This work uses deep sequencing of RNAs from maize endosperm and embryo to identify 54 maternally expressed genes and 46 paternally expressed genes and then examines genome-wide DNA methylation and gene expression, finding hypomethylation of the maternal allele and endosperm-specific expression for many of the imprinted genes.
Abstract
Imprinting describes the differential expression of alleles based on their parent of origin. Deep sequencing of RNAs from maize (Zea mays) endosperm and embryo tissue 14 d after pollination was used to identify imprinted genes among a set of ~12,000 genes that were expressed and contained sequence polymorphisms between the B73 and Mo17 genotypes. The analysis of parent-of-origin patterns of expression resulted in the identification of 100 putative imprinted genes in maize endosperm, including 54 maternally expressed genes (MEGs) and 46 paternally expressed genes (PEGs). Three of these genes have been previously identified as imprinted, while the remaining 97 genes represent novel imprinted maize genes. A genome-wide analysis of DNA methylation identified regions with reduced endosperm DNA methylation in, or near, 19 of the 100 imprinted genes. The reduced levels of DNA methylation in endosperm are caused by hypomethylation of the maternal allele for both MEGs and PEGs in all cases tested. Many of the imprinted genes with reduced DNA methylation levels also show endosperm-specific expression patterns. The imprinted maize genes were compared with imprinted genes identified in genome-wide screens of rice (Oryza sativa) and Arabidopsis thaliana, and at least 10 examples of conserved imprinting between maize and each of the other species were identified.
INTRODUCTION
During sexual reproduction, both parents contribute one haploid genome to their offspring, and for the majority of genes, the alleles inherited from the mother and father exhibit similar expression levels. However, a small proportion of genes exhibit parent-of-origin differences in the expression of the maternal and paternal alleles, which is termed imprinting (Huh et al., 2008). Imprinting reflects epigenetic control of two identical, or nearly identical, alleles that are present in the same nucleus. In plants, most examples of imprinting have been observed in the endosperm tissue and not in vegetative tissues (reviewed in Gehring et al., 2004; Huh et al., 2008; Jullien and Berger, 2009; Springer and Gutierrez-Marcos, 2009), although there is evidence that imprinting may also occur in embryos (Jahnke and Scholten, 2009). The endosperm is a triploid (2 maternal:1 paternal genomes) product of the double fertilization event in plant reproduction, and there is evidence that some imprinted genes play a role in regulation of endosperm development (Grossniklaus et al., 1998; Kinoshita et al., 1999; Luo et al., 2000; Köhler et al., 2003; Jullien et al., 2006). The kinship theory (Haig and Westoby, 1989) has been proposed as a model to explain why imprinting may provide adaptive value for parental control over allocation of resources to offspring but does not provide a mechanism for imprinted gene expression (Haig and Westoby, 1989). The kinship theory proposes that imprinting would be maintained when a gene’s expression level in one individual affects the fitness of other individuals who have different probabilities for carrying the maternal and paternal alleles (Haig, 2004).
Several lines of evidence suggest that DNA methylation and chromatin changes are important components of the imprinting mechanism. Genome-wide hypomethylation of the maternal genome in endosperm tissue has been observed in multiple plant species (Lauria et al., 2004; Gehring et al., 2009; Hsieh et al., 2009; Zemach et al., 2010). Endosperm hypomethylation in Arabidopsis thaliana is at least partially an active process controlled by demeter, a 5-methylcytosine DNA glycosylase that is expressed in the central cell before fertilization (Gehring et al., 2006). Many regions of endosperm hypomethylation correspond to fragments of transposable elements (Gehring et al., 2009; Zemach et al., 2010). In addition to being important for establishing imprinted expression, endosperm hypomethylation has been proposed to provide a mechanism to reinforce silencing of transposable elements in the adjacent embryo tissue (Springer, 2009; Köhler and Weinhofer-Molisch 2010; Mosher and Melnyk, 2010; Bauer and Fischer, 2011). In addition to the potential involvement of DNA methylation, there is also evidence that chromatin changes mediated by Polycomb group proteins contribute to imprinted regulation (Köhler and Weinhofer-Molisch, 2010; Raissig et al., 2011). At several imprinted maize (Zea mays) genes, there is evidence for both DNA methylation and chromatin modification differences between the maternal and paternal alleles (Haun and Springer 2008).
A handful of imprinted genes were discovered based on parent-of-origin–specific phenotypes associated with mutant alleles (reviewed in Gehring et al., 2004; Huh et al., 2008; Jullien and Berger, 2009; Springer and Gutierrez-Marcos, 2009; Raissig et al., 2011). Several groups have used modified differential display or single nucleotide polymorphism (SNP)–based assays to screen allelic expression to identify additional imprinted genes (Guo et al., 2003; Gutiérrez-Marcos et al., 2004; Stupar et al., 2007; McKeown et al., 2011). Gehring et al. (2009) profiled DNA methylation patterns in endosperm tissue and identified numerous regions of endosperm-specific hypomethylation. A number of additional candidate imprinted genes were identified by finding examples of genes with endosperm-specific expression located near regions of endosperm hypomethylation (Gehring et al., 2009). The development of RNA-seq technologies has enabled several recent genome-wide scans of imprinting in Arabidopsis (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011) and rice (Oryza sativa; Luo et al., 2011). These genome-wide scans can survey allelic expression patterns in the ~10,000 genes that contain polymorphisms between the two parental alleles and are expressed in endosperm. In each case, there have been roughly 100 maternally expressed genes (MEGs) or paternally expressed genes (PEGs) identified. The exact tissue, parental genotypes, and informatics filters used appear to substantially affect the identification of imprinted genes as the genes identified in the three Arabidopsis surveys differ substantially (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011). Generally, there is evidence for imprinting at a number of genes with putative regulatory or signaling roles.
Maize provides an excellent system to study parent-of-origin effects. The maize endosperm is relatively large and persists throughout seed development. In addition, the maize genome has a complex organization of interspersed genes and transposable elements. Given the hypothesis that imprinting may frequently arise via influences of nearby transposable elements (Köhler and Weinhofer-Molisch, 2010; Mosher and Melnyk, 2010; Bauer and Fischer, 2011), there may be numerous imprinted genes in the maize endosperm. In this study, very deep sequencing of transcripts in maize embryo and endosperm tissue was used to identify genes with parent-of-origin effects. A set of 100 putatively imprinted genes was identified. The identification of a large set of imprinted maize genes provides opportunities to further understand the mechanisms and role of imprinting in plants and to further understand seed development.
RESULTS
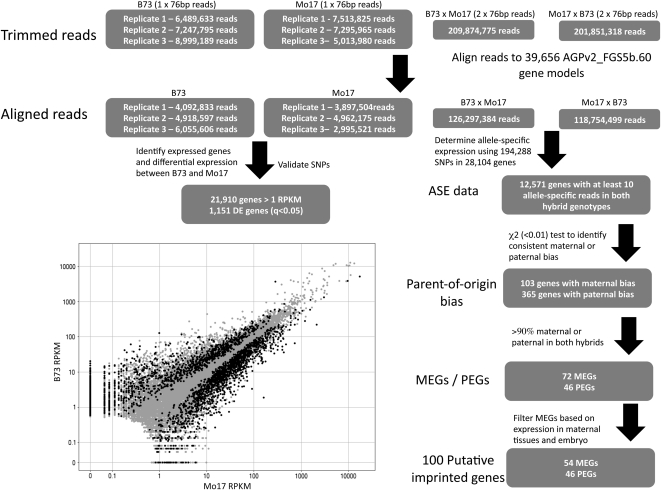
High-throughput sequencing of RNA provides an opportunity to study both gene expression and allele-specific expression levels in hybrid samples. We collected three biological replicates of B73 and Mo17 endosperm tissue 14 d after pollination (DAP), sequencing a relatively small number of single-end 76-bp reads (~5 to 9 million per sample) (Figure 1). In addition, we pooled tissue from three biological replicates of B73 × Mo17 and Mo17 × B73 reciprocal hybrids. We performed deep sequencing of hybrid tissues from pooled replicates instead of from biological replicates because the relevant comparison is between alleles and not between samples. Therefore, our statistics are applied to the sampling of reads for two alleles in the same sample and not to reads between samples. Substantially more sequence reads were recovered from the hybrid samples (Figure 1). In total, we recovered ~28 billion base pairs from >270 million reads of RNA-seq data from 14-DAP endosperm tissue (see Supplemental Table 1 online). The sequence reads were mapped to the 39,656 filtered gene models derived from the AGPv2 reference genome (AGPv2_FGSv5b) (Figure 1). These alignments were used to develop the estimates of relative gene expression levels based on the reads per kilobase per million reads (RPKM) using Tophat-Cufflinks (Trapnell et al., 2010) (RPKM values for all genes in embryo and endosperm tissue were uploaded to the Dryad Data Repository, http://datadryad.org/). Slightly over half (21,910 out of 39,656) of the genes have a RPKM value over 1 in both reciprocal hybrids and were retained as expressed genes for further analyses. We also gathered parental and reciprocal hybrid RNA-seq data from 14-DAP embryo tissue. The analysis of genes with endosperm-specific and embryo-specific gene expression patterns revealed that we could find examples of genes with moderate to high levels of expression in embryo but without transcripts in endosperm. However, we did not find genes with endosperm expression but no transcripts in embryo (see Supplemental Figure 1 online).
Figure 1.
Flow-Chart for Analyses of Endosperm RNA-Seq Data.
The number of trimmed reads is shown for each of the sequenced samples. These reads were aligned to the AGPv2_FGS5b gene models. The data from the inbreds were used to confirm SNPs and to evaluate differential expression in B73 relative to Mo17. The graph shows the RPKM values for the two genotypes, and the black data points indicate genes with significantly (q < 0.05) different expression in the two genotypes. The RNA-seq data from the hybrids were used for allele-specific expression analyses to identify imprinted genes.
Identification of Imprinted Maize Genes
Allele-specific gene expression was determined by mapping the sequence reads to a set of 1.55 million B73-Mo17 SNPs. Over 70% (28,104/39,656) of the Zm5a.60 FGS sequences have at least one B73-Mo17 SNP, and 58% (22,916) genes have at least three SNPs. The number of reads that correspond to the B73 and Mo17 alleles were determined for each SNP position and were summed across the gene to determine the number of maternal and paternal reads per gene (see Supplemental Data Set 1 online). In endosperm, there are 12,571 genes for which there are at least 10 reads that could be assigned to a specific allele in both reciprocal hybrids (Figure 1). This analysis enabled us to assess allele-specific expression for 57% of the genes that are expressed in endosperm tissue. Similar criteria identified 11,606 genes in embryo tissue with robust allele-specific expression data.
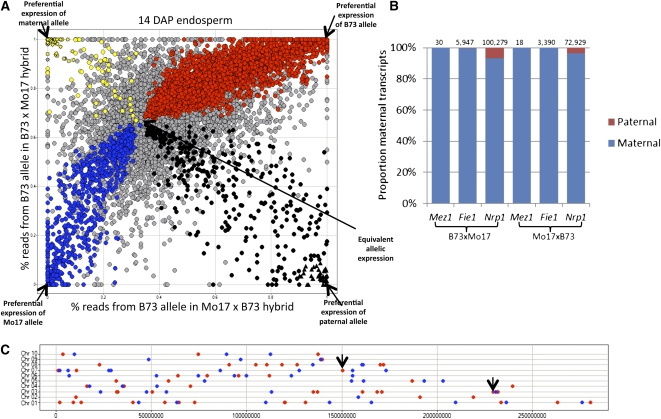
The relative allele-specific expression levels from the reciprocal hybrids can be used to assess the impact of allelic variation (B73 versus Mo17) (see Supplemental Figure 2 online) and parent-of-origin effects (maternal versus paternal) (Figure 2A). If the two alleles are equivalently expressed in endosperm tissue, we would expect to observe 66% of the transcripts derived from the B73 allele in the B73 × Mo17 endosperm tissue and 33% of the transcripts derived from the B73 allele in Mo17 × B73 endosperm tissue. In cases where one allele (B73 or Mo17) is preferentially expressed, presumably due to cis-regulatory variation, it is expected that expression would be biased toward the same allele in both reciprocal hybrids and will follow a curve toward complete B73 or Mo17 allele expression. By contrast, examples of parent-of-origin effects would be expected to exhibit a bias toward one allele in one hybrid and bias toward the other allele in the reciprocal hybrid. The observed data (Figure 2A) show that most genes are distributed along the curve, indicating common allelic variation for gene expression. A contrast of the allelic expression bias in hybrid samples with differential expression in the parental genotypes (see Supplemental Figure 2 online) provides further evidence for common cis-regulatory variation in B73 and Mo17 alleles. It should be noted that there is some evidence for preferential detection of the B73 allele (center of the distribution is shifted up and to the right) that is likely the result of more efficient mapping of sequence reads to the reference B73 allele than to the Mo17 allele rather than an actual expression bias. The genes were categorized based on whether they exhibit biased allelic expression in both hybrids (χ2 < 0.01) relative to the expectation of 2:1 maternal-to-paternal transcripts (see Supplemental Data Set 1 online). Nearly 5000 genes showed a consistent bias toward the B73 or Mo17 allele in both reciprocal hybrids (Figure 2A). A smaller number of genes showed evidence for consistent maternal (n = 103) or paternal (n = 365) bias in both reciprocal hybrids (Figure 2A; see Supplemental Data Set 1 online).
Figure 2.
Identification of Imprinted Genes in Maize Endosperm.
(A) The relative proportion of transcripts derived from the B73 allele was determined for both reciprocal hybrids (limited to only genes with at least 10 reads assigned to one allele in both hybrids). Genes indicated with a color are those that exhibit a significant (χ2 < 0.01) bias from the expected 2:1 ratio in both hybrids. Red coloration indicates genes that are biased toward higher expression of the B73 allele, while blue indicates genes that are biased toward higher expression of the Mo17 allele. The yellow and black spots represent genes with maternal or paternal bias, respectively. The triangles represent the 46 genes that have at least 90% of the transcripts derived from the paternal allele, and the star symbols represent the 72 genes with at least 90% of the transcripts derived from the maternal allele.
(B) The proportion of maternal (blue) and paternal (red) transcripts in both hybrids are shown for three genes previously identified as imprinted genes. The total number of allele-specific reads for each gene in each sample is indicated above the bar.
(C) Chromosomal distribution of imprinted genes. The chromosomal positions are shown for each of the 100 imprinted genes (red spots indicate maternal expression and blue spots indicate paternal expression). The black arrows indicate two loci where adjacent genes are both expressed from the maternal allele.
We focused on the relative expression of the maternal and paternal alleles of genes with parent-of-origin effects. The implementation of χ2 tests to find consistent maternal or paternal bias provides one mechanism to identify MEGs or PEGs (Figure 1). However, these criteria can identify genes with high expression levels that have parental effects of small magnitude. Therefore, we further filtered the list of MEGs and PEGs by requiring at least 90% of the transcripts to be maternal or paternal in both reciprocal hybrids (Figure 1). This resulted in the identification of 72 MEGs and 46 PEGs (see Supplemental Data Set 1 online). A similar set of allele-specific expression analyses were performed on the RNA-seq data obtained from embryo tissue to identify MEGs or PEGs in embryo. These analyses identified 29 MEGs and nine PEGs. However, all of these genes were also identified as imprinted in endosperm and all show much higher levels of expression in endosperm than in embryo. This finding has several potential explanations. First, it is possible that there is endosperm contaminating our embryo samples. However, while it is relatively easy to separate endosperm and embryo at 14 DAP, we can’t rule out contamination. A second explanation for our finding is that there is trafficking of transcripts produced in the endosperm to the embryo tissue. This would result in apparent parent-of-origin bias for these transcripts without active expression in embryos. A third explanation is that these transcripts exhibit active parent-of-origin–specific transcription in both embryo and endosperm, similar to mee1 (Jahnke and Scholten, 2009). A fourth possible explanation is that these are relatively stable transcripts that are inherited from the sperm or egg. With information only on steady state transcript levels, rather than on active transcription in embryo tissue, it is difficult to separate these potential explanations. We thus focused the remaining analyses on the endosperm-imprinted genes.
Genes could be identified as MEGs for several reasons in addition to true imprinted expression, including contamination by maternal tissues and the presence of stable maternally inherited transcripts. By contrast, PEGs could be due to the presence of transcripts provided by the male gamete or due to imprinted expression in endosperm. While it is possible that some of the MEGs and PEGs are the result of stable transcripts provided by the parental gametes, these are likely to be rare at 14 DAP. To assess whether some of the MEGs might result from contamination with maternal tissues, we assessed the relative expression patterns for the 58 MEGs that are represented in a maize gene expression atlas (Sekhon et al., 2011) (the remaining 14 MEGs were not assessed in Sekhon et al., 2011). Hierarchical clustering for these genes was performed in a series of tissues representing different developmental stages of embryo, endosperm, whole seed, and pericarp (see Supplemental Figure 3 online). A subset of the MEGs exhibited at least fivefold higher expression in whole seeds relative to endosperm tissue (18/58), and 13 of the 18 genes were not highly expressed in endosperm tissue. Thus, the maternal bias for these genes may be due to presence of small amounts of contamination from maternal tissues (see Supplemental Figure 3 online). The 18 genes were therefore omitted from further analyses, leaving 54 MEGs that were classified as putatively imprinted genes. Expression data indicated higher levels of expression in endosperm tissue than in whole seeds for all 40 of the remaining genes with expression atlas data (Sekhon et al., 2011).
The 54 MEGs and 46 PEGs resulted in 100 genes classified as putatively imprinted (see Supplemental Data Set 2 online). There are three well-characterized imprinted genes that are polymorphic in B73 and Mo17: Fie1 (Danilevskaya et al., 2003; Gutierrez-Marcos et al., 2003), Nrp1 (Guo et al., 2003), and Mez1 (Haun et al., 2007). Imprinting for each of these three loci was well supported by the RNA-seq data (Figure 2B). The cross between B73 and Mo17 did not include polymorphic or imprinted alleles for some other known imprinted loci; therefore, we could not assess imprinting at R (Kermicle, 1970), B (Selinger and Chandler, 2001), mee1 (Jahnke and Scholten, 2009), or Meg1 (Gutiérrez-Marcos et al., 2004). Stupar et al. (2007) reported imprinting for two other EST sequences in B73 × Mo17. Only one of these genes, GRMZM2G099295, was within the filtered gene set, and although this gene did not contain enough allele-specific reads to pass our filtering criteria, the observed reads, the presence of nine maternal reads, and zero paternal reads did support imprinting.
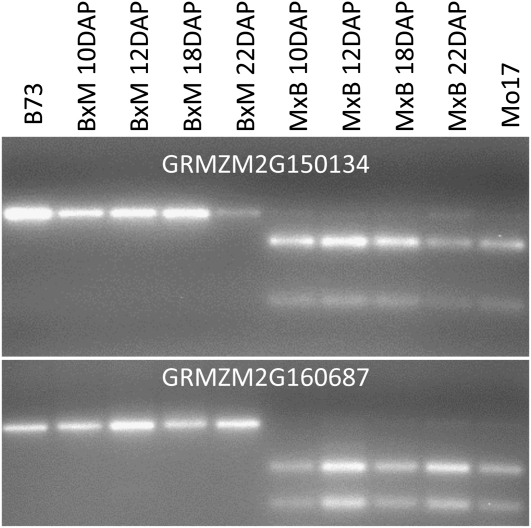
We proceeded to confirm the allele-specific expression pattern observed in the RNA-seq analysis for a number of the novel imprinted genes. The imprinted expression of 24 of the novel genes was assessed by either cleaved amplified polymorphic sequence (CAPS) assays (Figure 3) or sequencing of RT-PCR products (see Supplemental Figure 4 online) using an independent biological replicate sample of hybrid 14-DAP endosperm tissue. Imprinted expression was confirmed for 23 of the 24 genes that were tested. Together with the three previously identified examples of imprinting, we confirmed imprinting for 27 of the genes. For several of these genes (Figure 3), we further confirmed the imprinting at additional time points earlier and later in endosperm development.
Figure 3.
Persistent Imprinting throughout Endosperm Development.
For several genes, we developed CAPS assays to assess allele-specific expression at multiple stages of endosperm development including 10, 12, 18, and 22 DAP. The outer lanes provide the expected patterns for the B73 and Mo17 alleles (data are from 14-DAP B73 and Mo17 tissue). The inner lanes show the digests of products derived from reciprocal hybrids. The BxM samples represent hybrids in which B73 was the maternal parent and the MxB samples had Mo17 as the maternal parent.
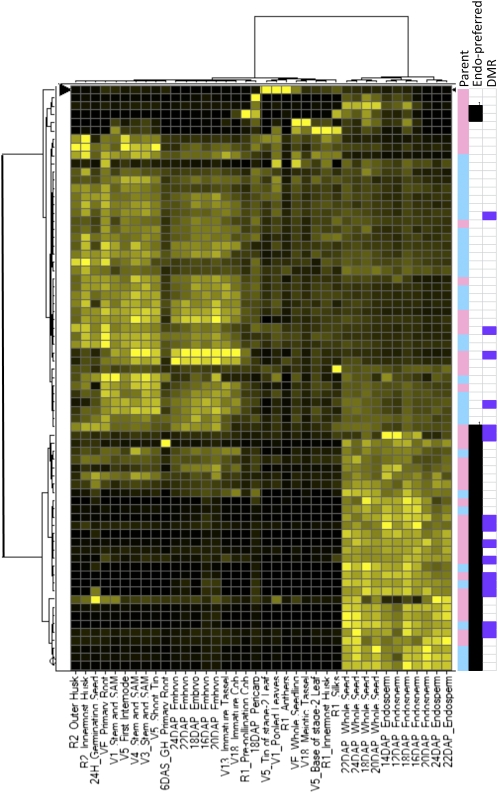
Previous studies of small numbers of imprinted genes have noted that many are expressed exclusively in endosperm tissue (Danilevskaya et al., 2003; Kinoshita et al., 2004). Many of our putative imprinted genes (71/100) were surveyed in a recent maize expression atlas (Sekhon et al., 2011). The normalized expression levels in all of these tissues were compared to identify genes with endosperm-preferred expression (Figure 4). The average expression in endosperm was compared with the average expression level in all other tissues (excluding whole seeds that include endosperm tissue) to identify genes with high levels of endosperm expression relative to other tissues. Nearly half of the genes (31/71) exhibit at least fivefold higher expression in endosperm than in other tissues (Figure 4). The genes with endosperm-preferred expression include both MEGs (20) and PEGs (11). The remaining genes exhibit moderate levels of expression in other tissues or developmental stages.
Figure 4.
Developmental Expression Pattern of Imprinted Genes.
Expression atlas data (Sekhon et al., 2011) were available for 71 of the 100 imprinted genes. The expression atlas data were normalized across all tissues. These patterns of expression were then assessed by hierarchical clustering using Ward’s method. The resulting heat map illustrates expression of each gene across the different tissues, with black indicating low expression and yellow indicating higher levels of expression. The three columns to the right of the dendrogram illustrate features of each imprinted gene. The first column indicates whether the gene is a MEG (pink) or a PEG (blue). The second column indicates which genes were classified as having endosperm-preferred expression (black). The final column indicates the genes that had a DMR in purple.
Potential Examples of Allele-Specific Imprinting
Imprinting describes unbalanced expression of two alleles based on their parent of origin. In our identification of MEG and PEG genes, we required >90% uniparental transcripts in both reciprocal hybrids samples. This requires that both the B73 and Mo17 alleles exhibit imprinted expression patterns. Given the hypothesis that imprinting may target transposable element sequences (Köhler and Weinhofer-Molisch, 2010; Mosher and Melnyk, 2010; Bauer and Fischer, 2011), there may be instances of imprinting in which certain alleles are imprinted but other alleles at the same locus do not exhibit imprinting due to epigenetic or genetic variation (allele-specific imprinting). Indeed, there are examples of allele-specific imprinting at the R, B, and dzr loci of maize (Kermicle, 1970; Chaudhuri and Messing, 1994; Selinger and Chandler, 2001), and several putative examples of allele-specific imprinting have been identified in Arabidopsis (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011). We identified potential allele-specific imprinting candidates as genes with at least 20 allele-specific reads in both reciprocal hybrids that had >98% of the reads from one parent in one of the hybrids but biallelic (40 to 80% maternal expression) in the other hybrid (see Supplemental Data Set 1). A total of 53 potential allele-specific imprinting candidates were identified in our data. These results would need to be tested in additional crosses to further evaluate allele-specific imprinting in maize.
Endosperm-Specific Hypomethylation at Many Imprinted Maize Genes
For many previously identified imprinted plant genes, there is evidence for reduced DNA methylation of the maternal allele (Jullien and Berger, 2009; Raissig et al., 2011). We generated whole-genome methylation profiling data for embryo, endosperm, and leaf tissue for both B73 and Mo17. The methylation profiling was performed by immunoprecipitation with a 5-methylcytosine antibody followed by hybridization to a 2.1-million-feature-long oligonucleotide microarray that contains probes every ~200 bp across the low-copy space of the maize genome. This array platform included 3058 probes within or in regions surrounding the 100 imprinted genes. We searched for examples of multiple adjacent probes near the imprinted genes that showed lower levels of DNA methylation in endosperm tissue relative to embryo and leaf in both B73 and Mo17. There are 19 genes that contain a region of endosperm-specific hypomethylation (see Supplemental Data Set 2 online). The 19 regions of reduced endosperm methylation include 12 genes with preferred maternal expression and seven genes with preferred paternal expression.
The reduced methylation in endosperm tissue is hypothesized to be the result of demethylation of the maternal allele while the paternal allele remains methylated. Indeed, one of these hypomethylated regions was found at the same site as the previously identified differentially methylated region (DMR) of maize Fie1 (Gutiérrez-Marcos et al., 2006; Hermon et al., 2007) (see Supplemental Figure 5A online). A previously identified DMR at Mez1 (Haun et al., 2007) was not identified using these criteria. However, closer inspection found that the single probe at the DMR showed evidence for lower methylation levels in endosperm (see Supplemental Figure 5B online). The Mez1 DMR is a near-repetitive sequence, so there are no other probes covering this region. It is likely that some other genes may also have DMRs that were not identified since they were not interrogated by multiple adjacent probes and did not fit our stringent criteria for DMR identification.
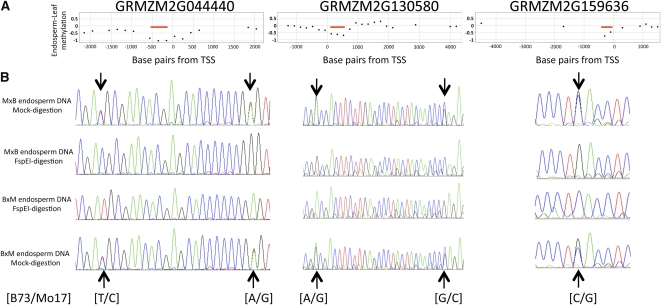
We proceeded to assess the allele-specific methylation patterns at several of the DMRs identified by the MeDIP-chip analysis. These assays were limited to nine DMR regions that include SNPs that allow for discrimination of the maternal and paternal alleles. For each region, we amplified undigested genomic DNA and genomic DNA that had been subjected to a restriction digest using the methylation-dependent restriction enzyme FspEI. The amplified products were then sequenced to assess the relative abundance of the two alleles. In all cases, the undigested DNA revealed a mixture of the maternal and paternal alleles (Figure 5B; see Supplemental Figure 6 online). However, following digestion with the methylation-dependent enzyme, only the maternal SNP was observed. This provides evidence for differential methylation of the paternal allele and maternal allele at these regions with lower levels of methylation at the maternal allele. In all nine examples tested (six MEGs and three PEGs), we found evidence for maternal hypomethylation.
Figure 5.
Identification of DMRs at Imprinted Genes.
(A) The panels display array-based methylation profiling data for three genes. The y axis represents the relative differences in methylation levels in endosperm and leaf tissue (negative values indicate lower methylation in endosperm). The x axis indicates the base pair distance of the probe from the transcription start site (TSS). Each data point represents a single probe, and the values are determined from the average of three replicates of B73 and Mo17 tissues. The red lines indicate regions that were identified as hypomethylated in endosperm.
(B) For each of these three genes, we identified SNPs within the hypomethylated region (listed under traces in [B73/Mo17] format). The SNP-containing regions were amplified and sequence in both reciprocal hybrids. The sequencing plots from the undigested DNA from the reciprocal hybrids (first and last traces) reveal presence of both alleles. However, following digestion with the methylation-dependent restriction enzyme FspEI (middle traces), only the maternal allele is detected confirming specific hypomethylation of the maternal allele.
Our data provided evidence for DMRs near some imprinted genes but not others. We hypothesized that DMRs may be more prevalent near imprinted genes that exhibit endosperm-preferred expression. These genes could be silenced by DNA methylation in other tissues but then exhibit increased expression in endosperm concomitant with reduced methylation. Expression atlas data (Sekhon et al., 2011) were available for 14/19 genes that contain DMRs. The majority of these genes (10/14) have at least fivefold higher expression levels in endosperm tissue than in other plant tissues (Figure 4; see Supplemental Data Set 2 online). This provides evidence for prevalent DMRs in imprinted genes with endosperm-preferred expression.
Characterization of Imprinted Maize Genes
The chromosomal positions of the remaining 100 imprinted genes were examined (Figure 2C). While clustering of imprinted genes is prevalent in mammals, there is little evidence of clustering in plant species. The imprinted genes were distributed across all 10 maize chromosomes. We found two examples in which adjacent genes show maternal-specific expression, including GRMZM2G354558/GRMZM2G354579 on chromosome 3 and GRMZM2G144594/GRMZM2G569356 on chromosome 7. In both cases, the two genes show primarily maternal expression and are expressed from the same strand. Other than these two examples, we did not observe evidence for clusters of multiple imprinted genes located within 250 kb of each other.
BLASTP alignments of the protein sequences from the maize imprinted genes identified a related Arabidopsis protein for 40/54 of the MEGs and 40/46 of the PEGs (see Supplemental Data Set 2 online). The annotations of these Arabidopsis genes were analyzed to identify functional categories of genes (based on GO_slim plant annotations) that are overrepresented using BinGO (Maere et al., 2005). The PEGs are enriched for binding activities based on nucleic acid and DNA binding proteins. In addition, these genes are enriched for kinase activities and for genes involved in flower development (see Supplemental Figure 7 online). The MEGs are enriched for several processes, including multicellular development and response to stimulus (Table 1). In addition, when the Gene Ontology (GO) biological process annotations of the imprinted genes were assessed, there are enrichments for chromatin modification and organization terms in both MEGs and PEGs. In particular, maize orthologs of Variant in Methylation1 (VIM1) (Woo et al., 2007), Vernalization5 (Greb et al., 2007), Fertilization Independent Endosperm (FIE) (Ohad et al., 1999), and Curly Leaf (Goodrich et al., 1997) exhibit imprinted expression patterns. The overrepresentation of chromatin proteins and transcription factors suggest that many targets of imprinting have important regulatory roles.
Table 1.
GO Slim Enrichments Imprinted Genes
| GO_ID | Description | Genes | P Valuea |
| PEGsb | |||
| 3677 | DNA binding | 11 | 0.01 |
| 9908 | Flower development | 4 | 0.01 |
| 3676 | Nucleic acid binding | 14 | 0.02 |
| 16301 | Kinase activity | 7 | 0.02 |
| 5488 | Binding | 24 | 0.02 |
| MEGsb | |||
| 7275 | Multicellular organismal development | 11 | 0.00 |
| 32501 | Multicellular organismal process | 11 | 0.00 |
| 50896 | Response to stimulus | 13 | 0.02 |
| 50789 | Regulation of biological process | 11 | 0.04 |
False discovery rate–corrected P value.
n = 40 genes with Arabidopsis homologs.
Several recent studies in Arabidopsis and rice have used RNA-seq to identify imprinted genes (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Wolff et al., 2011). We sought to determine whether similar genes are imprinted in multiple species. This comparison is complicated by the fact that different sets of genes were assessed in each species due to SNP availability and read coverage. Imprinting was assessed for roughly one-half to one-third of the genes in each of the species being studied. The 100 maize imprinted genes were used as queries against all rice or Arabidopsis protein sequences, and up to three close matches in the target species were identified. There are 11 examples of maize genes where one of the closest matches in rice exhibits imprinting (Luo et al., 2011) (Table 2). Luo et al. (2011) did not report which rice genes contained polymorphisms between the parental lines; therefore, it is not clear whether the absence of detected imprinting was due to lack of imprinting or lack of polymorphisms. Therefore, it is possible that the rice ortholog for some of the other imprinted maize genes is actually imprinted but could not be surveyed by Luo et al. (2011). The best Arabidopsis matches (up to three) were identified for 80 of the 100 imprinted maize genes and were compared with data from Arabidopsis studies. Similar to our study, each of the Arabidopsis imprinting studies identified a larger nonstringent list of imprinted genes and a subsequent filtered list that aimed to remove genes due to potential maternal tissue contamination or other false positives. We identified 10 maize imprinted genes for which Arabidopsis homologs were stringently identified as imprinted (Table 3). We also noted instances in which some of these genes were identified in the stringent list for one study but in the nonstringent list for another study and genes for which Gehring et al. (2009) identified endosperm DMRs near these Arabidopsis genes. There are two examples in which two different maize genes are both related to the same imprinted Arabidopsis genes. There are two genes for which imprinting is detected in all three species, AC19534.3FG003/Os04g22240/AT1G57820 (VIM1) and GRMZM2G365731/Os10g30944/AT4G11400 (ARID/BRIGHT DNA binding domain). Given that <1% of the genes in any one species were identified as imprinted, the overlap of imprinting among maize and Arabidopsis or rice was highly significant (P < 0.001). In addition, many examples that lack conservation for imprinting may reflect false-negatives due to lack of ability to assess imprinting in all three species.
Table 2.
Examples of Conserved Imprinting in Maize and Rice
| Maize Gene ID | Chromosome | Maternal Read No. | Paternal Read No. | Maize Pattern | Rice Gene ID | Rice Description | Luo et al., 2011 | Arabidopsis Gene ID |
| AC191534.3_FG003 | chr7 | 23 | 2397 | PEG | Os04g22240 | C3HC4-type zinc-finger | PEG | AT1G57820 |
| GRMZM2G000404 | chr7 | 0 | 160 | PEG | Os09g28940 | Ubiquitin-specific protease | PEG | |
| GRMZM2G028366 | chr5 | 15 | 13763 | PEG | Os02g12840 | DEAD-box ATP-dependent RNA helicase | PEG | AT1G54270 |
| GRMZM2G073700 | chr6 | 15858 | 592 | MEG | Os06g11730 | RNA binding (RRM/RBD/RNP motifs) | MEG | AT1G78260 |
| GRMZM2G110306 | chr1 | 0 | 33 | PEG | Os08g27240 | ARID/BRIGHT DNA binding domain | PEG | AT3G43240 |
| GRMZM2G118205 | chr4 | 9320 | 17 | MEG | Os08g04290 | OsFIE | MEG | AT3G20740 |
| GRMZM2G170099 | chr3 | 2891 | 20 | MEG | Os01g18810 | Hypothetical protein | MEG | |
| GRMZM2G365731 | chr1 | 2 | 396 | PEG | Os10g30944 | ARID/BRIGHT DNA binding domain | PEG | AT4G11400 |
| GRMZM2G379898 | chr1 | 72 | 0 | MEG | Os03g01320 | Protease inhibitor/seed storage/LTP | MEG | AT1G62500 |
| GRMZM2G447406 | chr8 | 100 | 3155 | PEG | Os01g70060 | Hypothetical protein | PEG | |
| GRMZM5G845175 | chr10 | 24 | 708 | PEG | Os04g32880 | CBS domain-membrane protein | PEG | AT1G09020 |
Table 3.
Examples of Conserved Imprinting in Maize and Arabidopsis
| Maize Gene ID | Chromosome | Maternal Read No. | Paternal Read No. | Maize Pattern | Arabidopsis Gene ID | Arabidopsis Description | Gehring et al. (2011)a | Hsieh et al. (2011)a | Wolff et al. (2011)a |
| AC191534.3_FG003 | chr7 | 23 | 2397 | PEG-DMR | AT1G57820 | VARIANT IN METHYLATION1 (VIM1) | PEG-DMRb | MEGfil | |
| GRMZM2G064905 | chr8 | 3 | 62 | PEG | AT5G53150 | DNAJ heat shock domain | PEG | MEGfil | |
| GRMZM2G127160 | chr6 | 1354 | 84630 | PEG | AT1G70560 | Trp aminotransferase of Arabidopsis1 (TAA1) | PEG-DMR | PEG | |
| GRMZM2G365731 | chr1 | 2 | 396 | PEG | AT4G11400 | ARID/BRIGHT DNA binding domain | DMR | PEG | |
| GRMZM2G014119 | chr6 | 6869 | 10 | MEG-DMR | AT5G03240 | Polyubiquitin3 (UBQ3) | MEG | MEGfil | MEGfil |
| GRMZM2G044440 | chr8 | 2795 | 9 | MEG-DMR | AT5G03240 | Polyubiquitin3 (UBQ3) | MEG | MEGfil | MEGfil |
| GRMZM2G103247 | chr9 | 98 | 2 | MEG-DMR | AT2G28890 | Poltergeist like4 (PLL4) | MEG-DMR | MEGfil | |
| GRMZM2G112925 | chr1 | 492 | 49 | MEG | AT2G28890 | Poltergeist like4 (PLL4) | MEG-DMR | MEGfil | |
| GRMZM2G370991 | chr5 | 15027 | 22 | MEG-DMR | AT5G03280 | ETHYLENE INSENSITIVE2 (EIN2) | MEG-DMR | MEG | MEGfil |
| GRMZM2G088020 | chr8 | 106 | 0 | MEG | AT5G53870 | Early nodulin-like protein1 (ENODL1) | MEGfil | MEG | MEGfil |
The genes listed as MEG or PEG passed all filters in the cited reference. Genes listed as MEGfil were filtered due to potential seed coat expression or because they did not pass stricter statistical cutoffs for imprinting in these studies but may actually be imprinted.
The presence of a DMR from Gehring et al. (2009) is indicated.
DISCUSSION
We used RNA-seq to identify MEGs and PEGs that likely represent examples of imprinting in maize. The use of genome-wide allele-specific expression profiling greatly increased the number of putatively imprinted maize genes. Previous studies documented ~10 maize imprinted genes (Springer and Gutierrez-Marcos, 2009; Raissig et al., 2011). Our analysis uncovered several of these same loci and identified many novel imprinted genes (the others did not have SNPs in B73/Mo17 and could not be assessed). Some known imprinted genes exhibit parent-of-origin effects during early endosperm development but show biallelic expression at later stages (Danilevskaya et al., 2003; Grimanelli et al., 2005). We intentionally profiled a relatively late stage of endosperm development to identify genes with consistent imprinting throughout endosperm development rather than delayed paternal activation. Indeed, each of the genes that we tested exhibited imprinting from 10 to 22 DAP. These genes are putatively classified as imprinted based on parent-of-origin bias in gene expression. However, it is formally possible that some of these allelic biases may represent stable transcripts inherited from parental cells rather than active monoallelic transcription in the F1 (Raissig et al., 2011). At least 30 of 71 genes surveyed in the expression atlas exhibit increasing transcript levels during endosperm development, which supports the classification of imprinted expression. The analysis of the expression patterns and DNA methylation patterns for the imprinted maize genes provided an opportunity to test several hypotheses about the expression patterns of imprinted genes and mechanisms of imprinting. We were also able to compare imprinting in maize and other species to identify examples of conserved imprinting between species.
Imprinting Is More Prevalent in the Endosperm Than in the Embryo
The majority of examples of imprinting in plants are documented in endosperm tissue (Huh et al., 2008; Jullien and Berger, 2009; Springer and Gutierrez-Marcos ,2009). There is one documented example of a maize gene (mee1) that is maternally expressed in both embryo and endosperm (Jahnke and Scholten, 2009). Several of the recent RNA-seq–based analyses of imprinting have compared embryo and endosperm tissue and have found few or no examples of imprinting in the embryo of Arabidopsis (Gehring et al., 2011; Hsieh et al., 2011) and rice (Luo et al., 2011). In our study, we assessed allele-specific expression in both endosperm and embryo tissue of maize. There were substantially fewer examples of MEGs or PEGs in embryo compared with endosperm. We also noted that each MEG or PEG identified in embryo was also observed in endosperm. This may reflect low levels of contamination of our embryo tissue with endosperm or maternal tissues, movement of transcripts from endosperm to embryo, or active imprinted transcription in both endosperm and embryo. It is difficult to fully evaluate these three potential explanations using data on steady state transcript levels. However, we did not observe evidence for imprinting of genes in embryo that are not imprinted in endosperm. Further studies, such as allele-specific occupancy for RNA polymerase II or nuclear run-on assays in embryo tissue, will be necessary to evaluate whether there is active allele-specific transcription for these genes.
Multiple Classes of Imprinted Genes in Plants
Previous studies in Arabidopsis have provided evidence for multiple molecular mechanisms of imprinting in plants. There is evidence that both DNA methylation and chromatin modifications can both play roles in imprinting, and there are likely additional mechanisms as well (Hsieh et al., 2011; Wolff et al., 2011). Our analysis of expression patterns and methylation levels provides further evidence for a role for DNA methylation regulating imprinting for a subset of imprinted genes. We noted that some of the maize-imprinted genes exhibit endosperm-preferred expression such that expression is low in all tissues except for endosperm. This same set of genes is highly enriched for DMRs near the imprinted genes. DNA methylation may play an important role in regulating expression and imprinting for this set of genes. It is noteworthy that the set of genes that have DMRs include both MEGs and PEGs and in both cases the maternal allele exhibits hypomethylation. The specific role of DNA methylation in regulating imprinting at PEGs is not clear but has been noted in Arabidopsis as well (Gehring et al., 2009, 2011; Hsieh et al., 2011; Wolff et al., 2011). Hsieh et al. (2011) provided evidence that maternal demethylation at PEGs may be important for proper silencing of the maternal allele. Maternal demethylation at PEGs may allow for targeting of PcG proteins to the maternal allele (Weinhofer et al., 2010; Hsieh et al., 2011).
Conservation of Imprinting among Species
High-resolution allele-specific expression profiling has now identified imprinted genes in three species. In all three species, there are ~50 to 150 imprinted genes identified from a set of 10,000 to 15,000 testable genes. It seems that imprinting is relatively infrequent in plants but does affect several hundred genes in each species, similar to the frequency of imprinted genes in mammals (Feil and Berger, 2007). Given these low rates, we would expect to observe very few examples of imprinting of the same genes in both species by chance. However, there are ~10 examples of conserved imprinting between each of the species. It should be noted that comparing imprinted genes between species is complicated by the requirement for SNPs between the two alleles; therefore, the set of genes tested is not the same between species. For example, the conservation for imprinting between GRMZM2G356731 and AT4G11400 was identified in one Arabidopsis study (Wolff et al., 2011) but could not be tested in the other studies due to lack of polymorphisms in AT4G11400 in the Landsberg erecta and Columbia ecotypes. Interestingly, we noted that the genes with conserved imprinting between species tend to be genes with endosperm-preferred expression. There are 11 examples of conserved imprinting that show endosperm-preferred expression in maize and only two examples of conserved imprinting that do not have endosperm-preferred expression (the other four genes with conserved imprinting do not have expression atlas data). This provides evidence that evolutionarily conserved imprinting is primarily restricted to genes that are regulated by DNA methylation and are exclusively expressed in the endosperm.
Genes with conserved imprinting in maize and either rice or Arabidopsis are potentially key regulators of seed development. The analysis of the GO annotations for genes that are imprinted in both maize and Arabidopsis identified overrepresentation for a number of biological process annotations. Half of these genes are annotated as being involved in regulation of biological processes. There are a number of genes with putative chromatin or DNA binding annotations as well. For example, genes related to VIM proteins, which are involved in regulating DNA methylation patterns in Arabidopsis (Woo et al., 2007), are imprinted in all three species. It is quite likely that the genes with conserved imprinting play important roles in regulation of seed development. Further functional analyses of imprinted plant genes will be necessary to shed light on the functional consequences of imprinting in plants.
METHODS
RNA-Seq Analysis
B73 and Mo17 plants were grown in Saint Paul at the University of Minnesota Agricultural Experiment Station during the summer of 2010. Reciprocal crosses and self-pollinations for all genotypes were performed on August 2nd, 3rd, and 4th, and several ears representing each cross were harvested 14 DAP. The endosperm and embryo tissue were dissected from at least five ears for each genotype and were pooled together and frozen in liquid nitrogen. RNA was isolated using Qiagen RNAeasy kits and subsequently purified by LiCl precipitation. These RNA samples were submitted to the University of Minnesota Biomedical Genomics Center for sequencing using the Illumina GAIIX (inbred samples) or HiSeq (hybrid samples) platforms.
Prior to alignment to the reference genome, the nucleotides of each raw read were scanned for low-quality bases and individual bases with PHRED quality values <15 out of 40 (Ewing and Green, 1998; Ewing et al., 1998) or having an error rate of ≤0.03% were removed by our trimming pipeline, which is a variant of previously published software but adapted for next-generation sequencing data (Li and Chou, 2004). Expression levels for three biological replicates of endosperm and embryo tissue from B73 and Mo17 were developed independently using the Tophat-Cufflinks transcript assembly pipeline (Trapnell et al., 2010). Reads were aligned to the B73 AGPv2 reference assembly using Tophat. Transcript quantification and expression levels in the form of RPKM for the B73 working gene set were calculated using Cufflinks under default parameters.
Trimmed reads were aligned to the reference genome using GSNAP (Wu and Nacu, 2010), and reads mapping uniquely or to single loci (less than or equal to two mismatches every 36 bp and less than or equal to three bp tails allowed) were used for subsequent analysis. Polymorphisms detected from the alignment of B73, Mo17, and reciprocal hybrid samples against the reference B73 genome were used to develop read counts of B73 and Mo17 alleles using previously identified SNP sites.
The number of reads containing the B73 or Mo17 allele was summed for all SNPs within the same gene. The 12,571 genes that contain at least 10 reads that could be assigned to a particular allele were used to perform χ2 tests (relative to an expected 2 maternal:1 paternal) ratio. The 468 genes that exhibit a significant (χ2 <0.01) difference from the 2:1 ratio were further filtered to identify genes for which at least 90% of the reads were derived from one parent in both reciprocal hybrids.
CAPS and RT-PCR Sequencing Validation Assays
Imprinted expression patterns were validated and further characterized using sequencing or CAPS assays for several genes. RT-PCR was performed on first-strand cDNA that was derived from an independent set of 14-DAP RNA samples or from a developmental series of endosperm time points (Haun et al., 2007) using the primers listed in Supplemental Table 2 online. In some cases, the RT-PCR products were directly sequenced to assess the contribution of the parental alleles in the hybrid samples. For other genes, the amplification products were digested with restriction enzymes that differentiate the B73 and Mo17 alleles (specific enzymes are listed in Supplemental Table 2 online).
DNA Methylation Profiling
DNA methylation was profiled according to the same methods described by Eichten et al. (2011). Endosperm DNA was isolated from three replicates of 14-DAP endosperm from both B73 and Mo17 (same tissue used for RNA-seq) using DNAeasy kits from Qiagen. Methylated DNA was immunoprecipitated with an anti-5- methylcytosine monoclonal antibody from 400 ng of sonicated DNA using the Methylated DNA immunoprecipitation (IP) kit (Zymo Research). For each replication and genotype, whole-genome amplification was conducted on 50 to 100 ng IP DNA and 50 to 100 ng of sonicated DNA (input control) using a whole-genome amplification kit (Sigma-Aldrich). For each amplified IP input sample, 3 μg amplified DNA was labeled using the dual-color labeling kit (Roche NimbleGen) according to the array manufacturer’s protocol (Roche NimbleGen Methylation User Guide v7.0). Each IP sample was labeled with Cy5, and each input/control sonicated DNA was labeled with Cy3. Samples were hybridized to the custom 2.1M probe array (Gene Expression Omnibus Platform GPL13499) for 16 to 20 h at 42°C. Slides were washed and scanned according to NimbleGen’s protocol for the GenePix4000B scanner. Images were aligned and quantified using NimbleScan software (Roche NimbleGen), which produced raw data reports for each probe on the array.
Normalization and Linear Modeling for DNA Methylation Data
Pair files exported from NimbleScan were imported into the Bioconductor statistical environment (http://bioconductor.org/). Sample-dependent MeDIP enrichments were estimated for each probe by fitting a fixed linear model accounting for array, dye, and sample effects to the data using the Limma package (Smyth, 2004). The following contrasts were then fit: B73 IP versus B73 input; Mo17 input versus B73 input; Mo17 IP versus (Mo17 input versus B73 input); B73 IP versus (Mo17 IP versus [Mo17 input versus B73 input]). Moderated t-statistics and the log-odds score for differential MeDIP enrichment was computed by empirical Bayes shrinkage of the standard errors with the false discovery rate controlled to 0.05.
Identification of DMRs
Putative DMRs were identified by comparing DNA methylation levels in endosperm and leaf tissue (Eichten et al., 2011). All probes within 3 kb of the imprinted genes were identified and the examples in which multiple adjacent probes had significantly lower (q < 0.05) methylation levels in endosperm relative to leaf tissue. The allele-specific methylation difference was tested for eight of these regions that included SNPs by amplification with and without digestion with the DNA methylation–dependent enzyme FspEI (New England Biolabs). One microgram of genomic DNA was digested for 16 h with FspEI according to the manufacturer’s recommendations. Mock digestions were performed substituting glycerol for restriction enzyme. Following digestion, PCR amplification of the target regions was performed (primers are listed in Supplemental Table 3 online). The amplified products were sequenced using dideoxy sequencing at the University of Minnesota Biomedical Genomics Center.
Informatics
The protein sequences from all of the filtered genes from annotation AGPv2_FGSv5b.60 were used as queries against the Arabidopsis thaliana (TAIR.10) or rice (Oryza sativa) Osa1 (Ouyang et al., 2007) protein sequences. The closest match that had an e-value < 1e-20 was retained for each of the maize genes. Annotations for maize genes were inferred based on the annotations for the closest sequence in Arabidopsis using The Arabidopsis Information Resource annotations.
Accession Numbers
The RNA-seq reads used for this study are deposited at the National Center for Biotechnology Information Short Read Archive under accession number SRA048055.2. The DNA methylation profiling microarray results are deposited at National Center for Biotechnology Information Gene Expression Omnibus under series accession number GSE33730. RPKM values for all genes in embryo and endosperm tissue are deposited in the Dryad Repository (http://dx.doi.org/10.5061/dryad.rb3b0d2p).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Relative Expression Levels in Endosperm and Embryo Tissue.
Supplemental Figure 2. Regulatory Variation in Allelic Expression in Embryo and Endosperm.
Supplemental Figure 3. Relative Expression Levels in Different Seed Tissues for Genes with Maternal Bias in Expression.
Supplemental Figure 4. Validation of Imprinted Expression Patterns.
Supplemental Figure 5. Analysis of DMRs at Fie1 and Mez1.
Supplemental Figure 6. Hypomethylation of the Maternal Allele at DMRs.
Supplemental Figure 7. Annotation Enrichments for MEGs and PEGs.
Supplemental Table 1. RNA-seq Reads for B73 and Mo17 Endosperm.
Supplemental Table 2. Primers Used for Validation of Imprinting.
Supplemental Table 3. Primers Used for Allele-Specific Methylation Analysis.
Supplemental Data Set 1. Allele-Specific Expression Counts for All Genes.
Supplemental Data Set 2. Allele-Specific Expression Details for 100 Imprinted Genes.
Acknowledgments
The authors acknowledge the Texas Advanced Computing Center at the University of Texas at Austin for providing high-performance computing and storage resources that have contributed to the research results reported in this article. Peter Hermanson assisted with the crosses and tissue collections for this study. The University of Minnesota Biomedical Genomics Center performed the Illumina sequencing for this study. The Minnesota Supercomputing Institute provided access to software and user support for data analyses. We thank Robert Stupar and William Haun for useful comments and suggestions on this study. The research was supported by a grant from the National Science Foundation to N.M.S. and M.W.V. (IOS-0922095).
AUTHOR CONTRIBUTIONS
R.A.S.-W., M.W.V., M.G., and N.M.S. designed the research. A.J.W., I.M., S.R.E., R.A.S.-W., and C.-T.Y. performed research. W.X., C.-T.Y., and P.S.S. contributed analytic/computational tools. A.J.W., I.M., S.R.E., C.-T.Y., P.S.S., M.W.V., M.G., and N.M.S. analyzed data. N.M.S. wrote the article.
References
- Bauer M.J., Fischer R.L. (2011). Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant Biol. 14: 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Messing J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. USA 91: 4867–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Hermon P., Hantke S., Muszynski M.G., Kollipara K., Ananiev E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2011). Heritable epigenetic variation among maize inbreds. PLoS Genet. 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feil R., Berger F. (2007). Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Choi Y., Fischer R.L. (2004). Imprinting and seed development. Plant Cell 16(suppl.): S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Greb T., Mylne J.S., Crevillen P., Geraldo N., An H., Gendall A.R., Dean C. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17: 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Guo M., Rupe M.A., Danilevskaya O.N., Yang X., Hu Z. (2003). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36: 30–44 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Biderre-Petit C., Khbaya B., O’Sullivan D.M., Wormald M., Perez P., Dickinson H.G. (2004). maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Dal Prà M., Scholten S., Kranz E., Perez P., Dickinson H.G. (2006). Epigenetic asymmetry of imprinted genes in plant gametes. Nat. Genet. 38: 876–878 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J.F., Pennington P.D., Costa L.M., Dickinson H.G. (2003). Imprinting in the endosperm: A possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. (2004). Genomic imprinting and kinship: How good is the evidence? Annu. Rev. Genet. 38: 553–585 [DOI] [PubMed] [Google Scholar]
- Haig D., Westoby M. (1989). Parent-specific gene-expression and the triploid endosperm. Am. Nat. 134: 147–155 [Google Scholar]
- Haun W.J., Laoueillé-Duprat S., O’connell M.J., Spillane C., Grossniklaus U., Phillips A.R., Kaeppler S.M., Springer N.M. (2007). Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J. 49: 325–337 [DOI] [PubMed] [Google Scholar]
- Haun W.J., Springer N.M. (2008). Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J. 56: 903–912 [DOI] [PubMed] [Google Scholar]
- Hermon P., Srilunchang K.O., Zou J., Dresselhaus T., Danilevskaya O.N. (2007). Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol. Biol. 64: 387–395 [DOI] [PubMed] [Google Scholar]
- Hsieh T.F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M.J., Hashimoto M., Kirkbride R.C., Harada J.J., Zilberman D., Fischer R.L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh J.H., Bauer M.J., Hsieh T.F., Fischer R.L. (2008). Cellular programming of plant gene imprinting. Cell 132: 735–744 [DOI] [PubMed] [Google Scholar]
- Jahnke S., Scholten S. (2009). Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 19: 1677–1681 [DOI] [PubMed] [Google Scholar]
- Jullien P.E., Berger F. (2009). Gamete-specific epigenetic mechanisms shape genomic imprinting. Curr. Opin. Plant Biol. 12: 637–642 [DOI] [PubMed] [Google Scholar]
- Jullien P.E., Katz A., Oliva M., Ohad N., Berger F. (2006). Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Kermicle J.L. (1970). Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66: 69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S.E., Fischer R.L., Kakutani T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Yadegari R., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Weinhofer-Molisch I. (2010). Mechanisms and evolution of genomic imprinting in plants. Heredity 105: 57–63 [DOI] [PubMed] [Google Scholar]
- Lauria M., Rupe M., Guo M., Kranz E., Pirona R., Viotti A., Lund G. (2004). Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chou H.H. (2004). LUCY2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics 20: 2865–2866 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Taylor J.M., Spriggs A., Zhang H., Wu X., Russell S., Singh M., Koltunow A. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- McKeown P.C., et al. (2011). Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W. (2010). siRNAs and DNA methylation: Seedy epigenetics. Trends Plant Sci. 15: 204–210 [DOI] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., et al. (2007). The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 35(Database issue): D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Baroux C., Grossniklaus U. (2011). Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Selinger D.A., Chandler V.L. (2001). B-Bolivia, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol. 125: 1363–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3 [DOI] [PubMed] [Google Scholar]
- Springer N.M. (2009). Small RNAs: How seeds remember to obey their mother. Curr. Biol. 19: R649–R651 [DOI] [PubMed] [Google Scholar]
- Springer N.M., Gutierrez-Marcos J. (2009). Imprinting in maize. In The Maize Handbook, S. Hake and J. Bennetzen, eds (New York: Springer), pp. 429–440 [Google Scholar]
- Stupar R.M., Hermanson P.J., Springer N.M. (2007). Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 145: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M.T., Spillane C., Nordborg M., Rehmsmeier M., Köhler C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Pontes O., Pikaard C.S., Richards E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Silva P., Rodrigues J.A., Dotson B., Brooks M.D., Zilberman D. (2010). Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]