This work reports on a comprehensive systems biology analysis of maize leaves, concluding that C4 photosynthesis is established from sink tissue without an intermediate phase of C3 or C2 photosynthesis.
Abstract
We systematically analyzed a developmental gradient of the third maize (Zea mays) leaf from the point of emergence into the light to the tip in 10 continuous leaf slices to study organ development and physiological and biochemical functions. Transcriptome analysis, oxygen sensitivity of photosynthesis, and photosynthetic rate measurements showed that the maize leaf undergoes a sink-to-source transition without an intermediate phase of C3 photosynthesis or operation of a photorespiratory carbon pump. Metabolome and transcriptome analysis, chlorophyll and protein measurements, as well as dry weight determination, showed continuous gradients for all analyzed items. The absence of binary on–off switches and regulons pointed to a morphogradient along the leaf as the determining factor of developmental stage. Analysis of transcription factors for differential expression along the leaf gradient defined a list of putative regulators orchestrating the sink-to-source transition and establishment of C4 photosynthesis. Finally, transcriptome and metabolome analysis, as well as enzyme activity measurements, and absolute quantification of selected metabolites revised the current model of maize C4 photosynthesis. All data sets are included within the publication to serve as a resource for maize leaf systems biology.
INTRODUCTION
The mechanisms underlying organ development and function are fundamental questions of biology. In plants, grass leaves represent an excellent model in which the establishment of various functions can be followed in a base-to-tip developmental gradient in a single leaf. Cells at the tip of the leaf are the oldest and most mature cells, while cells at the base are the youngest (Nelson and Langdale, 1992). We chose maize (Zea mays) to follow the establishment of photosynthetic functions during leaf development. Maize employs the highly efficient C4 type of photosynthesis, which concurrently evolved in multiple seed plant families ~15 to 40 million years ago, long after C3 photosynthesis had been established (Edwards and Smith, 2010). It has been previously proposed that the evolutionary progression from C3 to C4 can also be detected in maize leaves along a spatial gradient (Nelson and Langdale, 1992, and references therein), very much like Haeckel suggested that ontogeny recapitulates phylogeny during embryo development in animals (Haeckel, 1866).
C4 photosynthesis has been considered a possible route for spawning a second green revolution in C3 crop plants, such as rice (Oryza sativa) (Hibberd et al., 2008). Plants using C4 photosynthesis are capable of producing biomass at faster rates than C3 plants, or, alternatively, these plants can inhabit harsher habitats with limited resources (Sage, 2004, and references therein). The key limitation for more productive photosynthesis is the concentration of carbon dioxide at the site of its assimilation, the reductive pentose phosphate pathway (rPPP) in plant chloroplasts. The enrichment of carbon dioxide around ribulose-1,5-bisphosphate carboxylase/oxygenase minimizes the oxygenation of ribulose-1,5-bisphosphate, which leads to a reduced rate of photorespiration. From an engineering standpoint, C4 photosynthesis, similar to a supercharged combustion engine, enriches the limiting factor, carbon dioxide, via a biochemical cycle operating between the site of initial fixation and final assimilation. Although the C4 cycle as described below appears deceptively simple, differences between C3 and C4 photosynthesis go beyond just the addition of the C4 cycle on top of the rPPP and include photorespiration, protein translation, cellular and tissue architecture, electron transfer adaptations, cell–cell connections, and likely other still unknown adaptations (Bräutigam et al., 2011; Gowik et al., 2011). In C4 plants, carbon dioxide is enriched by affixing it to an acceptor, transferring it to the site of final assimilation, liberating it, and returning and recycling the acceptor for a new round. This system is referred to as the C4 cycle. To avoid a futile cycle, the site of initial fixation, the mesophyll, is spatially separated from the site of assimilation, the bundle sheath. Canonically, maize operates a linear C4 cycle (Hatch, 1987; Furbank, 2011): In the compartment of initial fixation, the mesophyll, the carbon dioxide acceptor phosphoenolpyruvate (PEP) is formed in the chloroplast from pyruvate by pyruvate:phosphate dikinase (PPDK) and then exported to the cytosol. There, carbon dioxide in the form of bicarbonate is fixed by PEP carboxylase (PEPC), creating the dicarboxylic C4 acid oxaloacetate (OAA) from PEP. OAA is subsequently transferred to the chloroplast and reduced to malate, which is then exported to the cytosol of mesophyll cells. Malate is transported by mass flow to the bundle sheath, the compartment of final assimilation, where it is imported into chloroplasts and decarboxylated by the NADP-dependent malic enzyme (NADP-ME), yielding carbon dioxide, pyruvate, and NADPH. Pyruvate is exported from the chloroplast and returned to the mesophyll for regeneration of the acceptor PEP (Hatch, 1987). Whereas this canonical model of NADP-ME C4 photosynthesis is depicted in many textbooks, several reports question its simplicity; however, an alternative model has not yet been formulated. For example, bundle sheath strands can efficiently decarboxylate not only malate but also the amino acid Asp (Chapman and Hatch, 1981). Older maize leaves, at least, harbor a second decarboxylation enzyme, PEP carboxykinase (PEP-CK), which releases carbon dioxide from OAA, producing PEP (Wingler et al., 1999). Furthermore, approximately one-quarter of radioactively labeled carbon dioxide that was fed to maize leaves was found to be rapidly incorporated into Asp (Hatch, 1971). Such side routes to the canonical NADP-ME C4 pathway would require alternative transfer metabolites between mesophyll and bundle sheath cells, such as Asp or Ala, and alternative decarboxylation pathways would alter the demands on the remaining enzymes and the intracellular (Bräutigam and Weber, 2011a) and intercellular (Sowínski et al., 2008) transport systems. Understanding both the intracellular transport system between chloroplasts and cytosol and the intercellular transport between the mesophyll and bundle sheath cells is still in its infancy (Bräutigam et al., 2008; Sowínski et al., 2008; Bräutigam and Weber, 2011a, 2011b; Weber and von Caemmerer, 2010; Weber and Linka, 2011). Finally, understanding the regulatory circuits controlling C4 photosynthesis is an ongoing quest in plant biology. Although limited information is available, such as the light dependence of C4 enzyme expression (Chollet et al., 1996), the transcription factors mediating the abundant, cell-specific expression patterns remain unknown.
Recent work demonstrates that the maize leaf displays a gradient with regard to proteins (Majeran et al., 2010) and that large-scale transcriptional changes between four leaf areas can be detected (Li et al., 2010). In this work, we set out to generate a comprehensive systems level picture of the changes in metabolite, enzyme activity, and transcript amounts occurring along a developmental gradient of a growing maize leaf. Using this systems biology data set, we addressed the questions of (1) how photosynthesis is organized along the developmental gradient of the light-exposed leaf with special regard to the presence of C3 photosynthesis, (2) whether the biochemistry of the C4 cycle changes along this developmental gradient, and (3) which regulatory modules define the developmental progression in the gradient.
RESULTS AND DISCUSSION
Organization of the Light-Exposed Third Maize Leaf
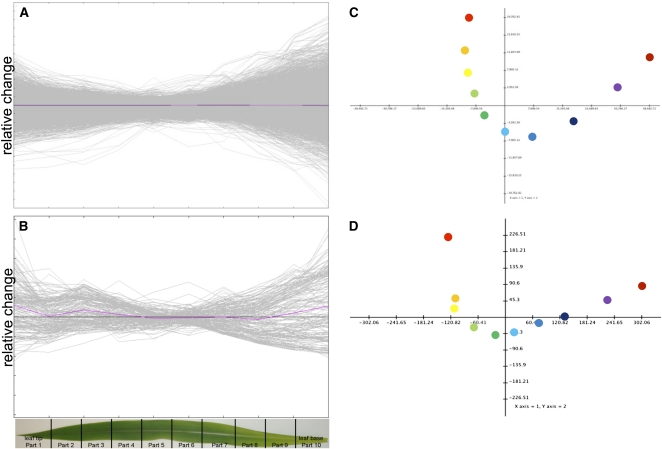
The transcript and metabolite amounts, as well as protein and chlorophyll contents, displayed characteristic and continuous changes along a tip-to-base gradient of the light-exposed part of the third leaf of maize (Figures 1A and 1B). The relative expression or metabolite contents were most distinct at the distal parts of the leaf compared with relatively minor changes in the center of the leaf. A principal components analysis of transcript and metabolite amounts along the leaf gradient demonstrated clear separation of the leaf slices. The principal components determining this pattern were the distance from the leaf base (component 1) and the distance from the leaf center (component 2). The complete data set is available in readable form as Supplemental Data Set 1 online.
Figure 1.
Relative Transcript and Metabolite Levels Are Organized along the Developmental Gradient of the Leaf.
Relative transcript abundance (A), relative metabolite abundance (B), and principal component analysis (C) of transcript levels. The first two components explain 83.5% of the variation (D) principal component of the metabolite levels. The first two components explain 85.5% of the variation.
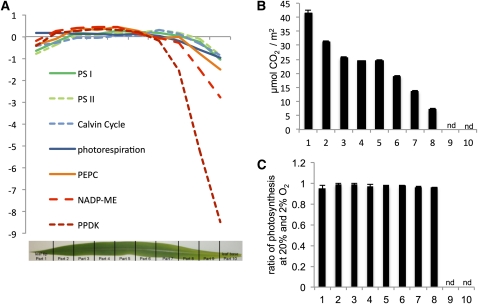
If the leaf was divided from top to bottom into slices, with slice 1 being the tip, gene expression patterns reflecting biochemical pathways could be followed through the development of the leaf (Figure 2A). Since previous work has demonstrated good correlation between transcript and protein abundance in maize (Li et al., 2010), we took transcript amounts as proxies for the corresponding protein amounts. Steady state amounts of transcripts encoding the classical NADP-ME C4 proteins PEPC, PPDK, and NADP-ME were low toward the leaf base and increased until they reached a maximum around slice 2 or 3 for PEPC and slice 10 for the decarboxylation enzymes (Figure 2A). Transcripts representing subunits of photosystems I and II and of the rPPP had a similar pattern, but their increase was much less pronounced than that of the C4 transcripts. The pattern of the photorespiratory transcripts mirrored that of the photosystems and of the rPPP (Figure 2A). No peak of photorespiratory transcripts was observed where expression of the C4 transcripts was low. Photosynthesis, measured as carbon fixation per leaf area, steadily increased between the bottom and the top of the leaf (Figure 2B). Finally, the oxygen sensitivity of photosynthesis was measured to determine whether C3 photosynthesis or inefficient C4 photosynthesis would occur in the light-exposed leaf, which should be reflected by a major increase in the apparent photosynthetic rate at low oxygen partial pressure. However, the ratio of photosynthetic rates measured at high and low oxygen partial pressures did not change along the leaf gradient (Figure 2C). Maize leaves were previously hypothesized to undergo a C3-to-C4 transition. That is, the program initiating C4 photosynthesis was proposed being switched on in a particular region of the leaf (summarized in Nelson and Langdale, 1992). This switch, if existent, was an important target for understanding C4 genesis and thus replicating it in making C4 rice. Our systems-level analysis does not support this hypothesis: Photorespiratory transcripts do not peak in the presumed area of C3-ness (Figure 2A,) and there is no evidence for C3 photosynthesis or leaky C4 photosynthesis, as oxygen sensitivity of photosynthesis did not change along the leaf gradient (Figure 2C). We thus conclude that the maize leaf undergoes a gradual sink-to-source transition without a distinct intermediary C3 phase.
Figure 2.
Photosynthetic Transitions in the Maize Leaf.
(A) Average relative expression levels for the transcripts encoding photosystem I, photosystem II, the RPPP and three key C4 proteins.
(B) Photosynthetic rate along the light-exposed leaf. Error bars depict sd of three biological replicates. nd, not determined since not exposed to light.
(C) Ratio of photosynthesis at 20% and 2% O2 concentration along the leaf gradient. Error bars depict sd; nd, not determined since not exposed to light, two technical replicates.
Metabolite Clusters
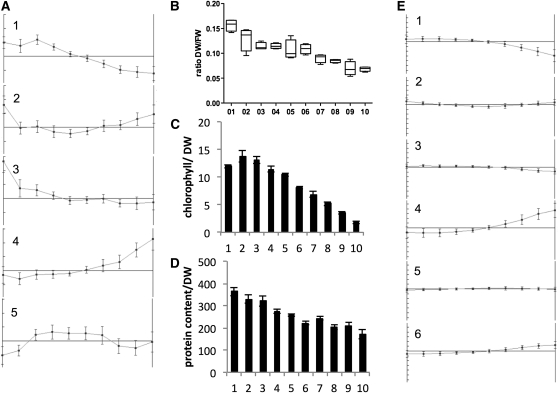
Although the leaf did not contain a zone of C3 photosynthesis, it clearly displayed a gradient along its length (Figure 1; Majeran et al., 2010). To investigate the nature of the gradient in detail, extractable metabolites were analyzed by clustering algorithms. For K-means clustering, a figure of merit analysis determined five clusters as the best compromise between cluster formation with limited information loss (Friedman and Stuetzle, 1981) (see Supplemental Figure 1 online). Metabolites in the pyruvate cluster with 23 members, cluster 1, were low at the bottom of the leaf and increased until slice 3 where the increase leveled off (Figure 3A). Cluster 2, with 15 members, contained metabolites that were high at the very bottom and at the tip, while cluster 3, with 10 members, contained metabolites that were level until the middle of the leaf and then increased toward the tip. The building block cluster, cluster 4, was the largest cluster with 60 members. The metabolites in this cluster started high at the bottom and decreased toward the middle of leaf from where the level stabilized. Cluster 5 was the malate cluster whose metabolites had the highest level between slice 3 and slice 7 and lower levels at the tip and the bottom (Figure 3A; condensed list of metabolites in Table 1).
Figure 3.
Changes along the Leaf Gradient.
(A) K-means clusters of metabolites. Cluster 1 is the pyruvate cluster with 23 members; cluster 2 contains 15 metabolites; cluster 3 is the raffinose cluster with 10 members; cluster 4 is the building block with 60 members; cluster 5 is the malate cluster with 10 members.
(B) The fresh weight (FW)–to–DW ratio indicating a low water content at the leaf tip.
(C) The chlorophyll content.
(D) and (E) The protein content (D) and K-means clustering of transcripts (E). Error bars indicate sd of four biological replicates.
Table 1.
Condensed List of Metabolites within Each Cluster
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 |
| Pyruvate | Man | Raffinose | Coumaric acid | Asp |
| Ala | Galactitol | Galactinol | Ferulic acid | Malate |
| Glycerate | Diethylene glycol | Stachyose | Phosphate | Fumarate |
| Glu | Salicylic acid | Tryptamine | Glucosephosphates | Citrate (additional: isocitrate) |
| Citrulline | Ribonic acid | Trp | Four sugars | Glyoxylate |
| Galactose, lipid fraction | Cys | α-Ketoglutarate | 15 proteinogenic amino acids | γ-Tocopherol |
| Glycerol, lipid fraction | NAD | Nicotinamide | 10 sphingolipids | Threonic acid |
| Digalactosylglycerol | Five fatty acids | Gluconic acid | Four sterols | 3-O-galactosylglycerol |
| 3-O-galactoglycerolipids | Three other metabolites | Myristic acid | Six fatty acids | Two other metabolites |
| Five carotenoids | UDP-glucose | Four precursors | ||
| Four fatty acids | 12 other metabolites | |||
| Five other metabolites |
Metabolites were K-means clustered. A figure of merit analysis determined five clusters as the best compromise between cluster formation with limited information loss (Friedman and Stuetzle, 1981).
Apart from pyruvate, cluster 1 contained Ala, glycerate, Glu, and citrulline. Five carotenoids, α-tocopherol, glycerol, and Gal of the lipid fraction as well as digalactosylglycerol and 3-O-galactoglycerolipids, four fatty acids, and five other metabolites were also members of cluster 1 (Table 1). Surprisingly, the C4 acids formed a distinct cluster, the malate cluster 5, with Asp, fumarate, citrate, glyoxylate, γ-tocopherol, 3-O-galactosylglycerol, and three other metabolites. Since the C3 and C4 acids clearly separated into distinct clusters, there must be a major shift in the C4 cycle. One likely explanation was the observed gradient in PEPC activity. At the point where PEPC activity started to decrease in slice 3 (Figure 2A), the C4 acid pool sizes also sloped downwards. This may indicate that the balance between C3 and C4 acid pools shifted toward C3 acids because carboxylation activity decreased while the sum of decarboxylation activities increased relative to each other (Figure 2A). Some of the pool sizes of tricarboxylic acid (TCA) cycle acids connected to malate also shifted with alterations in PEPC activity. The malate pattern extended to fumarate, citrate, and isocitrate but not to succinate and 2-oxoglutarate. The pool sizes of TCA cycle intermediates were thus only partially isolated from the C4 cycle. The remaining metabolites shadow the buildup of the chloroplasts and thylakoids, including their pigments. The galactolipids, which dominate the chloroplast membranes (Dörmann and Benning, 2002), were mostly members of the pyruvate cluster, as were the accessory pigments carotenoids that can diffuse excess light energy via the xanthophyll cycle (Bilger and Bjorkman, 1990).
Cluster 2 contained Man, galactitol, diethylene glycol, salicylic acid, five fatty acids, and six other metabolites, while the raffinose cluster 3 contains raffinose, stachyose, galactinol, and seven other metabolites (Table 1). Metabolites from both clusters elevated toward the tip, although metabolites in cluster 2 also elevated at the very bottom (Figure 3A). The abundance of metabolites of the raffinose family from cluster 2 and cluster 3 (myo-inositol-2-P, raffinose, stachyose, and galactinol) pointed toward a drought response (Seki et al., 2007) at the tip, but, strikingly, cluster 2 and cluster 3 did not include amino acids such as Pro. The dry weight–to–fresh weight ratio increased toward the tip, with only half the water content at the tip (Figure 3B). However, the third maize leaf analyzed in this study did not show any apparent signs of cell death at the tip (see Supplemental Figures 2A and 2D online). We thus hypothesize that despite the low water content at the tip (Figure 3B), the accumulation of compatible solutes at the tip allows photosynthesis to operate efficiently (Figure 2B). On mature field-grown maize plants, the majority of leaf tips are completely dry with only dead cells remaining. We hypothesize that maize leaves undergo a constitutive innate drought response toward the tip of each leaf to continue photosynthesizing (Figure 2B) until water content gets too low to maintain metabolism and cells undergo cell death. Considering the parallel venation pattern of grasses, any drought stress will likely initially manifest in the leaf tip. In leaf tips of the third maize leaf, chlorophyll content was already reduced (Figure 3C); however, the tissue was likely not senescent since the protein content was high (Figure 3D), photosynthesis was highly efficient (Figure 2B), and senescence markers were not highly expressed (see Supplemental Data Set 1 online).
Cluster 4 was termed the building block cluster. It contained 15 proteinogenic amino acids but not Asp, Ala, and Glu, which were part of the pyruvate and malate clusters. In addition to the amino acids, four precursors (shikimate, quinate, homoserine, and S-adenosylhomoserine) were part of this cluster. The major sugars Glc and Fru as well as the minor sugars Rib and Fru had elevated amounts at the leaf base. Ten sphingolipids, four sterols, and six fatty acids were part of cluster 4. Finally, coumaric and ferulic acid, isopentenylpyrophosphate, glucosephosphates, free phosphate, and 12 other metabolites finished the cluster (Table 1). The leaf base represents a sink tissue (Evert et al., 1996) with minimal photosynthetic activity (Figure 2B). Chloroplasts began to develop at the leaf base (Evert et al., 1996) and chlorophyll content increased (Figure 3C). Genes encoding components of the rPPP and the electron transfer chain were highly expressed. Consequently, proteinogenic amino acids were in high demand and thus present in large amounts. The major sugars likely reflected transferred carbon, while the minor sugars and lignin precursors pointed to active cell wall synthesis. Membrane buildup was in process. Transcript analysis of four distinct zones of the maize leaf found increased transcript amounts for cell wall, lipids, secondary metabolism, and chloroplast targeting for the area at the bottom of the gradient (Li et al., 2010), thereby corroborating the analysis of metabolites. Proteins involved in lipid synthesis also peak toward the base of the gradient (Majeran et al., 2010).
In summary, four clusters defined the leaf gradient, the building block cluster defined by elevated metabolites at the bottom end of the gradient, which was followed by the C4 malate cluster with increased C4 acids and TCA cycle intermediates, which in turn was followed by the C4 pyruvate cluster with high C3 acids, carotenoids, and galactolipids. The tip of the leaf contained elevated amounts of drought-related metabolites of the raffinose family, which are included in clusters 2 and 3 (Figure 3E).
Modules of the Leaf: Transcripts
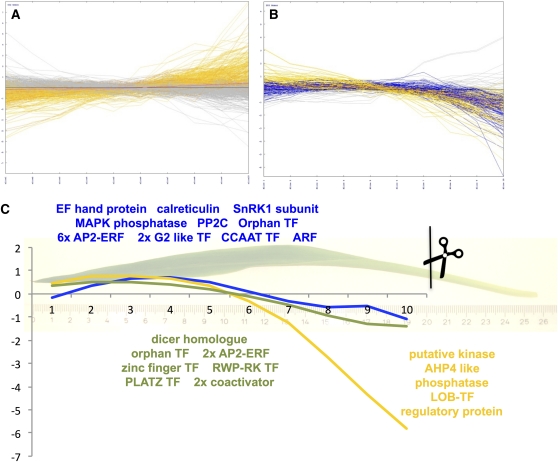
The changes along the maize leaf were recently investigated using four leaf segments sampled from different parts of the leaf (Li et al., 2010). Li et al. identified 938 transcription factors that showed a differential expression pattern between at least two of the segments. In our study, we followed three different strategies to assess the dynamics and extent of the reprogramming of the transcriptome along the maize leaf developmental gradient: (1) K-means clustering to identify patterns of expression along the leaf, (2) hierarchical clustering to identify transcripts with similar patterns as the C4 transcripts, and (3) a comparison of this leaf gradient with previously published data from four distinct leaf segments (Li et al., 2010).
The K-means clustering was prefaced by a figure of merit analysis, which prompted us to choose six clusters as a good solution (see Supplemental Figure 3 online). Four distinct patterns were evident in the clusters: Clusters 1 and 3 contained transcripts that are either very low (1450) or low (8521) toward the bottom of the gradient, clusters 4 and 6 contained transcripts that are either very high (1067) or high (4287) at the bottom, cluster 2 transcripts (4459) were high at the bottom and the top, while the largest cluster 5 (10,935) displayed little change (Figure 3E). The rate of change in transcript abundance (Figures 1A and 3E) agrees with those published earlier for differences between noncontinuous leaf segments (Li et al., 2010) and is comparatively modest, especially for the average changes of each cluster (Figure 3E). Our continuous gradient revealed that virtually all changes in transcript abundance changes were gradual along the gradient. That is, no binary switches, which would manifest in extreme changes (Figure 1A), and consequentially no regulons of genes with sudden onset (Figure 3E) could be detected. Transcripts with changes in the transition zone (Li et al., 2010) corresponding to slice 10 in this study showed steady declines or increases throughout the remainder of the gradient rather than an on–off behavior. These patterns indicated that it is highly likely that the transcriptional changes and functional changes were set up by a morphogradient along the leaf, which may be defined either by one or several metabolites (such as those in the pyruvate cluster) or one or more transcripts (Figure 3E). Although light is necessary for transcription of C4 genes (Langdale et al., 1988), neither the emergence of the leaf into the light at the border between slices 9 and 10 (see Supplemental Figure 2A online) nor the beginning of a planar leaf surface at slices 7 and 8 (see Supplemental Figure 2D online) lead to marked changes in gene expression in the leaf (Figures 1A and 3E). Thus, light was a necessary (Langdale et al., 1988) but not a sufficient cue to alter the gene expression program abruptly, since its availability to the leaf did not cause marked changes. The C4-related transcripts were members of clusters 1 and 3. In comparison to the metabolite clusters, no cluster resembling the malate cluster, which is elevated in the middle of the gradient, was detected. Thus, the factor determining the metabolite accumulation pattern was likely of posttranscriptional nature.
A large number of transcripts of both transcription factors and other functions increased or decreased consistently along the gradient. To narrow down consistent changes, the continuous gradient was compared with the noncontinuous segmental results from Li et al. (2010). Three patterns were originally defined, high at tip and low at the bottom, a group high in the transition zone and low at the tip and very bottom, and a group high at the bottom and low at the tip. Since the bottom of the continuous gradient corresponded to the transition zone in the earlier experiment, the second and third groups merged into one for the purpose of the comparison.
Of the group that was low at the tip, 632 of 725 (87%) were detected reliably on the microarray and 529 (73%) were significantly changed along the gradient based on analysis of variance. Of the significantly changed ones, 276 (52%) decreased toward the tip. For 11%, various expression patterns were detected, while 37% showed a pattern opposite to expectations (Figure 4A).
Figure 4.
Targeted Expression Analysis of Regulatory Functions.
(A) and (B) Expression pattern of transcripts detected as low at the tip (A) and high at the tip (B) in a previous analysis. Patterns in orange confirm the expectation based on Li et al. (2010), patterns in blue partially confirm, and patterns in gray have different patterns.
(C) Transcripts coexpressed with PEPC (blue), NADP-ME (green), and PPDK (yellow) major isoforms.
Of the group that was high at the tip, 203 of 213 (95%) were detected reliably on the microarray and 186 (92%) were significantly changed along the gradient based on analysis of variance. Of those significantly changed transcripts, 68 (37%) continuously increased in expression toward the leaf tip. The plurality, 52%, did increase in expression but dipped slightly at the tip similar to chlorophyll content and PEPC activity (Figure 4B). This change in pattern was visible only with a continuous gradient and cannot be detected with segmental analysis. Only 10% showed patterns that were not congruous with earlier data. If the morphogradient was set up by transcripts that reflected positional information, only 68 transcription factors increasing in expression would be on the short list of candidate transcription factors at or near the core of the morphogradient. By contrast, 276 transcription factors decrease more or less continuously. Adding this second analysis reduced the list of potential transcription factor from 938 in the earlier study down to 344 in our work. Additional analyses have the potential to reduce the list to the point where single-gene functional analyses become feasible. Three important pieces of information are missing: (1) Which of these factors, if any, display a similar gradient in older and bigger leaves, (2) does this gradual behavior extend throughout the leaf to the point of emergence from the apical meristem, and (3) which factors have a similar gradient in other grass species? In older maize leaves of 40-cm length, enzyme activity measurements clearly show gradients for the C4 marker enzymes (see Supplemental Figure 4 online), which are similar to those in the young leaves. This at least indicated that older, more mature leaves still display a gradient.
Systematic analyses of dicot C4 species showed that all C4 enzyme activities except for malate dehydrogenase are, at least to some degree, regulated at the transcriptional level (Bräutigam et al., 2011; Gowik et al., 2011). Even if the C4-related transcripts are piggybacking on the developmental gradient, the direct regulators of their transcription would be expected to be coexpressed with or just predating their targets. We identified transcripts encoding putative regulators that were tightly coexpressed with the major C4 transcripts. The transcript for the major isoform of PEPC accumulated slowly throughout the gradient, reached a plateau between slices 3 and 5, and dipped toward the tip (Figure 4C). In the hierarchical clustering (see Supplemental Data Set 2 online, readable with MeV, www.tm4.org/mev), 16 transcripts representing regulatory functions were identified (see Supplemental Table 1 online). Two of these transcripts related to calcium signaling, one EF hand protein, and a calreticulin. An SnRK1 subunit implicated in sugar and nitrogen signaling (Rolland et al., 2006) had the same pattern as PEPC. In addition, a mitogen-activated protein kinase phosphatase and PP2C, which is involved in abscisic acid (ABA) and drought signaling in Arabidopsis thaliana (Kuhn et al., 2006), were coregulated with PEPC. Finally, 13 transcription factors, one orphan, four APETALA2 (AP2)-ETHYLENE RESPONSE FACTORS (ERFs), two G2-like myb transcription factors, one Auxin Response Factor (ARF), and one CCAAT-type transcription factor tightly correlate with PEPC throughout the gradient. The Arabidopsis homologs of these transcription factors are involved in ABA signal transduction and ethylene signal transduction (see Supplemental Table 1 online). Possibly, an ABA and/or ethylene-driven regulon was used in evolution of C4 photosynthesis. Neither DOF1 nor DOF2, which are known to bind the PEPC promoter region (Yanagisawa and Sheen, 1998), are tightly coexpressed with PEPC. For maize nuclear factor and PEP-I, no sequences were deposited at the National Center for Biotechnology Information (NCBI); hence, they could not be compared with the current data. Notably, none of the tightly correlated transcripts are known to be involved in light signaling, underscoring that light is necessary but not sufficient to drive expression (see above). The two major NADP-ME isoforms showed the same pattern as PEPC up to slice 3 but lacked the dip at the tip. Only nine transcripts encoding regulatory functions tightly correlate (see Supplemental Table 1 online): a dicer homolog, one orphan transcription factor, two AP2-ERFs, a zinc-finger transcription factor, a PLATZ transcription factor, and two coactivators. The PPDK transcript behaved quite differently; it accumulated from very low levels toward slice 5 and then mirrored NADP-ME. Comparatively few regulatory transcripts mirror this more extreme pattern (see Supplemental Table 1 online): a kinase, one phosphatase, a phosphorelay transmitter similar to AHP4 of Arabidopsis, a LOB-type transcription factor, and a regulatory protein similar to a flowering regulator from Arabidopsis. Selected transcript abundance patterns were confirmed by quantitative RT-PCR (see Supplemental Figure 5 online). The differences in pattern between the three key C4 transcripts pointed to the fact that a simple generic C4 regulon may not exist. Rather, additional data sets taken during leaf development of maize and other grasses will increase the resolution of covariation analyses and lead to the identification of the leaf morphogradients and ultimately of regulons that induce expression of the separate C4 genes.
C4 Photosynthesis along the Developmental Gradient of the Leaf
It was recently proposed that C4 plants undergo changes in their mode of C4 photosynthesis based on developmental stage and in response to environmental cues (Furbank, 2011). The well-defined maize leaf developmental gradient analyzed in our study represented a unique opportunity to test this hypothesis.
The classical C4 genes PEPC, PPDK, and NADP-ME were identified from the literature, and their abundance and expression pattern was used to identify transcripts with similar abundance and pattern. The C4 genes were among the transcripts that occupy more than one per thousand of the total (see Supplemental Data Set 3 online). Aside from PEPC, PPDK, and NADP-ME, the list of abundant transcripts contained mainly transcripts that encode the chloroplast electron transfer chain and the rPPP (see Supplemental Data Set 3 online). Surprisingly, genes for a plastid-localized Asp aminotransferase (AspAT) and PEP-CK were also members of this group of 147 transcripts. Coexpressed transcripts frequently act in the same or in connected pathways (Eisen et al., 1998; Reumann and Weber, 2006). Hence, all transcripts were clustered to identify transcripts that are coexpressed with known C4 transcripts. The known C4 transcripts are low toward the bottom of the leaf and increase toward the tip. The very tip portion is slightly lower in expression compared with middle of the leaf blade (Figure 2A). If transcripts, which might be active in any of the C4 types, were plotted, one Ala aminotransferase (AlaAT) and a plastidic AspAT as well as a PEP-CK would display a comparable pattern (see Supplemental Figure 6 online). Taken together with the observation that in maize, 25% of the carbon label initially was located in Asp (Hatch, 1971) and the observation that Asp is a carbon donor to the bundle sheath (Chapman and Hatch, 1981), we decided to investigate the seemingly simple C4 cycle of maize at the levels of transcripts, metabolites, and enzyme activity.
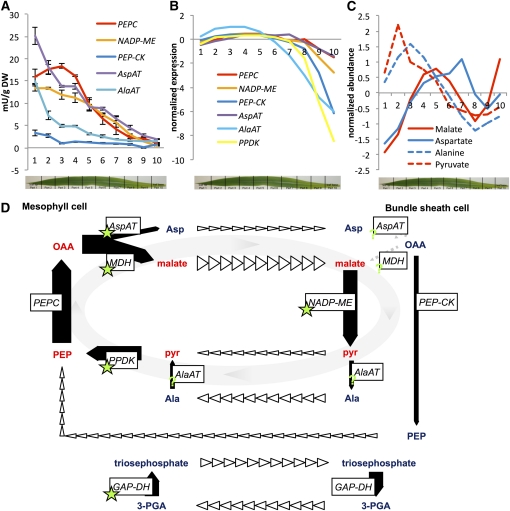
We initiated the analysis by testing whether the leaf had reached C4 configuration at the point where the analysis commenced. In slice 10, the bottom of the gradient, the leaf was already differentiated into a vein, bundle sheath, mesophyll, mesophyll, bundle sheath, vein configuration (see Supplemental Figure 2 online). PEPC activity increased from the bottom, reached a maximum at slice 3, and decreased only slightly toward the tip of the leaf. Maximal activity was 18 milli units/mg dry weight (DW). The major decarboxylation enzyme NADP-ME increased from the bottom toward the top and reached its maximal activity at the leaf tip with close to 15 mU/mg DW. Both AspAT and AlaAT had similar patterns compared with NADP-ME and reached activities of 25 mU/mg and 15 mU/mg DW. Although PEP-CK activity was only a quarter of NADP-ME, it had a comparable pattern and reached up to 4 mU/mg DW (Figure 5A). The pattern for all enzymes except PEPC was similar; that is, the activity was low at the bottom of the leaf and increased toward the tip.
Figure 5.
Selected Enzyme Activities and Metabolite Contents along a Maize Leaf.
(A) Enzyme activities. Error bars indicate sd of four biological replicates.
(B) Expression pattern of the major isoform for each enzyme.
(C) Normalized abundance of four C4 cycle metabolites, with absolute values in slice three: malate, 13.5 mmol/mg DW; Asp,4 mmol/mg DW; Ala, 4 mmol/mg DW.
(D) Model of the C4 cycle in maize. Arrow widths equal approximate flows, bold arrows represent enzyme activities, open arrowheads indicate transport between the cells, and green stars denote plastid-localized steps. pyr, pyruvate; 3-PGA, 3-phosphoglycerate; MDH, malate dehydrogenase; GAP-DH, glycerinaldehyde-3-phosphate dehydrogenase.
On the basis of transcript abundance, the major enzyme isoform of PEPC mirrored the pattern of PEPC activity in the leaf and also peaked around slice 3 (Figure 5B). Transcripts of the major isoforms of NADP-ME, PEP-CK, AspAT, AlaAT, and PPDK, displayed a pattern comparable to that of PEPC but different than the extractable activities of the enzymes. Hence, the total activity was likely composed of multiple isoforms of NADP-ME, PEP-CK, AspAT, and AlaAT and/or subject to posttranscriptional regulation. Indeed, there were other isoforms that were of appreciable transcript abundance (see Supplemental Data Set 3 online) and patterns unlike that displayed by the major isoform (Figure 5B). The enzyme activity of AspAT was sufficient to support the carboxylation and decarboxylation activity, while AlaAT fell short for the majority of the leaf. PEP-CK activity was appreciable. While certainly not the major decarboxylation activity, its activity was high enough to catalyze at least one-fifth of the decarboxylation reactions. This was almost certainly an underestimation since PEP-CK was assayed in the unfavorable reverse reaction (Ashton et al., 1990). If amino acids carried part of the carbon flow in the C4 cycle, their abundance should mirror that of the canonical C4 cycle acids malate and pyruvate. Ala and Asp mirrored the accumulation pattern of pyruvate and malate, respectively (Figure 5C). In addition, not only their pattern but also their absolute abundance should be comparable to that of malate. The absolute abundance of Asp and Ala were about one-fourth of the abundance of malate (see Supplemental Figure 7 online).
Taken together, these results suggested a revised model of the C4 cycle in maize (Figure 5D): After PEP is carboxylated to OAA, it is moved to the chloroplast, either in exchange with malate through DiT1 (Kinoshita et al., 2011) or in exchange with Asp through DiT2 (Renne et al., 2003), which are produced by malate dehydrogenase and AspAT, respectively, in the chloroplast. The major AspAT in maize is predicted to be chloroplast localized. Labeling experiments by Hatch (1971) indicated that as much as 25% of the carbon initially labels Asp, not malate. Both C4 acids diffuse to the bundle sheath, reducing the necessary mass flow compared with either C4 acid carrying the full load. In the bundle sheath, malate is taken up by a currently unknown mechanism into the chloroplast and decarboxylated. Asp may have two fates: It may be transaminated to OAA and decarboxylated by PEP-CK to PEP, or it may enter the chloroplast by an as yet unknown mechanism and be transformed via OAA to malate to serve as the substrate for NADP-ME. It has long been assumed that malate transfer is preferable to Asp transfer since malate carries a reducing equivalent while Asp does not. However, it has been shown that triosephosphate translocator is one of the most abundant chloroplast envelope proteins (Bräutigam et al., 2008) and that the reduction of 3-phosphoglycerate is almost entirely located in the mesophyll (Majeran et al., 2005), making the generation of reducing equivalents unnecessary in the bundle sheath. The regeneration of the carbon acceptor and its transfer to the mesophyll may be dissected by analysis of metabolite compartmentation, flux, and gradients and may occur as PEP, pyruvate, or Ala (Figure 5D).
We propose that independent of environmental or developmental cues, the core C4 cycle in maize is set up already as a branched rather than a linear cycle. In addition to the scheme presented (Figure 5D), the branched core C4 cycle is also connected to basal metabolism (for example, see Leegood and von Caemmerer, 1988). A distribution of carbon between two C4 acids and three C3 acids reduces the diffusion requirements for any one molecule between mesophyll and bundle sheath. This distribution becomes especially important considering that distribution by diffusion is by no means proven (Sowínski et al., 2008; Bräutigam and Weber, 2011a). It remains to be investigated whether the distribution of carbon to malate and Asp is fixed at 3:1 as reported (Hatch, 1971) or whether this ratio is adjusted by the plant during its life cycle (Furbank, 2011). Within the age gradient in a single leaf, there is no evidence in the enzyme activities, transcript abundance, or metabolite accumulation pattern to suggest that operation of the cycle switches from one transfer acid to another (Figure 5). The presence of higher PEP-CK activity in older maize plants with older leaves (Wingler et al., 1999), however, points to a developmental regulation between leaves rather than within a leaf, similar to what has been recently observed in the dicotyledonous C4 plant Cleome gynandra (Sommer et al., 2012). Environmental adaptation of Asp metabolism based on N availability in maize leaves with regard to pool size and turnover has also been demonstrated (Khamis et al., 1992). Hence, the C4 cycle is apparently quite flexible.
Conclusion
On the basis of a comprehensive systems biology data set, we conclude that C4 photosynthesis is established from sink tissue without an intermediate phase of C3 or C2 photosynthesis. That is, the likely evolutionary events are not recapitulated during ontogeny. No binary on–off switches were detected within the leaf gradient, pointing to gradual onset of features and, therefore, morphogradients as the determinants for leaf development. Finally, the biochemistry of C4 photosynthesis is more complex than anticipated but stays constant throughout the leaf.
METHODS
Plant Growth and Harvest
Maize (Zea mays) plants of the ecotype B73 were grown in the greenhouse for 14 to 15 d in clay pots in Floraton soil. Natural light, a shading system, and artificial light were used to extend the daylight period to 16 h at a photon flux density of ~500 μmol m−2 s−1. The humidity in the greenhouse was between 75 and 90%. The greenhouse’s ventilation system kept the temperature at 24°C.
The third leaf was harvested at 18-cm length measured from tip to emergence from the stem. Leaves were harvested by placing them atop a custom-made leaf guillotine where they were snap frozen (see Supplemental Figure 8 online). By closing the lid, the leaf is cut into 10 pieces of 2-cm width each, the last of which had not yet emerged (see Supplemental Figure 2 online). Twenty plants were pooled for each biological replicate. The sections were ground to a fine powder in a porcelain mortar cooled with liquid nitrogen. Frozen powder was used for enzyme assays and metabolomics and transcriptomics analysis. For the DW–to–fresh weight ratio, 30 mg of ground and frozen plant material was dried in a vacuum dryer overnight, and the weight was recorded before and after drying. Δ-13C values were determined according to Coplen et al. (2006). Chlorophyll was determined according to Porra et al. (1989), and protein content was measured with the BCA method (Thermo Fisher Scientific). Enzymatic activities were determined as summarized by Ashton et al. (1990). Photosynthetic rate was measured with a LI-6400XT portable photosynthesis analyzer (LI-COR Environmental) under greenhouse conditions at the time of sampling for the invasive experiments. Oxygen sensitivity of photosynthesis was measured according to Dai et al. (1996). Oxygen partial pressure was controlled by a custom-built gas exchange system. Four biological replicates were measured in all analyses except where otherwise noted.
Metabolite Profiling
Lyophilized tissue equivalent to 200 mg of fresh weight was used for metabolite profiling. Metabolites were extracted with the use of accelerated solvent extraction with polar (methanol + water, 80 + 20 by volume) and nonpolar (methanol + dichlormethan, 40 + 60 by volume) solvents. Subsequent analyses of metabolites by gas chromatography–mass spectrometry (GC-MS) were performed as described elsewhere (Roessner et al., 2000; Walk et al., 2007). In addition, liquid chromatography–tandem mass spectrometry (Niessen, 2003) analyses were performed with the use of an Agilent 1100 capillary LC system (Agilent Technologies) coupled with an Applied Biosystems/MDS SCIEX API 4000 triple quadrupole mass spectrometer (AB Sciex). After reverse-phase HPLC separation, detection and quantification of metabolites were performed in the multiple reaction monitoring and full scan mode (Gergov et al., 2003). Absolute Ala, Asp, and malate contents were estimated by GC-MS (Fiehn et al., 2000), which included an external complex standard and were quantified by coupled enzymatic assays (Bergmeyer, 1974).
Transcript Profiling
The mRNA was isolated after the method of Logemann et al. (1987) and from the same plant material in which the enzyme activities and metabolites were measured. The isolated RNA was purified with the RNeasy purification kit according to the manufacturer’s instructions (Qiagen). The quality was checked with the Agilent 2100 Bioanalyzer using the RNA 6000 Nano kit. The cDNA and following antisense cRNA synthesis was performed according the one-color microarray-based gene expression analysis protocol (Agilent Technologies). An aliquot of 1.65 μg of this RNA was loaded on one-color microarrays with custom-designed oligonucleotide probes (Agilent 025271). Transcripts were normalized to the 75th percentile within each array using the Agilent Gene spring program. Arrays can be accessed under submission number GSE33861 in the NCBI Gene Expression Omnibus database. Quantitative RT-PCR was performed with three biological replicates using the SYBR-green technique (MESA GREEN qPCR MasterMix Plus; Eurogentec) and gene-specific primers (see Supplemental Table 2 online) as described by Schmittgen and Livak (2008). Relative expression values were calculated with the 2−ΔΔct (cycle threshold) method after Pfaffl (2001) with threshold values normalized to expression of 18S rRNA.
Data Analysis
For data analysis, the maize transcript list was downloaded from www.maizesequence.org. For each transcript, a best BLAST hit was produced with Sorghum bicolor and Arabidopsis thaliana as databases (Altschul et al., 1997). Gene Ontology terms were added based on the S. bicolor annotation. Information about putative and known transcription factors (Pérez-Rodríguez et al., 2010) and transport proteins (http://membranetransport.org/) were added based on the Arabidopsis annotation. A Mapman annotation was downloaded from (http://mapman.gabipd.org; Thimm et al., 2004). Protein localization was predicted based on amino acid sequence (Emanuelsson et al., 2000). For each maize transcript, an annotation was created based on the Arabidopsis TAIR10 description (Swarbreck et al., 2008) and, if not available, manually added based on the Sorghum data. Transcripts without known or predicted functions were labeled POUF (for protein of unknown function). Based on all information, transcripts were grouped into classes in a hierarchical manner. Arabidopsis information was given precedence over other information given that Arabidopsis annotations are currently the best within the plant genomes. Group and functional assignments throughout the publication are based on this annotation table. The complete annotation table, including all raw data, can be accessed as Supplemental Data Set 1 online. The major isoform of C4 enzymes were determined by read mapping of raw data from Li et al. (2010) on the maize transcriptome since analyzed data were not included in the original publication. Read mappings were normalized to reads per million without any further correction factors applied. The data are included in Supplemental Data Set 1 online.
All large-scale data analyses were performed with the MultiExperiment Viewer (http://www.tm4.org/mev/; Saeed et al., 2003). Average metabolite contents were expressed as z-scores (the number of standard deviations the value is different from the mean of all values), resulting in mean centered values. Only metabolites detectable in all biological replicates of eight or more slices were analyzed. Transcripts were normalized to the 75th percentile within each array; the mean of replicates was calculated for each slice, followed by mean centering along each row. For K-means cluster analysis, the ideal number of clusters was determined by figure of merit analysis as implemented in MeV (Saeed et al., 2003). Metabolites and transcripts were clustered by Euclidian average linkage clustering and visualized in MeV. For comparison with the Li et al. (2010) data set, transcription factors of different groups were extracted from Li et al. (2010) supplemental data and visualized in MeV (see Supplemental Data Sets 4 and 5 online). Transcripts coexpressed with major C4 enzymes were determined by hierarchical clustering followed by list extraction from MeV. All raw data, including the MeV readable files, are provided as supplemental material accompanying the publication (see Supplemental Data Sets 1 and 2 online).
Accession Numbers
Microarray data from this article can be found in the NCBI Gene Expression Omnibus database under accession number GSE33861.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Figure of Merit Analysis of Metabolite Clustering.
Supplemental Figure 2. Configuration of the Leaf Anatomy along the Stem.
Supplemental Figure 3. Figure of Merit Analysis of Transcript Clustering.
Supplemental Figure 4. Enzyme Activity for Three C4 Marker Enzymes in Maize Leaves of 40-cm Length.
Supplemental Figure 5. qRT-PCR Results for Selected Regulatory Transcripts.
Supplemental Figure 6. Expression Pattern for Enzymes Likely Involved in C4 Photosynthesis.
Supplemental Figure 7. Absolute Concentrations of Malate, Aspartate, and Alanine along the Leaf Gradient.
Supplemental Figure 8. The Guillotine Used for Sampling the Gradient.
Supplemental Table 1. List of Maize Identifiers of Regulatory Transcripts Coregulated with PEPC, PPDK, or NADP-ME.
Supplemental Table 2. Primers Used for qRT-PCR.
Supplemental Data Set 1. The Complete Data Set in Human-Readable Form.
Supplemental Data Set 2. MeV Readable Hierarchical Clustering of All Data to Identify Transcripts Coregulated with Major C4 Enzymes.
Supplemental Data Set 3. Maize Transcripts with the Highest Number of Read Mappings Based on SRR039507 and SRR039508 Originally Published by Li et al. (2010).
Supplemental Data Set 4. MeV Readable Microarray Data for G1 and G2 Upregulated Transcription Factors Published as a Supplemental File by Li et al. (2010).
Supplemental Data Set 5. MeV Readable Microarray Data for G3 Upregulated Transcription Factors Published as a Supplemental File by Li et al. (2010)
Acknowledgments
This study was supported by a grant of the German Federal Ministry of Education and Research (BioEnergy 2021, OPTIMAS) and grants from the German Research Foundation (IRTG 1525 and FOR 1186 PROMICS to A.P.M.W.). We thank Katrin L. Weber for support with GC-MS analyses.
AUTHOR CONTRIBUTIONS
T.R.P. sampled the gradient, measured all parameters except transcripts and relative metabolite content, analyzed the data, and cowrote the article. A.B. designed the research, produced the custom annotation, analyzed data, and wrote the article. U. Schlüter measured transcripts. A.K.D. measured absolute metabolite contents and photosynthesis rates. C.C. and U. Scholz provided tables for the custom annotation. H.F. measured relative metabolite contents. R.P. and U.R. supported the oxygen sensitivity measurements. U. Sonnewald designed the research and analyzed data. A.P.M.W. designed the research, analyzed data, and cowrote the article.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton A.R., Burnell J.N., Furbank R.T., Jenkins C.L.D., Hatch M.D. (1990). The enzymes in C4 photosynthesis. In Enzymes of Primary Metabolism. Methods in Plant Biochemistry, P.M. Dey and J.B. Harborne, eds (London: Academic Press), pp. 39–72 [Google Scholar]
- Bergmeyer H.U. (1974). Methoden der Enzymatischen Analyse, 3rd ed. (Weinheim/Bergstrasse, Germany: Verlag Chemie) [Google Scholar]
- Bilger W., Bjorkman O. (1990). Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbancy changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth. Res. 25: 173–185 [DOI] [PubMed] [Google Scholar]
- Bräutigam A., Hoffmann-Benning S., Weber A.P.M. (2008). Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148: 568–579 Erratum. Plant Physiol. 148: 1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155: 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Weber A.P.M. (2011a). Transport processes – Connecting the reactions of C4 photosynthesis. In Advances in Photosynthesis and Respiration, 1st ed, Vol. 32, A.S. Raghavendra and R.F. Sage, eds (Dordrecht, The Netherlands: Springer), pp. 199–219 [Google Scholar]
- Bräutigam A., Weber A.P.M. (2011b). Do metabolite transport processes limit photosynthesis? Plant Physiol. 155: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.S.R., Hatch M.D. (1981). Aspartate decarboylation in bundle sheath cells of Zea mays and its possible contribution to C4 photosynthesis. Aust. J. Plant Physiol. 8: 237–248 [Google Scholar]
- Chollet R., Vidal J., O’Leary M.H. (1996). Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Coplen T.B., Brand W.A., Gehre M., Gröning M., Meijer H.A.J., Toman B., Verkouteren R.M. (2006). New guidelines for delta13C measurements. Anal. Chem. 78: 2439–2441 [DOI] [PubMed] [Google Scholar]
- Dai Z., Ku M.S.B., Edwards G.E. (1996). Oxygen sensitivity of photosynthesis and photorespiration in different photosynthetic types in the genus Flaveria. Planta 198: 563–571 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Edwards E.J., Smith S.A. (2010). Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl. Acad. Sci. USA 107: 2532–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Evert R.F., Russin W.A., Bosabalidis A.M. (1996). Anatomical and ultrastructural changes associated with sink-to-source transition in developing maize leaves. Int. J. Plant Sci. 157: 247–261 [Google Scholar]
- Fiehn O., Kopka J., Dörmann P., Altmann T., Trethewey R.N., Willmitzer L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Friedman J.H., Stuetzle W. (1981). Projection pursuit regression. J. Am. Stat. Assoc. 76: 817–823 [Google Scholar]
- Furbank R.T. (2011). Evolution of the C(4) photosynthetic mechanism: Are there really three C(4) acid decarboxylation types? J. Exp. Bot. 62: 3103–3108 [DOI] [PubMed] [Google Scholar]
- Gergov M., Ojanperä I., Vuori E. (2003). Simultaneous screening for 238 drugs in blood by liquid chromatography-ion spray tandem mass spectrometry with multiple-reaction monitoring. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 795: 41–53 [DOI] [PubMed] [Google Scholar]
- Gowik U., Bräutigam A., Weber K.L., Weber A.P.M., Westhoff P. (2011). Evolution of C4 photosynthesis in the genus flaveria: How many and which genes does it take to make C4? Plant Cell 23: 2087–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E. (1866). Generelle Morphologie. I: Allgemeine Anatomie der Organismen. II: Allgemeine Entwicklungsgeschichte der Organismen. (Berlin: Verlag Georg Reimer) [Google Scholar]
- Hatch M.D. (1971). The C4 pathway of photosynthesis. Evidence for an intermediate pool of carbon dioxide and the identity of the donor C4 dicarboxylic acid. Biochem. J. 125: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D. (1987). C-4 photosynthesis - A unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895: 81–106 [Google Scholar]
- Hibberd J.M., Sheehy J.E., Langdale J.A. (2008). Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr. Opin. Plant Biol. 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Khamis S., Lamaze T., Farineau J. (1992). Effect of nitrate limitation on the photosynthetically active pools of aspartate and malate in maize, a NADP malic enzyme C4 plant. Physiol. Plant. 85: 223–229 [Google Scholar]
- Kinoshita H., Nagasaki J., Yoshikawa N., Yamamoto A., Takito S., Kawasaki M., Sugiyama T., Miyake H., Weber A.P.M., Taniguchi M. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65: 15–26 [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J.A., Zelitch I., Miller E., Nelson T. (1988). Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 7: 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood R.C., von Caemmerer S. (1988). The relationship between contents of photosynthetic metabolites and the rate of photosynthetic carbon assimilation in leaves of Amaranthus edulis L. Planta 174: 253–262 [DOI] [PubMed] [Google Scholar]
- Li P.H., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Majeran W., Cai Y., Sun Q., van Wijk K.J. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., Ponnala L., Connolly B., Huang M.S., Reidel E., Zhang C.K., Asakura Y., Bhuiyan N.H., Sun Q., Turgeon R., van Wijk K.J. (2010). Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Langdale J.A. (1992). Developmental genetics of C-4 photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 25–47 [Google Scholar]
- Niessen W.M. (2003). Progress in liquid chromatography-mass spectrometry instrumentation and its impact on high-throughput screening. J. Chromatogr. A 1000: 413–436 [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P., Riaño-Pachón D.M., Corrêa L.G.G., Rensing S.A., Kersten B., Mueller-Roeber B. (2010). PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38(Database issue): D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll b extracted with 4 different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Renné P., Dreßen U., Hebbeker U., Hille D., Flügge U.I., Westhoff P., Weber A.P.M. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35: 316–331 [DOI] [PubMed] [Google Scholar]
- Reumann S., Weber A.P.M. (2006). Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim. Biophys. Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. (2000). Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sage R.F. (2004). The evolution of C-4 photosynthesis. New Phytol. 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Sommer M., Bräutigam A., Weber A.P.M. (2012). The dicotyledonous NAD-malic enzyme C4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biol. (vol.) 14: in press [DOI] [PubMed] [Google Scholar]
- Sowiński P., Szczepanik J., Minchin P.E.H. (2008). On the mechanism of C4 photosynthesis intermediate exchange between Kranz mesophyll and bundle sheath cells in grasses. J. Exp. Bot. 59: 1137–1147 [DOI] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36(Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Weber A.P.M., Linka N. (2011). Connecting the plastid: Transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62: 53–77 [DOI] [PubMed] [Google Scholar]
- Weber A.P.M., von Caemmerer S. (2010). Plastid transport and metabolism of C3 and C4 plants—Comparative analysis and possible biotechnological exploitation. Curr. Opin. Plant Biol. 13: 257–265 [DOI] [PubMed] [Google Scholar]
- Walk, T., et al. (2007). Means and methods for analyzing a sample by means of chromatography–mass spectrometry. International Patent WO 2007/012643.
- Wingler A., Walker R.P., Chen Z.H., Leegood R.C. (1999). Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol. 120: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S., Sheen J. (1998). Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10: 75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]