and fewer low-expressed loci in our data.
It was also proposed that patterns of RNA decay intermediates across an miRNA precursor (degradome patterns) can be useful to distinguish miRNA loci from siRNA loci (German et al., 2008; Addo-Quaye et al., 2009). Since the processing of primary miRNA transcripts by DCL enzymes leaves polyadenylated remnants with a 5′-monophosphate, such cleavage products are highly represented on 5′ ends of mature miRNAs compared with others. Indeed, the recent analysis of rice miRNA precursors based on degradome patterns adds further strength to our results (Li et al., 2010). That study suggested that several small RNAs, including miR439, miR442, miR445, miR813, miR815, miR818, and miR819, are siRNA-like miRNAs as our small RNA data have demonstrated.
Although most of the siRNA-like miRNA loci delineated in our analysis were derived from repetitive sequence regions or transposable elements (TEs), the possibility that one or more may be newly evolved miRNAs (Voinnet, 2009) cannot be entirely ruled out. It was proposed that some transposable elements (TEs) can encode both siRNAs and miRNAs in plants (Piriyapongsa and Jordan, 2008). These dual-coding TEs can fold to form the stem-loop structures to produce miRNAs along with long double-stranded RNAs that typically are processed as siRNAs. These authors proposed that 38 TE-derived rice MIR genes may be dually encoding siRNAs and miRNAs. However, our data showed that most of those putative miRNAs were not detected or were expressed at a very low level, while heterogeneous siRNAs were highly represented on both strands. In our analysis, only two MIR531 loci have the potential to be dual-coding MIR genes, in which another small RNA is more abundant rather than annotated miRNA. In addition, we cannot rule out the other possibility that some siRNA-like miRNAs might be functional miRNAs only in specific cells or in certain conditions, while expressed or processed as siRNA loci in a larger number of cells in the surrounding tissues or other conditions. In this case, some siRNA-like miRNAs may be classified later as typical miRNAs or marginal miRNAs depending on the sampling of the source. In the future, more data from different tissues or conditions could be useful to reevaluate siRNA-like miRNAs and expand the list of rice miRNAs that are likely to be functional. Even if we cannot formally rule out that no young miRNA is amid the more than one-third of the annotated miRNAs that we concluded to be siRNA-like miRNAs, it seems worthwhile to point out these classifications. Since rice is a system that serves as a model for other Poaceae and also is amenable to large-scale functional studies, our analysis could help to avoid time-consuming work in other grasses based on what we as ascertained as misannotated miRNAs in rice.
New Variants of Mature miRNAs
We proposed the most abundant small RNAs to be the more reliable miRNAs rather than those annotated miRNAs observed only at low abundances. These most reliable miRNAs differ from the annotated sequences by minor shifts of one to four nucleotides, different lengths, and small RNAs unrelated to annotated miRNAs, or in some cases they were originally annotated as the miRNA*s. Newly proposed miRNAs, which are the most abundant small RNAs from annotated MIR genes, are also supported by mapping of target cleavage sites. We showed target cleavage guided by the most abundant small RNAs rather than low-expressed annotated miRNAs in miR397b.2, miR529a-5p, mIR171g.2, and miR171h.2 (Figures 5 and 9). In addition, the MIR397b foldback structure seems to be more favorable to processing the one that generates the most abundant sequence (miR397b.2) rather than the annotated miR397b (see Supplemental Figure 5A online). The base pair properties and bulge structure within the lower stem region below the miR/miR* duplex region are proposed to have an important role in miRNA precursor processing (Cuperus et al., 2010a; Mateos et al., 2010; Meyers et al., 2010; Song et al., 2010; Werner et al., 2010). Consistent with this, the 5′ end of the annotated Zma-miR397b extends beyond the stem structure, while Zma-miR397b.2, a homolog of Os-miR397b.2, is incorporated in a stem structure (see Supplemental Figure 5B online). Interestingly, this conservation between rice and maize is not found in Arabidopsis, in which a different stem structure of Ath-miR397b favors processing of the miRNA to generate the annotated mature miRNA sequence.
Variable Size of miRNAs
In plants, the majority of miRNAs are 21 nucleotides in length. However, out of our set of rice miRNAs, including typical annotated and newly identified miRNAs, only about half (124/244) were 21 nucleotides in length, while 30% were 24 nucleotides, 4.5% were 20 nucleotides, and 12% were 22 nucleotides (see Supplemental Table 6 online). Generally, 21-nucleotide canonical miRNAs have less asymmetric bulges within miRNA:miRNA* duplexes. However, more bulges in the miRNA* tends to produce 20-nucleotide miRNAs, such as Os-miR156 and Os-miR395.2, whereas more bulges in miRNAs tend to produce 22- nucleotide miRNAs, such as Os-miR397b.2 (Cuperus et al., 2010b).
Interestingly, a substantial number of 24-nucleotide miRNAs (30%) were identified in the set of typical annotated and newly identified miRNAs. Additionally, we found that many 24-nucleotide miRNAs are highly expressed in panicle tissues. Although most plant miRNAs are processed by DCL1 activity to produce 21-nucleotide miRNA:miRNA* duplexes, another member of the Dicer family, DCL3, is known to produce 24-nucleotide siRNAs and miRNAs (Wu et al., 2010). In rice, DCL1a, DCL2, DCL3a, and DCL4 are ubiquitously expressed in different tissues during development, whereas DCL1b and DCL3b exhibit panicle preferential expression (Kapoor et al., 2008). Since DCL3a is expressed at a much higher level than DCL3b, DCL3a might act to produce 24-nucleotide miRNAs in most tissues. Some 24-nucleotide miRNAs might be panicle-preferential either by transcriptional regulation of the corresponding MIR genes or by the activity of DCL3b.
We presented evidence of target cleavage by 24-nucleotide miR812q (Figure 5), although we failed to validate target cleavages by some other newly identified 24-nucleotide miRNAs. However, given that 24-nucleotide miRNAs were absent in AGO1 complexes that direct target mRNA cleavage, it might be possible that they enter alternative AGO complexes (Wu et al., 2009). It has been proposed that these 24-nucleotide miRNAs are mostly recruited by AGO4 to induce DNA methylation of their target genes (Wu et al., 2010). Like AGO4 in Arabidopsis, AGO4a and AGO4b in rice showed enhanced expression during early stages of floral development (Kapoor et al., 2008). These observations, when taken together with our data, imply that panicle-preferential DCL3b, AGO4a, and AGO4b might have important roles in the production and function of panicle-preferential 24-nucleotide miRNAs.
Environmental Stress–Regulated miRNAs
Perhaps the most well-studied miRNAs that are regulated by and function during environmental stress are miR399 and miR827, both of which are induced by phosphate starvation (Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006; Duan et al., 2008; Pant et al., 2009; Lin et al., 2010; Lundmark et al., 2010; Kant et al., 2011). Our study confirmed this regulation (Figure 6E) as well as the sulfur starvation induction of miR395 (Jones-Rhoades and Bartel, 2004) that, to our knowledge, prior to this work had not been reported in rice. These confirmations imply the functional conservation of the corresponding miRNAs between Arabidopsis and rice.
We also report regulation of known miRNAs, such as miR827, miR397b, miR1425, and miR820.2. We found that phosphate starvation–responsive miR827 was also upregulated by potassium starvation under salt stress conditions. Recently, miR827 was also reported to be induced by H2O2 treatment in rice (Li et al., 2011). Together, these findings support a role of miR827 in oxidative stress tolerance that is common to all the aforementioned stress conditions. Targets of miR827, the SPX domain–encoding genes, have been shown to be involved in phosphate-related signal transduction and regulation pathways, from yeast to plants (Duan et al., 2008; Hürlimann et al., 2009; Wang et al., 2009). In rice, the majority of SPX genes were also upregulated under cold stress (Zhao et al., 2009b). However, the role of SPX domain proteins under potassium starvation and salt stress conditions has yet to be studied. The importance of studying combinations of stresses is emphasized by the finding that neither salt or potassium stress alone induced miR827 (Figure 6) and because plants often face multiple simultaneous stresses in nature
In Arabidopsis, salt stress induction of miR397 has been reported (Sunkar and Zhu, 2004). However, our results showed that Os-miR397b.2 was induced by heat stress. This kind of differential regulation of a miRNA between different plant species was also reported in the case of miR398 (Jia et al., 2009). In poplar, miR398 was induced by ABA treatment or salt stress. However, the expression of miR398 in Arabidopsis was decreased by salt stress or by ABA treatment (Jagadeeswaran et al., 2009; Jia et al., 2009).
Previously, we confirmed that fertility restorer (Rf-1) gene families were targets for miR1425 (Lu et al., 2008a). Rf-1 is known to confer cold tolerance to hybrid rice at the booting stage by increasing the number of potentially fertile pollen grains (Komori and Imaseki, 2005). Our gene expression data support this and can explain this tolerance mechanism because the suppression of miR1425 during cold stress would in turn induce Rf-1 genes.
In the case of Os-miR820.2 and Os-DRM2, we observed an unexpected relationship between their expression patterns under drought stress. In general, if miRNAs guide the cleavage and degradation of the target mRNA transcripts, their expression levels should be inversely correlated. However, miR820.2 is downregulated during drought stress, while its target DRM2 also decreased in abundance. This incoherent case implies that miR820.2 may provide the ability to fine-tune the spatiotemporal expression of DRM2 in response to drought. DRM2 is implicated in DNA de novo methylation; thus, miR820.2 may play a role in buffering and increasing the precision of this process under drought conditions. Epigenetic regulation is thought to play important roles in plant responses to environmental stress (Vernhettes et al., 1998; Kim et al., 2010). The regulation of DRM2 by miR820.2 may contribute to our understanding of the crosstalk between miRNA and epigenetic regulation during environmental stress.
Two of the new miRNAs identified in our study were also found to be environmental stress–regulated miRNAs. miR812q was dramatically induced by cold stress. Its target RNA is inversely regulated and encodes CIPK10. CIPK (for Calcineurin B-Like [CBL] protein interacting protein kinase) proteins contain a Ser/Thr protein kinase domain that is activated through interaction with CBL, which contains four EF hands for Ca2+ binding (Nagae et al., 2003; Batistic and Kudla, 2009). The CBL-CIPK signaling system is known to be a Ca2+-dependent network mediating environmental stress tolerance. In rice, expressions of CIPK genes are regulated by various stresses, such as drought, salt, and cold (Xiang et al., 2007). Thus, miR812q may be required to modulate CBL-CIPK signaling in response to cold stress. The other example of a newly identified stress-regulated miRNA is miR3979, which is repressed in roots under nitrogen depletion. Nitrogen is a crucial plant macronutrient that is needed in the greatest amount among all mineral elements required by plants (Frink et al., 1999). In land plants, nitrogen deficiency results in reduced growth, chlorosis of leaves, anthocyanin accumulation, and increased root surfaces (Lian et al., 2006; Zhang, 2007). Intriguingly, one of predicted targets of nitrogen starvation–responsive miR3979 encodes an anthranilate phosphosribosyltransferase, which is involved in Trp biosynthesis. The Trp pathway in plants leads to the biosynthesis of not only Trp but also secondary compounds, such as the plant hormone auxin and the phytoalexin camalexin (Kutchan, 1995; Radwanski and Last, 1995). In Arabidopsis, Trp biosynthetic genes are induced by amino acid starvation, oxidative stress, and an abiotic elicitor (Zhao et al., 1998). Thus, it is plausible that under nitrogen starvation conditions, downregulation of miR3979 induces Trp biosynthesis, followed by increased auxin production, resulting in lateral root initiation to absorb more nitrogen from the soil (Linkohr et al., 2002; Laskowski et al., 2006; Osmont et al., 2007).
Significance of Differentially Expressed miRNA Family Members
We demonstrated that out of 148 rice miRNA families, 24 miRNA families (~16%) consist of more than two distinct sequences often encoded by multiple genes. Interestingly, six families showed differential expression patterns of distinct miRNA family members. Moreover, two miRNA families, miR156/529 and miR171, showed specialization of their cleavage function between or among members as a consequence of sequence and expression differences.
Differential expression of members of miRNA families, such as miR164, miR159, miR169, and miR319, was also reported in Arabidopsis, but the methodology did not focus on the mature miRNAs (Palatnik et al., 2007; Sieber et al., 2007; Li et al., 2008; Nag et al., 2009). In the Arabidopsis studies, expression patterns of distinct miRNA sequences were analyzed at the primary MIR transcript level using promoter:β-glucuronidase fusions or real-time quantitative PCR. Our analysis took advantage of two technical approaches to address specifically the expression patterns of the functional mature miRNAs. First, high-throughput sequencing gave us single-nucleotide resolution to distinguish distinct miRNA family members as well as their expression levels represented by sequencing frequency. This was especially useful for defining the most prominent miRNA(s) from each precursor for analysis of the expression pattern. Focusing on these rather than on poorly expressed miRNAs simply because they were annotated offers an advantage because of the variable criteria used for earlier annotations. Second, to validate differential expression patterns of closely related miRNAs, we used the splinted-ligation-mediated miRNA detection method, which is more sensitive than standard RNA gel blots. This is particularly effective at distinguishing differences in 3′ end nucleotides or length.
Most members of miRNA families share considerable sequence identity, and their sequence differences were mostly found within the last two nucleotides of the 3′ end, a region not considered significant for target RNA selection (Mallory et al., 2004). Although differential expression should be enough to confer functional specificity of miRNA family members, diversity both in expression and target selection may provide even more functional significance. The closely related miRNAs miR159 and miR319 in Arabidopsis represent a case for which functional specificity was established by both sequence and expression differences among distinct miRNAs (Palatnik et al., 2007). Mature miRNAs produced from the MIR159 and MIR319 loci share sequence identity at 17 out of 21 nucleotides and represent the same family. Highly expressed miR159 family members mainly target MYB mRNAs, but the miRNA sequences limit their ability to guide cleavage of TCP mRNAs. In a different manner, MYB targeting by miR319 is restricted by their relatively lower expression levels. This case showed that the functions of miR159 and miR319 are exclusive to different targets. This is in contrast with the results of our analysis of distinct members of the miR156/miR529 and the miR171 families. Our data indicate that sequence variants within these families share common targets yet also have their own targets.
In case of the miR171 family, we showed that three GRAS genes can be potentially targeted by four distinct miR171 sequence variants, two GRAS genes by two miR171 sequence variants, and another GRAS gene by two variants (Figure 9). Interestingly, the selectivity of these regulatory interactions was further mediated by tissue-preferential expression of the distinct miRNA family members involved: Preferential cleavage of the GRAS13 mRNA is guided by mi171i in the panicle and by miR171g.2 and miR171 h.2 in root. Cleavage of GRAS8 is mediated by these miR171 family members and also the ubiquitous miR171b-f. These results argue that single-nucleotide sequence differences can be sufficient for distinct miRNA family members to target different sets of mRNAs and another mechanism for ensuring tight regulation of target gene families in a selected tissue is the differential expression of individual family members.
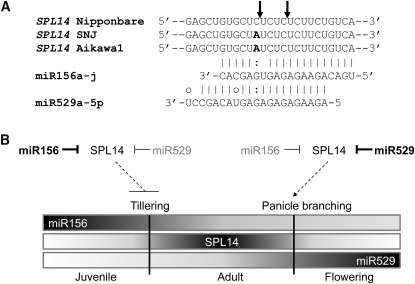
These points are further substantiated by the activity of members of the miR156/529 family in a manner that also likely has agricultural significance. We showed that seedling-preferential miR156 regulates 12 SPL genes, whereas panicle-preferential miR529-5p is predicted or known to guide cleavage of seven of these, including SPL14. It is notable that SPL14 is encoded by a rice grain yield-associated quantitative trait locus, IDEAL PLANT ARCHITECTURE1/WEALTHY FARMER’S PANICLE (IPA1/WFP), in which enhanced expression of SPL14 generates an ideal plant architecture with high yield potential (Jiao et al., 2010; Miura et al., 2010). Higher SPL14 expression represses shoot branching and promotes panicle branching. Intriguingly, japonica cultivar Shaoniejing (SNJ) and Aikawa1 plant lines with IPA1/WFP phenotypes harbor a common single point mutation, which is a C-to-A mutation in the miR156 target site in SPL14. The mutated position corresponds to the 15th nucleotide from the 5′ end of miR156a-j. This led to the suggestion that the point mutation in SNJ and Akiwa1 perturbs the cleavage of SPL14 transcripts by miR156a-j in the vegetative stage, resulting in higher expression of SPL14 and consequent repression of shoot development (Jiao et al., 2010; Miura et al., 2010). However, the mechanism of increased panicle branching was not clear from previous work. In light of our data, SPL14 regulation by miR529a-5p might be critical for this mechanism, as evidenced by three pieces of data. First, cleavage of SPL14 in panicles primarily mapped to a location regulated by miR529a-5p rather than by miR156a-j (Figure 9). Second, the point mutation in SNJ and Akiawa1 alters the 11th nucleotide from the 5′ end of miR529a-5p, exactly at the cleavage site (Figure 10A). Third, it was reported that SPL14 is downregulated during panicle development, when miR156a-j was weakly expressed (Xie et al., 2006). Thus, as illustrated in Figure 10B, rather than miR156a-j alone, it seems more likely that panicle-preferential miR529 or both miRNAs cause downregulation of SPL14 during panicle development and consequent repression of panicle branching. Family members of the miR529 sequence type are not found in Arabidopsis but are present in other grasses. Therefore, miR156/529 sequence variants may contribute to the complex regulation of plant architecture and yield in other important crops as well. Moreover, the miR156/529 and miR171 families emphasize the potential impact that miRNA sequence variants may have on the broader array of plant processes that are regulated by miRNAs.
Figure 10.
miR529 May Be Critical for Regulation of SPL14 in Panicle.
(A) The SNJ and Akiawa1 sequence difference is adjacent to the miR529 cleavage site of Os-SPL14.
(B) Model for regulation of tillering and panicle branching by miR156 and miR529.
METHODS
Plant Growth and Stress Treatment
Rice seeds (Oryza sativa ssp japonica cv Nipponbare) were husked, surface-sterilized with 3% sodium hydrochlorite, washed with sterile water three times, and germinated on half-strength Murashige and Skoog (MS) medium containing 0.2% (w/v) Phytagel in a sterile plastic box. Plants were grown in a growth chamber under conditions of 12 h light at 28°C and 12 h dark at 25°C.
Nine-day-old seedlings were acclimated for 5 d by opening the plastic cap and adding tap water onto the MS agar medium. Five days later, 2-week-old seedlings were moved to tap water in a glass tube and used for stress treatments and controls. For drought treatment, seedlings were air-dried for 8 h. For salt stress treatment, seedlings were transferred to 300 mM NaCl solution for 8 h. For cold stress treatment, seedlings were transferred to 4°C in a cold room under continuous light for 8 h. For heat stress treatment, seedlings were moved to 42°C for 8 h. Dual stresses were also treated under the same conditions. Control plants were sampled at the same time. All the collected tissues were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
For nutrient starvation treatments, the endosperm was first removed from 9-d-old seedlings to avoid nutrient transport. Seedlings were then transferred to MS nutrient-deficient media or MS media (for control). For MS nutrient-deficient media, specific components of MS were omitted or substituted with others as follows: For nitrogen-deficient medium, NH4NO3 was omitted and KCl was substituted for KNO3. For phosphate-deficient medium, KH2PO4 was omitted. For potassium-deficient medium, KNO3 was omitted. For sulfate-deficient medium, all SO4 was substituted with Cl2. After 5 d of treatment, shoots and roots were separated for independent analysis.
Mature plants at the booting stage were treated with various environmental stresses under the same conditions as described above, but for 3 d, with the exception of the drought treatment. For drought treatment, pots were removed from the water, holes were punched in the pots to help draining, and pots were dehydrated for 7 d. After stress treatment, samples were taken of mature panicles, which were ~15 cm and wrapped with panicle sheath.
Small RNA Library Construction and Sequencing
The small RNA libraries were constructed as described (Lu et al., 2007). Briefly, total RNA was isolated with Trizol reagent (Molecular Research Center). Low molecular weight RNA was isolated from 200 μg of total RNAs by PEG8000/NaCl precipitation. Small RNAs in the size range of 20 to 30 nucleotides were purified from 15% denaturing PAGE gels and ligated first with the 5′ RNA adaptor and then with the 3′ RNA adaptor. In each step, the ligated products were PAGE gel purified. After first-strand synthesis and 18 cycles of PCR amplification, the final bands were PAGE gel purified and submitted for sequencing on an Illumina 1G or IIGX analyzer. Sequencing was performed at Illumina, National Center for Genome Research, Cold Spring Harbor Laboratory, or the Delaware Biotechnology Institute.
Computational Analysis of Sequencing Data
The raw sequencing data were trimmed by removing adaptor sequences and mapped to the rice genome (The Institute for Genomic Research [TIGR] release version 5; http://www.tigr.org/) using Bowtie (Langmead et al., 2009). Reads perfectly matching the rice genome, excluding those matching tRNAs, rRNAs, snRNA, and snoRNAs, were used for further analysis. Rice mature miRNAs and their precursors were retrieved from miRBase (version 14; http://www.mirbase.org/).
miRNA Pipeline
The miRNA prediction pipeline is outlined in Figure 4. Individual steps in the process were performed with Perl scripts (see Supplemental Data Set 1 online) combined with miREAP (https://sourceforge.net/projects/mireap/) and CentroidFold (Sato et al., 2009). We used miREAP to evaluate the pairing of the miRNA and miR* with the parameters set to allow a maximal distance of 400 nucleotides between miRNA and miRNA* (-d 400), extending 25 nucleotides at the end of precursor (-f 25), with filters optimized for animal miRNAs turned off and including minor tuning for plant miRNA characteristics (our modified version of miREAP is available upon request). In addition, we asked for two miRNA features: single-strand bias ≥0.9 and an abundance bias ≥0.7 based on the features of conserved miRNAs. CentroidFold was used with default settings to visualize the overall miRNA precursor structure for manual evaluation.
RNA Gel Blot Analysis
For small RNA gel blot analysis, low molecular weight RNA isolated from 150 μg of total RNA was separated on 15% PAGE gels with 7 M urea, transferred to Hybond N+ nylon membranes (Amersham Biosciences), and fixed on the membranes by baking at 80°C for 2 h. DNA oligonucleotide probes complementary to miRNA sequences were end labeled with [γ-32P]ATP and optiKinase (Amersham Biosciences) and purified by the QIAquick nucleotide removal kit (Qiagen). The membranes were prehybridized for at least 2 h and hybridized for at least 12 h in an Ultrahyb-Oligo hybridization buffer (Ambion) at 40°C. DNA oligonucleotides used as probes are listed in Supplemental Table 7 online. To detect mRNAs, 20 μg of total RNA was loaded on 1.5% formaldehyde agarose gels, transferred to Hybond N+ nylon membranes, and probed with 32P-labeled sequences following prehybridization in Church and Gilbert's hybridization buffer (Church and Gilbert, 1984). Prehybridization and hybridization were performed at 58°C. The probes were generated by PCR using primers described in Supplemental Table 7 online. Visualization of miRNA and mRNAs was performed with a Typhoon phosphor imager (GE Healthcare Life Sciences).
Splinted-Ligation-Mediated miRNA Detection
To detect miRNAs, splinted ligation was performed using the miRtect-IT miRNA labeling and detection kit (Affymetrix) as described (Maroney et al., 2008; Jeong et al., 2010). For each miRNA, we designed specific bridge oligonucleotides as shown in Supplemental Table 7 online. Briefly, miRNAs were captured from 1 to 16 μg of total RNA with a specific bridge oligonucleotide and ligated to a 32P-end-labeled detection oligo with T4 DNA ligase. Ligated products were separated on 15% PAGE gels with 7 M urea and visualized with a Typhoon phosphor imager.
miRNA Target Prediction and Validation
All the new miRNAs were used to predict targets from the rice TIGR genome cDNA release 5 using the prediction tool psRNATarget (http://bioinfo3.noble.org/psRNATarget/). To validate target cleavage, a modified procedure for RNA ligase–mediated (RLM) 5′-RACE was performed using the FirstChoice RLM-RACE kit (Ambion) as described (Jeong et al., 2010). One microgram of total RNAs from stress-treated tissues was ligated to 5′-RACE RNA adaptor without calf intestine alkaline phosphatase treatment. The gene-specific outer primers then were used for cDNA synthesis. Initial PCR was performed using the 5′-RACE outer primer and a gene-specific outer primer. Nested PCR was performed using 1/50 of the initial PCR, the 5′-RACE inner primer, and a gene-specific inner primer. The gene-specific primers are listed in Supplemental Table 7 online. RACE fragments were cloned into the pGEM T-easy vector after gel purification (Invitrogen) and sequenced.
Accession Numbers
Newly generated small RNA data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE32973. The data are also available at http://mpss.udel.edu/rice_sRNA2/.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Differential Accumulation of 20-, 21-, and 24-Nucleotide Small RNAs from Roots, Shoots, and Panicles in a Represented Region of Rice Chromosome 5.
Supplemental Figure 2. Change in miRNA Expression in the Os-DCL1 RNAi Lines.
Supplemental Figure 3. Comparison of the Relative Abundance of Each miRNA between Control and Stress Libraries.
Supplemental Figure 4. Heat Map and Cluster Analysis of Validated miRNAs from 62 Small RNA Libraries.
Supplemental Figure 5. Predicted Secondary Stem-Loop Structures of Os-MIR397b and ZMA-MIR397b.
Supplemental Table 1. Summary Statistics for Small RNA Libraries.
Supplemental Table 2. Evaluation of Known miRNA Precursors.
Supplemental Table 3. Reliable miRNAs from Known miRNA Precursors.
Supplemental Table 4. New miRNAs and Their Predicted Targets.
Supplemental Table 5. List of Predicted Targets of New miRNAs.
Supplemental Table 6. Length Distribution of miRNAs.
Supplemental Table 7. Oligomers Used in This Study.
Supplemental Table 8. Information of Target Genes for miR156 and miR171.
Supplemental Data Set 1. Perl Script of Modified miREAP.
Acknowledgments
We thank Bruce Kingham, Dick McCombie, and Greg May for assistance with sequencing; Kan Nobuta, Roli Shrivastava, and Mayumi Nakano for computational analysis; Mayumi Nakano for her work on the Web interface; Liang-Hu Qu and Chun-Long Chen for providing rice snoRNA sequences; XiaoFeng Cao for providing DCL1 RNAi seeds; and Hajime Sakai and Randy Parker for their assistance in growing and caring for the rice plants. This study was supported by USDA Grants 2007-01991 to P.J.G., 2011-67013-30036 to P.J.G. and B.C.M., and 2005-35604-15326 to B.C.M. and P.J.G. D.-H.J. was partially supported by a Korean Research Foundation Fellowship funded by the Korean Government (KRF-2006-214-C00085).
AUTHOR CONTRIBUTIONS
D.-H.J. and P.J.G. designed the research. D.-H.J. and S.P. performed the research. D.-H.J., J.Z., B.C.M., and P.J.G. analyzed the data. J.Z., S.G.R.G., E.D.P., and B.C.M. contributed new analytic and computational tools. D.-H.J. and P.J.G. wrote the article.
References
- Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43: 837–848 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Kudla J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793: 985–992 [DOI] [PubMed] [Google Scholar]
- Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. (2003). Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Cheng Y., Jia D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010b). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C. (2010a). Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P.X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39(Web Server issue) W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Yi K., Dang L., Huang H., Wu W., Wu P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frink C.R., Waggoner P.E., Ausubel J.H. (1999). Nitrogen fertilizer: Retrospect and prospect. Proc. Natl. Acad. Sci. USA 96: 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008). miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36(Database issue): D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. (2009). Uncovering small RNA-mediated responses to phosphate-deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.Q., Peng J., Qiu C.X., Yang Z.M. (2009a). Heavy metal-regulated new microRNAs from rice. J. Inorg. Biochem. 103: 282–287 [DOI] [PubMed] [Google Scholar]
- Huang Y., Ji L., Huang Q., Vassylyev D.G., Chen X., Ma J.B. (2009b). Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461: 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann H.C., Pinson B., Stadler-Waibel M., Zeeman S.C., Freimoser F.M. (2009). The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 10: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Running M.P., Meyerowitz E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G., Saini A., Sunkar R. (2009). Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Jeong D.H., German M.A., Rymarquis L.A., Thatcher S.R., Green P.J. (2010). Abiotic stress-associated miRNAs: Detection and functional analysis. Methods Mol. Biol. 592: 203–230 [DOI] [PubMed] [Google Scholar]
- Jia X., Wang W.X., Ren L., Chen Q.J., Mendu V., Willcut B., Dinkins R., Tang X., Tang G. (2009). Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 71: 51–59 [DOI] [PubMed] [Google Scholar]
- Jian X., Zhang L., Li G., Zhang L., Wang X., Cao X., Fang X., Chen F. (2010). Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 95: 47–55 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Arora R., Lama T., Nijhawan A., Khurana J.P., Tyagi A.K., Kapoor S. (2008). Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima C.G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y.N., Saito K., Takahashi H., Dalmay T. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Nishioka T., Seki M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U., Weinl S., Blazevic D., Batistic O., Kudla J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Imaseki H. (2005). Transgenic rice hybrids that carry the Rf-1 gene at multiple loci show improved fertility at low temperature. Plant Cell Environ. 28: 425–431 [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchan T.M. (1995). Alkaloid biosynthesis[mdash]The basis for metabolic engineering of medicinal plants. Plant Cell 7: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., et al. (2008). Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Biol. 8: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M., Biller S., Stanley K., Kajstura T., Prusty R. (2006). Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47: 788–792 [DOI] [PubMed] [Google Scholar]
- Li T., Li H., Zhang Y.X., Liu J.Y. (2011). Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 39: 2821–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X., Oono Y., Zhu J., He X.J., Wu J.M., Iida K., Lu X.Y., Cui X., Jin H., Zhu J.K. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M.J., Zhang W., Sunkar R. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62: 742–459 [DOI] [PubMed] [Google Scholar]
- Lian X., Wang S., Zhang J., Feng Q., Zhang L., Fan D., Li X., Yuan D., Han B., Zhang Q. (2006). Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol. Biol. 60: 617–631 [DOI] [PubMed] [Google Scholar]
- Lin S.I., et al. (2010). Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51: 2119–2131 [DOI] [PubMed] [Google Scholar]
- Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Lippman Z., Martienssen R. (2004). The role of RNA interference in heterochromatic silencing. Nature 431: 364–370 [DOI] [PubMed] [Google Scholar]
- Liu B., Li P., Li X., Liu C., Cao S., Chu C., Cao X. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139: 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., et al. (2008a). Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Meyers B.C., Green P.J. (2007). Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117 [DOI] [PubMed] [Google Scholar]
- Lu S., Sun Y.H., Chiang V.L. (2008b). Stress-responsive microRNAs in Populus. Plant J. 55: 131–151 [DOI] [PubMed] [Google Scholar]
- Lundmark M., Kørner C.J., Nielsen T.H. (2010). Global analysis of microRNA in Arabidopsis in response to phosphate starvation as studied by locked nucleic acid-based microarrays. Physiol. Plant. 140: 57–68 [DOI] [PubMed] [Google Scholar]
- Luo Y.C., Zhou H., Li Y., Chen J.Y., Yang J.H., Chen Y.Q., Qu L.H. (2006). Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 580: 5111–5116 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Vaucheret H. (2004). MicroRNAs: Something important between the genes. Curr. Opin. Plant Biol. 7: 120–125 [DOI] [PubMed] [Google Scholar]
- Malone C.D., Hannon G.J. (2009). Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney P.A., Chamnongpol S., Souret F., Nilsen T.W. (2008). Direct detection of small RNAs using splinted ligation. Nat. Protoc. 3: 279–287 [DOI] [PubMed] [Google Scholar]
- Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. (2010). Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 20: 49–54 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Simon S.A., Zhai J. (2010). MicroRNA processing: Battle of the bulge. Curr. Biol. 20: R68–R70 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Aksay G., Dolgosheina E., Ebhardt H.A., Magrini V., Mardis E.R., Sahinalp S.C., Unrau P.J. (2008). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M., Nozawa A., Koizumi N., Sano H., Hashimoto H., Sato M., Shimizu T. (2003). The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J. Biol. Chem. 278: 42240–42246 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Muller R., Nielsen T.H. (2010). Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant. 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., Carrington J.C., Weigel D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Pant B.D., Musialak-Lange M., Nuc P., May P., Buhtz A., Kehr J., Walther D., Scheible W.R. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J., Jordan I.K. (2008). Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski E.R., Last R.L. (1995). Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 7: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Sanan-Mishra N., Kumar V., Sopory S.K., Mukherjee S.K. (2009). Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Mol. Genet. Genomics 282: 463–474 [DOI] [PubMed] [Google Scholar]
- Sato K., Hamada M., Asai K., Mituyama T. (2009). CENTROIDFOLD: A web server for RNA secondary structure prediction. Nucleic Acids Res. 37(Web Server issue): W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P., Wellmer F., Gheyselinck J., Riechmann J.L., Meyerowitz E.M. (2007). Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Song L., Axtell M.J., Fedoroff N.V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Chinnusamy V., Zhu J., Zhu J.K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhou X., Zheng Y., Zhang W., Zhu J.K. (2008). Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernhettes S., Grandbastien M.A., Casacuberta J.M. (1998). The evolutionary analysis of the Tnt1 retrotransposon in Nicotiana species reveals the high variability of its regulatory sequences. Mol. Biol. Evol. 15: 827–836 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang C., Ying S., Huang H., Li K., Wu P., Shou H. (2009). Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang J.F., Zhou H., Chen Y.Q., Luo Q.J., Qu L.H. (2004). Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 32: 1688–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Wollmann H., Schneeberger K., Weigel D. (2010). Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20: 42–48 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. (2009). Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang Y., Xiong L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144: 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Wu C., Xiong L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.J., Zhang J.J., Xue H.W. (2009). Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 37: 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.F., Yuan L.J., Shao Y., Du W., Yan D.W., Lu Y.T. (2008). The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 31: 562–574 [DOI] [PubMed] [Google Scholar]
- Zhang Q. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA 104: 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ge L., Liang R., Li W., Ruan K., Lin H., Jin Y. (2009a). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354: 585–590 [DOI] [PubMed] [Google Scholar]
- Zhao J., Williams C.C., Last R.L. (1998). Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Liu F., Xu W., Di C., Zhou S., Xue Y., Yu J., Su Z. (2009b). Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol. J. 7: 550–561 [DOI] [PubMed] [Google Scholar]
- Zhou M., Lianfeng G., Li P., Song X., Wei L., Chen Z., Cao X. (2010). Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Front. Biol. 5: 67–90 [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J.K., Zhang W. (2008). Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 1779: 780–788 [DOI] [PubMed] [Google Scholar]
- Zhou X., Wang G., Zhang W. (2007). UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 3: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Spriggs A., Matthew L., Fan L., Kennedy G., Gubler F., Helliwell C. (2008). A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 18: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]